Abstract
Rifampicin (R) and isoniazid (H) are key first-line anti-tuberculosis drugs. Failure to detect resistance to these two drugs early results in treatment failure and poor clinical outcomes. The study purpose was to validate the use of the GenoType MTBDRplus line probe assay (LPA) to detect resistance to R and H in Mycobacterium tuberculosis strains directly from smear-positive sputum samples in India.
Method
Smear positive sputum specimens from 320 patients were subjected to LPA and results compared against those from conventional Lowenstein Jensen (LJ) culture and drug susceptibility testing (C&DST). All specimens with discordant R DST results were subjected to either sequencing of the rpoB gene and/or repeat DST on liquid culture (MGIT 960) at a National Reference Laboratory.
Results
Significantly higher proportion of interpretable results were observed with LPA compared to LJ C&DST (94% vs. 80%, p-value <0.01). A total of 248 patients had both LJ and LPA DST results available; 232 (93.5%) had concordant R DST results. Among the 16 discordant R DST results, 13 (81%) were resolved in agreement with LPA results. Final LPA performance characteristics were sensitivity 96% (CI: 90%–98%), specificity 99% (CI: 95%–99%), positive predictive value 99% (CI: 95%–99%), and negative predictive value 95% (CI: 89%–98%). The median turnaround testing time, including specimen transportation time, on LPA was 11 days as compared with 89 days for LJ C&DST.
Conclusions
LPA proved highly accurate in the rapid detection of R resistance. The reduction in time to diagnosis may potentially enable earlier commencement of the appropriate drug therapy, leading to some reduction of transmission of drug-resistant strains.
Introduction
The global threat of multidrug-resistant tuberculosis (MDR-TB; strains of Mycobacterium tuberculosis resistant to at least rifampicin and isoniazid) to TB control, underscores the importance of prompt and rapid identification of such resistant Mycobacterium tuberculosis (M.tb) strains. Isoniazid (H) and rifampicin (R) are the key first-line anti-tuberculosis drugs, and resistance to these drugs i.e. MDR-TB, is likely to result in treatment failure and poor clinical outcomes [1], [2]
India has the largest number of estimated MDR-TB cases amongst notified TB patients of any country [3]. Up to the time of the study, the Government of India's Revised National TB Control Programme (RNTCP) had relied on conventional Lowenstein Jensen (LJ) culture and drug susceptibility testing (C&DST) for the diagnosis of drug resistant TB cases. By December 2009, there were however only 14 such laboratories across the country validated and certified by the RNTCP for conducting LJ C&DST [4].The mean time to detect drug resistance on egg based LJ media is around 3–4 months [5]. Even using the more modern broth-based liquid culture systems, C&DST results from sputum specimens still takes several weeks [6]. However, newly developed molecular based methods have advantages over conventional phenotypic methods in terms of both accuracy and turnaround time. The GenoType MTBDRplus assay is a commercially available line probe assay (LPA) from Hain Lifescience, Nehren, Germany, and is designed to simultaneously detect the most important gene mutations conferring R (rpoB genes) and H (inhA, katG) resistance in M. tb isolates within 8 hours [7].
A 2008 meta-analysis found that the GenoType MTBDRplus assay and another similar commercial test had a pooled sensitivity of 98% for detecting R resistance and 89% for detecting H resistance and specificity of 99% for R and H [8]. Testing can be performed on culture isolates or acid fast bacilli (AFB) positive sputum specimens, and can provide results within 8 hours, making this a promising tool to accelerate the diagnosis of MDR-TB cases, and hence improve management of MDR-TB cases.
Although the GenoType MTBDRplus assay has been studied in several laboratories, there is a wide variation in circulating M.tb strains across the globe [9], [10], and false negative results can occur due to the presence of unique genetic mutations in the different settings [8], [11]–[16]. Hence validation in different settings is needed to ensure acceptable performance. With its large number of MDR-TB cases, validation in India was deemed necessary ahead of wide-scale introduction of LPA for the programmatic management of drug resistant TB (PMDT) in the country.
To address this issue, a cross sectional study to evaluate the assay directly on sputum specimens was conducted under programmatic conditions in India by the Foundation for Innovative Diagnostics (FIND) in 2008–09. Three laboratories located in different regions of India and certified by the RNTCP for performing LJC & DST, were selected as the sites for the study. The primary objective of the study was to evaluate the accuracy of rifampicin and isoniazid susceptibility results by LPA performed directly on AFB smear-positive sputum specimens, compared against LJ C&DST. The secondary objective was to evaluate the operational performance characteristics of LPA versus LJ C& DST, specifically the time to reporting of test results and the proportion of invalid tests results.
Materials and Methods
Setting
The study was conducted at the public sector RNTCP state level Intermediate Reference Laboratories (IRL) in Hyderabad (Andhra Pradesh State), Ahmadabad (Gujarat State), and the Mycobacteriology laboratory at SMS Medical College, Jaipur (Rajasthan State). Smear-positive sputum specimens of TB patients from surrounding districts, who were failing on first-line anti-TB treatment, were routinely transported to these IRLs for LJ C&DST. The Ahmedabad and Hyderabad laboratories had been validated by a National Reference Laboratory (NRL) for conducting LJ C&DST. The Jaipur laboratory was undergoing validation at the time of the study. Hence the LJ C&DST for all specimens from this site was conducted at the National JALMA Institute for Leprosy and Other Mycobacterial Diseases Agra Uttar Pradesh State (NRL).
Enrolment
Between November 2008 and January 2009, all patients submitting sputum specimens for LJ C&DST at the 3 study sites, were enrolled consecutively. As per RNTCP guidelines at the time of the study, MDR-TB suspects were defined as those TB patients who remained sputum smear positive after 4 months of treatment with an RNTCP Category 2 re-treatment regimen and who were not on treatment with any second-line anti-TB drugs (including any fluoroquinolone, ethionamide, cycloserine, PAS, or any second line injectable agent) at the time of specimen collection.
A cumulative sample size pre-specified for the study as per project protocol was approximately 250 specimens with both LPA and LJ C&DST results, across all sites. This was based on an estimated 30% prevalence of R resistance in tested specimens. The minimum acceptable performance parameter for LPA was pre-specified as detection of 95% of R resistance cases based on 5% precision.
Patients were excluded from enrolment into the study if specimens contained any preservative such as cetyl pyridinum chloride or if the sputum samples were from patients who had previously been confirmed as MDR-TB by any one of the standard laboratory procedures.
Ethical considerations
The study was conducted in India, based on a memorandum of understanding between FIND and Government of India for the introduction of rapid new TB diagnostics in RNTCP of India. Accordingly, patients were managed as per organisational policy which was based on the results of LJ C&DST, as per routine. As LPA had not been validated for patient care in India and was not used for patient care prior to this study under RNTCP, the LPA results were not made available and hence not considered for decision making on patient care. As per the project protocol, as testing was conducted on remnant anonymised specimens, which otherwise be discarded and did not influence in any way patient management, informed consent was not considered necessary. The study protocol, after detailed review, was initially approved by the National laboratory committee constituted under RNTCP. The study protocol was further reviewed and approved independently by the ethical committees at each of the three study sites (namely the Ethics committee of the SMS Medical College and attached Medical College, and the Institutional Review Boards of the RNTCP State TB & Demonstration centres of Ahmedabad and Hyderabad).
Laboratory procedures
Specimen collection and transportation
Two sputum specimens (one morning and one spot) were collected from all MDR-TB suspects in a pre-sterilised 50 ml centrifuge tube for LJ C& DST, in line with the programmatic guidelines. At the time of the specimen collection, a standard “request for C&DST form” was filled out by the respective laboratory staff. Specimens were transported within 7 days employing a cold chain to the reference laboratory without any preservative in the 50 ml centrifuge tube.
Specimen processing and LJ Culture and DST
Fresh sputum specimens were processed by N-acetyl-L-cystein-sodium hydroxide (Nalc-NaOH) method (with final NaOH concentration of 1%) as recommended by US Centers for Disease Control and Prevention CDC [17]. The concentrated sediment was re-suspended in 1–2 ml of phosphate buffer, subjected to Ziehl-Neelsen staining and inoculated on LJ media. After the growth on LJ slopes was obtained, isolates were subjected to M.tb complex identification by testing growth on para-nitro-benzoic acid medium, niacin test and nitrate reductase test [18]. All M.tb complex isolates were tested by 1% proportion method for drug susceptibility with critical drug concentration of isoniazid (0.2 µg/ml) and rifampicin (40 µg/ml) [19].
Line probe assay: DNA extraction
From each patient the specimen with the highest smear grading based on RNTCP guidelines [20] was tested by LPA. An aliquot of the processed sputum deposits was coded and assigned a unique study ID number for the LPA test. DNA extraction and amplification was performed as recommended by the manufacturer. Briefly, 0.5 ml of processed sputum deposit was centrifuged at 10,000 g for 15 minutes, re-suspended in 100 µl of molecular grade water, sealed and heated for 20 minutes at 95°C in a water bath followed by ultrasonication for 15 min at room temperature. This suspension was centrifuged at 13,000 g for 5 minutes, and the supernatant (DNA Extract) transferred by pipette to a fresh tube without disturbing the pellet. A 5 µL aliquot of this extracted DNA was used for amplification procedures. The Genotype MTBDRplus assay version.1 was performed as recommended by the manufacturer [7].
PCR amplification
Amplification was performed by combining 35 µL of primer nucleotide mix supplied by the manufacturer (PNM) with 5 µL of 10× PCR buffer (containing 15 mM MgCl2), 2 µL MgCl2 (25 mM MgCl2), 3 µL molecular grade H2O, 0.2 µL (1 unit) Hot-Star Taq polymerase (QIAGEN, Hilden, Germany), and 5 µL of the DNA for a total final volume of 50.2 µL. The amplification profile for direct patient material as described by the manufacturer was used for all sputum specimens. First, the template DNA was denatured for 15 minutes at 95°C, followed by 10 cycles consisting of 30 seconds at 95°C and 2 minutes at 58°C, with an additional 30 cycles consisting of 25 seconds at 95°C, 40 seconds at 53°C and 40 seconds at 70°C. The final cycle consisted of an 8 minute run at 70°C.
Hybridization and Detection
Hybridization was performed using the hybridization Kits, including reagents and 12 well plastic tray and instrument (Twincubator) as provided by the manufacturer [7]. Briefly, 20 µl of denaturation solution (DEN,blue) were mixed thoroughly in a plastic 12-well tray, with 20 µl of amplified sample (PCR product) and incubated at room temperature for 5 minutes. After denaturation, the biotin-labelled amplicons were hybridized (using HYB, green solution) to the single stranded membrane-bound probes. After a stringent washing (using STR-red solution), a streptavidinalkaline phosphatase conjugate (1∶100 dilution of Con-C with CON-D) was added to the strips and an alkaline phosphatase-mediated staining reaction (1∶100 dilution of SUB-C with SUB-D) was observed in the bands where the amplicon and the probe had hybridized [7].
Interpretation of results
The MTBDRplus assay strip contains 27 reaction zones; 21 of them are probes for mutations and 6 are control probes for verification of the test procedures. The six control probes include a conjugate control, and amplification control, an M.tuberculosis complex-specific control (TUB), an rpoB amplification control, a katG amplification control, and an inhA amplification control. For the detection of R resistance, the probes cover the rpoB gene, while the H resistance specific probes cover positions in katG and inhA. The absence of at least one of the wild-type bands or the presence of bands indicating a mutation in each drug resistance-related gene implies that the sample tested is resistant to the respective antibiotic. When all the wild-type probes of a gene stain positive and there is no detectable mutation within the region examined, the sample tested is susceptible to the respective antibiotic. In order to give a valid result, all six expected control bands should appear correctly. Otherwise, the result is considered invalid. [7].
Repeat testing
Sputum specimens resulting in inconsistent development of bands on the MTBDRplus strip and/or if no M.tb control band appeared, underwent repeat PCR and hybridization from the extracted DNA. Isolates with discordant results of the LPA and the LJ C&DST, were sent to a national reference laboratory (JALMA Institute, Agra) for sequencing of rpoB and repeat C& DST testing using liquid (MGIT 960) culture systems. Sequencing of discordant results sample was performed using BigDye Terminator v3.1 Cycle Sequencing Kit and analyzed on an ABI 3130xl genetic analyzer (Applied Biosystems, USA). Sequencing results were compared with the DNA sequences of wild type reference strain M. tuberculosis H37Rv using MegAlign program (DNASTAR, Madison, WI, USA).
Data Analysis
The sensitivity, specificity, predictive value positive (PPV), predictive value negative (NPV), and overall accuracy of LPA results were compared to the conventional LJ DST results for R and H, and the ability of R resistance alone to predict MDR. For calculation, the reference DST result for R was the LJ DST result for specimens with initially concordant LPA and LJ DST results, and the results of the repeat testing procedure described above for those specimens with initially discordant LJ and LPA DST results. An analysis of banding patterns associated with R and H resistance in MDR-TB and non MDR-TB strains was performed. Statistical tests to assess the test performance used under the study were odds ratio, sensitivity, specificity, positive predictive value, and negative predictive value. These were calculated using free online statistical calculators available at http://www.medcalc.org/calc/.
Results
Between November 2008 and January 2009, sputum specimens from 320 sputum smear positive MDR-TB suspects were received from the pre-identified districts as per the study protocol, and entered into the study. Only 1 specimen of the 2 received per patient was tested on LPA, with the specimen showing the better growth on LJ media tested for susceptibility to R and H.
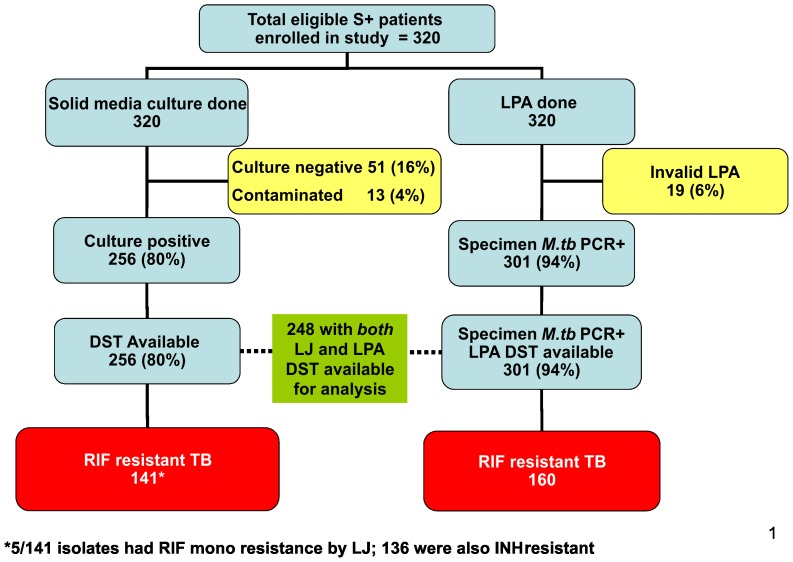
Of the 320 patients, 72% (230/320) were male, and the mean age was 37.1 years. Testing of all 640 samples by culture yielded at least one positive growth for M.tb complex in 256 (80%) patients. Samples from 51 (16%) patients had no growth and in 13 (4%) patients the cultures were contaminated. LPA gave interpretable results for 301 (94%) patients. LJ DST results were available for all 256 patients with positive culture results (table 1).
Table 1. Proportion of invalid LPA results in comparison to culture results.
| LJ Positive | LJ No Growth | LJ Contaminated | Total | |
| Valid LPA | 248 | 41 | 12 | 301 (94%) |
| Invalid LPA | 8 | 10 | 1 | 19 (5.9%) |
| Total | 256 (80%) | 51 (16%) | 13 (4%) | 320 |
Overall, a significantly higher proportion of patients had interpretable results from LPA as compared to LJ culture and DST (94% vs. 80%, p-value <0.01). Amongst the 51 culture negative patients, 41 (80%) had a valid LPA result, and 12 (92.3%) of the 13 patients with contaminated cultures had an interpretable LPA result (Table 1). There was a significantly higher likelihood of obtaining an interpretable MTBDRplus result from a specimen with a positive smear grade compared to those specimens with a scanty smear grading (p-value 0.012) (Table 2).
Table 2. Association between smear positivity grades and LPA test result.
| Smear Grade | ||||||||
| Scanty | 1+ | 2+ | 3+ | |||||
| No | % | No | % | No | % | No | % | |
| Valid LPA | 6 | 17 | 5 | 5 | 6 | 8 | 2 | 2 |
| Invalid LPA | 30 | 83 | 105 | 95 | 70 | 92 | 96 | 98 |
| Total | 36 | 110 | 76 | 98 | ||||
| p-value: 0.012 | ||||||||
DST results
The median time from the specimen collection to the LJ C&DST result being available was 87 days (range of 42 to 208 days). In comparison, the median time to obtain a LPA result was just 11 days (range of 1 to 76 days). This time included specimen shipment time which varied from 1 to 7 days.
In this patient population of consecutively-enrolled smear-positive TB patients suspected of having MDR-TB, the prevalence of rifampicin resistance was high. Among the 256 patients with available LJ DST results, 136 (53%) were resistant to both R and H i.e. MDR-TB, 5 (2%) were resistant to R and susceptible to H, 54 (21%) were resistant to H and susceptible to R and 61 (24%) were susceptible to both R and H. Among the 301 patients with interpretable LPA results, 127 (42%) were MDR-TB, 33 (11%) had R mono-resistance, 30 (10%) were resistant to H and susceptible to R, and 111 (37%) were susceptible to both R and H (Figure 1). Overall, a total of 141 patients were detected with R-resistance by LJ media, whereas 160 were detected by LPA i.e. an additional 19 (6%) R-resistant cases were identified by LPA.
Figure 1. Suspect enrolment and result summary.
Drug resistance associated mutations
The most common mutation detected by LPA in the rpoB gene was S531L (47%), diagnosed by the presence of MUT3 band. There was no significant difference in the prevalence of this mutation in MDR-TB specimen and R mono-resistance specimen (47% vs 46%). Four (2.7%) cases had multiple mutations - 2 had D516V and S531L mutations, one had H526Y and H526D mutations, and one had H526D and S531L mutations. Of the overall 160 patients in whom R-resistance was detected, 60 were on the basis of missing Wild type probes and did not have any positive mutant probe (table 3).
Table 3. Gene mutation patterns in resistant M.tb strains using Genotype MTBDRplus LPA.
| Gene | Band | Gene Region or Mutation | MDR (n = 127) | H monoresistance (n = 30) | R monoresistance (n = 33) |
| rpoB | WT1 | 506–509 | 126 (99) | 30 (100) | 33 (1 00) |
| WT2 | 510–513 | 121 (95) | 30 (100) | 31 (94) | |
| WT3 | 513–517 | 104 (82) | 30 (100) | 28 (85) | |
| WT4 | 516–519 | 104 (82) | 30 (100) | 29 (88) | |
| WT5 | 518–522 | 125 (98) | 30(100) | 33 (100) | |
| WT6 | 521–525 | 122 (96) | 30 (100) | 32 (97) | |
| WT7 | 526–529 | 101 (80) | 30 (100) | 27 (82) | |
| WT8 | 530–533 | 50 (39) | 30 (100) | 12 (36) | |
| MUT1 | D516V | 11 (9) | 0 (0) | 4 (12) | |
| MUT2A | H526Y | 8 (6) | 0 (0) | 2 (6) | |
| MUT2B | H526D | 4 (3) | 0 (0) | 0 (0) | |
| MUT3 | S531L | 60 (47) | 0 (0) | 15 (46) | |
| katG | WT | 315 | 19 (15) | 8 (27) | 33 (100) |
| MUT1 | S315T1 | 97 (76) | 15 (50) | 0 (0) | |
| MUT2 | S315T2 | 0 (0) | 0 (0) | 0 (0) | |
| inhA | WT1 | 0.9375 | 110 (87) | 28 (93) | 33 (100) |
| WT2 | −8 | 110 (87) | 26 (87) | 33 (100) | |
| MUT1 | C15T | 9 (7) | 3 (10) | 0 (0) | |
| MUT2 | A16G | 0 (0) | 0 (0) | 0 (0) | |
| MUT3A | T8C | 2 (2) | 0 (0) | 0 (0) | |
| MUT3B | T8A | 0 (0) | 1 (3) | 0 (0) |
Definition of abbreviations: H = isoniazid; MDR = multidrug-resistant; R = rifampicin.
Values are numbers, with percentages in parentheses.
Concordance between LJ DST and LPA DST
A total of 248 patients had both LJ and LPA DST results available. Initial analysis showed agreement of results in 232 (94%) patients, including 127 with R-resistance and 105 with R-susceptibility on both LPA and LJ media (initial concordance 94%; Sensitivity: 93% (CI: 88%–96%); Specificity: 94% (CI: 88%–97%) Positive Predictive value: 95% (CI: 90%–97%) Negative Predictive value: 92% (CI: 86%–96%) (Table 4). There were 16 (6%) specimens with discordant rifampicin DST results between LJ and LPA. These included 9 results that were R-resistant on LJ and susceptible on LPA, and 7 that were R-susceptible on LJ and resistant on LPA (table 4). Each of these 16 specimen were subjected to sequencing of rpoB gene and repeat Culture and DST at the national reference laboratory as per the repeat testing procedure. Of these, 11 specimens could be sequenced, and culture and DST results of 15 results were available at the end of the study. For specimen that could not be either sequenced or have a valid result on culture and DST at national reference laboratory, LJ C&DST being the pre-existing procedure, was presumed to be correct. The repeat testing procedure agreed with the LPA result in 12 instances (including one mixed infection) and the LJ media result in 3 instances. One test could not be performed due to heavy contamination found in the MGIT DST (table 5).
Table 4. Correlation of R and H resistance on LJ DST with LPA.
| Rifampicin Resistance | Isoniazid Resistance | ||||||
| LJ DST Rifampicin Resistant | LJ DST Rifampicin Sensitive | Total Results | LJ DST Isoniazid Resistant | LJ DST Isoniazid Sensitive | Total Results | ||
| LPA Rifampicin Resistant | 127 | 7 | 134 | LPA Isoniazid Resistant | 133 | 2 | 135 |
| LPA Rifampicin Sensitive | 9 | 105 | 114 | LPA Isoniazid Sensitive | 52 | 61 | 113 |
| Total Results | 136 | 112 | 248 | Total Results | 185 | 63 | 248 |
| Concordance: 94% | Concordance: 78% | ||||||
| Sensitivity: 93% (CI: 88%–96%) | Sensitivity: 72% (CI: 65%–78%) | ||||||
| Specificity: 94% (CI: 88%–97%) | Specificity: 97% (CI: 89%–99%) | ||||||
| Positive Predictive value: 95% (CI: 90%–97%) | Positive Predictive value: 99% (CI: 95%–99%) | ||||||
| Negative Predictive value: 92% (CI: 86%–96%) | Negative Predictive value: 54% (CI: 45%–63%) | ||||||
Table 5. Results of discordance resolution at national reference laboratory.
| Specimen Number | Rifampicin LJ C& DST | LPA result for Rifampicin Resistance | Sequencing result at National Reference Laboratory | Culture and DST at National Reference Laboratory | Final Interpretation |
| A | Resistant | Sensitive | - | Contaminated | Resistant |
| B | Resistant | Sensitive | - | Sensitive | Sensitive |
| C | Resistant | Sensitive | - | Sensitive | Sensitive |
| D | Resistant | Sensitive | Sensitive | Sensitive | Sensitive |
| E | Resistant | Sensitive | Sensitive | Sensitive | Sensitive |
| F | Resistant | Sensitive | - | Sensitive | Sensitive |
| G | Resistant | Sensitive | - | Sensitive | Sensitive |
| H | Resistant | Sensitive | Resistant | Resistant | Resistant |
| I | Resistant | Sensitive | Sensitive | Resistant | Resistant |
| J | Susceptible | Resistant | Resistant | Sensitive | Resistant |
| K | Susceptible | Resistant | Resistant | Resistant | Resistant |
| L | Susceptible | Resistant (Mixed Infection) | Sensitive | Resistant | Mixed Infection |
| M | Susceptible | Resistant | Resistant | Resistant | Resistant |
| N | Susceptible | Resistant | Resistant | Resistant | Resistant |
| O | Susceptible | Resistant | Resistant | Resistant | Resistant |
| P | Susceptible | Resistant | Resistant | Resistant | Resistant |
Based on the final DST results, i.e. the LJ DST adjusted for repeat testing conducted at national reference laboratory, LPA detected R-resistance with concordance 97%, sensitivity, 96% (CI: 90%–98%), specificity, 99% (CI: 95%–99%), positive predictive value, 99% (CI: 95%–99%) and negative predictive value, 95% (CI: 89%–98%). (Table 6)
Table 6. Final reconciled results of LPA-LJ Rifampicin resistance correlation in view of sequencing and Liquid Culture and DST at National reference laboratory.
| LJ C&DST reconciled with liquid culture & DST/sequencing at national reference laboratory | |||
| Resistant | Sensitive | Total | |
| LPA Rifampicin Resistant | 133 | 1 | 134 |
| LPA Rifampicin Sensitive | 6 | 108 | 114 |
| Total | 139 | 109 | 248 |
| Concordance: 97% | |||
| Sensitivity: 96% (CI: 90%–98%) | |||
| Specificity: 99% (CI: 95%–99%) | |||
| Positive Predictive value: 99% (CI: 95%–99%) | |||
| Negative Predictive value: 95% (CI: 89%–98%) | |||
In relation to H, the analysis showed agreement of results in 194 (78%) patients, including 133 with H-resistance and 61 with H-susceptibility on both LPA and LJ media (Sensitivity: 72% (CI: 65%–78%); Specificity: 97% (CI: 89%–99%); Positive Predictive value: 99% (CI: 95%–99%) Negative Predictive value: 54% (CI: 45%–63%). There were 54 (22%) specimens with discordant H DST results between LJ C&DST and LPA (table 4).
Discussion
The current study demonstrated that overall sensitivity and specificity of LPA assay for detection of R resistance was high at 96% and 99% respectively. This was similar to previously reported results in studies from South Africa, Germany, and Italy [21]–[28]. However, sensitivity and specificity of LPA assay for detection of H, resistance were 72% and 97% respectively. The sensitivity to detect H resistance is somewhat lower than previously reported results [8], [28].
The study findings suggest that LPA is suitable for routine use in settings where a standardised second line anti-TB drug regimen is provided to MDR-TB cases and where R-resistance is also treated with the same standardised MDR-TB regimen. Patients tested were more likely to have valid results available with LPA to guide clinical action, compared to LJ; this reflected reduced recovery of viable M. tuberculosis and culture contamination with LJ. This translated to increased detection of drug-resistant TB. With the observed 99% specificity of the LPA assay in detecting R-resistance in M.tb isolates, most of the patients would be appropriately treated with the standardised MDR-TB treatment regimen if this test were to be used for the routine and rapid diagnosis of R-resistance and MDR-TB. This is further reaffirmed by the results of LJ DST in the study, which showed that R mono-resistance was relatively rare.
As reported widely elsewhere, phenotypic rifampicin resistance was strongly associated with mutation in the 81 base pair region of rpoB targeted in the LPA assay [24], [25]. In this study, the most commonly observed mutations were in the region of rpoB 530–533, mostly S531L mutation. This is similar to the findings of in a South African study [15].
In this patient population of consecutively-enrolled smear-positive TB patients suspected of having MDR-TB, the prevalence of rifampicin resistance was unsurprisingly high. While using LPA in settings with significantly lower levels of MDR-TB, routine implementation of quality assurance guidance issued by agencies such as Global Laboratory Initiative (GLI) would be of paramount importance to address any concerns of false negative results.
The routine use of LPA can substantially reduce the time to diagnosis of R and/or H-resistant TB, and can hence potentially enable earlier commencement of appropriate drug therapy and thereby facilitate prevention of further transmission of drug-resistant strains. This confers a major advantage to this test. Until the time of the study, India had no access to newer diagnostic tests in its public funded TB laboratories and relied largely on LJ based solid C&DST. Unacceptable delays in obtaining both culture and DST results by these conventional methods were commonplace, specifically in drug resistant cases. A number of patients may be “lost” due to default and/or death whilst awaiting the availability of the DST results. The study also highlights that the availability of rapid diagnostics at central laboratories needs to be supplemented with rapid specimen transportation mechanisms.
Conclusion
The multi-centric study reported here evaluated the performance of the Genotype MTBDRplus for the detection of R and H resistance under routine conditions in 3 state level reference laboratories in India, and provided direct evidence on the accuracy and feasibility of LPA. The GenoType MTBDRplus LPA version 1 assay is a sensitive and specific tool for the detection of rifampicin resistance in AFB smear-positive sputum specimens. The relatively quick turnaround time and the potential avenue for rapid screening of a large number of specimens/patients make it suitable as a first-line molecular diagnostic test for rifampicin resistance in settings such as India. The results of this evaluation were presented to the National TB Laboratory Committee of India in July 2009, which endorsed the routine use of the GenoType MTBDRplus LPA in the national TB programme for the testing of MDR–TB suspects [29], [30].
Acknowledgments
The present study was a part of the LPA demonstration project undertaken by FIND as per their memorandum of understanding with Government of India. We wish to acknowledge the contributions of the Dr VM Katoch, Secretary, Department of Health Research Ministry of Health & Family Welfare, Government of India & Director-General Indian Council of Medical Research, the staff of the three project sites, District Tuberculosis Officers of all the districts covered under the project and the WHO-RNTCP consultants for their efforts in the introduction of rapid TB diagnostics.
Funding Statement
The authors have no support or funding to report.
References
- 1. Orenstein EW, Basu S, Shah NS, Andrews JR, Friedland GH, et al. (2009) Treatment outcomes among patients with multidrug-resistant tuberculosis: systematic review and meta-analysis. Lancet Infect Dis 9: 153–161. [DOI] [PubMed] [Google Scholar]
- 2. Johnston JC, Shahidi NC, Sadatsafavi M, Fitzgerald JM (2009) Treatment outcomes of multidrug-resistant tuberculosis: a systematic review and meta-analysis. PLoS One 4: e6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organisation: Global tuberculosis control - epidemiology, strategy, financing. WHO Report 2012 WHO/HTM/TB/2012.411. WHO Website. Available: http://www.who.int/tb/publications/global_report/2012/en/index.html. Accessed 2013 Dec 1
- 4.Revised National tuberculosis programme, India, document downloadable, RNTCP status report 2009. RNTCP website. Available: www.tbcindia.org/documents. Accessed 2013 Dec 1
- 5.Revised National TB Control Programme Guidelines for Programmatic Management of Drug Resistant TB (PMDT) in India, May 2012, page 19
- 6. Srisuwanvilai LO, Monkongdee P, Podewils LJ, Ngamlert K, Pobkeeree V, et al. (2008) Performance of the BACTEC MGIT 960 compared with solid media for detection of Mycobacterium in Bangkok, Thailand. Diagn Microbiol Infect Dis 2008 61 (4) 402–407. [DOI] [PubMed] [Google Scholar]
- 7.Genotype MTBDRplus™, version 1.0 [product insert]. Nehren, Germany: Hain Lifescience,GmbH. Hain lifescience website. Available: http://www.hainlifescience.com/pdf/304xx_pbl.pdf. Accessed 2013 Nov 1 [Google Scholar]
- 8. Ling DI, Zwerling AA, Pai M (2008) GenoType MTBDR assays for the diagnosis of multidrug-resistant tuberculosis: a meta-analysis. Eur Respir J 32: 1165–1174. [DOI] [PubMed] [Google Scholar]
- 9. Malik AN, Godfrey-Faussett P (2005) Effects of genetic variability of Mycobacterium tuberculosis strains on the presentation of disease. Lancet Infect Dis 5 (3) 174–183. [DOI] [PubMed] [Google Scholar]
- 10. Nicol MP, Wilkinson RJ (2008) The clinical consequences of strain diversity in Mycobacterium tuberculosis. Tran R Soc Trop Med Hyg 102 (10) 955–65. [DOI] [PubMed] [Google Scholar]
- 11. Hillemann D, Weizenegger M, Kubica T, Richter E, Niemann S (2005) Use of Genotype MTBDR assay for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis complex isolates. J Clin Microbiol 43 (8) 3699–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Makinen J, Marttila HJ, Marjamaki M, Viljanen MK, Soini H (2006) Comparison of two commercially available DNA line probe assays for detection of multidrug-resistant Mycobacterium tuberculosis . J Clin Microbiol 2006 44 (2) 350–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miotto P, Piana F, Penati V, Caducci F, Migloiori GB, et al. (2006) Use of Genotype MTBDR assay for molecular detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis clinical strains isolated in Italy. J Clin Microbiol 44 (7) 2485–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brossier F, Veziris N, Tuffot-Pernot C, Jarlier V, Sougakoff W (2006) Performance of the Genotype MTBDR line probe assay for detection of resistance to rifampin and isoniazid in strains of Mycobacterium tuberculosis with low- and high-level resistance. J Clin Microbiol 44 (10) 3659–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barnard M, Albert H, Coetzee G, O'Brien R, Bosman ME (2008) Rapid molecular screening for multidrug-resistant tuberculosis in a high-volume public health laboratory in South Africa. Am J Respir Crit Care Med 177: 787–792. [DOI] [PubMed] [Google Scholar]
- 16. Evans J, Stead MC, Nicol MP, Segal H (2009) Rapid genotypic assays to identify drug-resistant Mycobacterium tuberculosis in South Africa. J Antimicrob Chemother 63 (1) 11–16. [DOI] [PubMed] [Google Scholar]
- 17.Kent PT, Kubica GP (1985). Public health mycobacteriology. A guide for a level III laboratory. Centers for Disease Control, Atlanta, GA. [Google Scholar]
- 18. Vestal AL (1977) In: Procedures for isolation and identification of Mycobacteria, U.S. Dep. of Health, Educ. and Welf. CDC Atlanta, Georgia, Publication (77-8230) 15–19. [Google Scholar]
- 19. Canetti G, Fox W, Khomenko A, Mahler HT, Menon NK, et al. (1969) Advances in techniques of testing mycobacterial drug sensitivity, and the use of sensitivity tests in tuberculosis control programmes. Bull World Health Organ 4: 21–43. [PMC free article] [PubMed] [Google Scholar]
- 20.RNTCP website. Available: http://www.tbcindia.nic.in/pdfs/Module%20for%20Laboratory%20Technician.pdf. Accessed 2013 Dec 1
- 21. Hillemann D, Kubica T, Agzamova R, Venera B, Rusch-Gerdes S, et al. (2005) Rifampicin and isoniazid resistance mutations in Mycobacterium tuberculosis strains isolated from patients in Kazakhstan. Int J Tuberc Lung Dis 9 (10) 1161–1167. [PubMed] [Google Scholar]
- 22. Cavusoglu C, Turhan A, Akinci P, Soyler I (2006) Evaluation of the Genotype MTBDR assay for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis isolates. J Clin Microbiol 44 (7) 2338–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miotto P, Piana F, Penati V, Canducci F, Migliori GB, et al. (2006) Use of genotype MTBDR assay for molecular detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis clinical strains isolated in Italy. J Clin Microbiol 44 (7) 2485–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mäkinen J, Marttila HJ, Marjamaki M, Viljanen MK, Soini H (2006) Comparison of two commercially available DNA line probe assays for detection of multidrug-resistant Mycobacterium tuberculosis . J Clin Microbiol 44 (2) 350–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brossier F, Veziris N, Truffot-Pernot C, Jarlier V, Sougakoff W (2006) Performance of the genotype MTBDR line probe assay for detection of resistance to rifampin and isoniazid in strains of Mycobacterium tuberculosis with low- and high-level resistance. J Clin Microbiol 44 (10) 3659–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bwanga F, Hoffner S, Haile M, Joloba ML (2009) Direct susceptibility testing for multi-drug resistant tuberculosis: a meta-analysis. BMC Infect Dis 9: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Albert Heidi, Bwanga Fred, Mukkada Sheena, Nyesiga Barnabas, Ademu Patrick, et al. (2010) Rapid screening of MDR-TB using molecular Line Probe Assay is feasible in Uganda. BMC Infectious Diseases 10: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Telenti A, Imboden P, Marchesi F, Cole S, Colston MJ, et al. (1993) Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis . Lancet 341: 647–650. [DOI] [PubMed] [Google Scholar]
- 29.Minutes of 17th RNTCP National Laboratory Committee held on 7th July 2009 at New Delhi; RNTCP website. Available: http://www.tbcindia.nic.in/Pdfs/17th%20(July%202009).pdf. Accessed 2013 Dec 1
- 30.Minutes of 18th National Laboratory Committee Meeting, 12th February, 2010; RNTCP website. Available: http://www.tbcindia.nic.in/Pdfs/18th_National_Laboratory_Committee_Meeting_Minutes.pdf Accessed 2013 Dec 1