Abstract
As a major tumor suppressor gene, the role of PinX1 in breast cancer and its molecular mechanism remain unclear. In this study, overexpression of PinX1 was generated in 3 breast cancer cell lines, and knockdown of PinX1 was performed in a nontumorigenic breast cell line. The regulation of PinX1 on cell proliferation and cell cycle was observed. A microarray-based lncRNA and mRNA expression profile screening was also performed. We found a lower growth rate, G0/G1 phase arrest, and S phase inhibition in the PinX1 overexpressed breast cancer cells, while a higher growth rate, decreased G0/G1 phase, and increased S phase rate in the PinX1 knocked-down nontumorigenic breast cell. A total of 977 mRNAs and 631 lncRNAs were identified as differentially expressed transcripts between PinX1 overexpressed and control MCF-7 cells. Further analysis identified the involvement of these mRNAs in 52 cancer related pathways and various other biological processes. 11 enhancer-like lncRNAs and 25 lincRNAs with their adjacent mRNA pairs were identified as coregulated transcripts. Our results confirmed the role of PinX1 as a major tumor suppressor gene in breast cancer cell lines and provided information for further research on the molecular mechanisms of PinX1 in tumorigenesis.
1. Introduction
The potent tumor suppressor PinX1 was originally isolated as one of the Pin2/TRF1 interaction proteins. Unlike other telomere-associated proteins, PinX1 is unique because it can directly interact with the telomerase catalytic component TERT and inhibit telomerase activity [1]. Previous studies determined that PinX1, recruited to the telomeres by TRF1, provided a critical link between TRF1 and telomerase inhibition to help maintain telomere homeostasis [2]. The inhibition of endogenous PinX1 in human cancer cells increases the telomerase activity and elongates the telomeres, whereas overexpression of PinX1 inhibits telomerase activity and induces cell crisis [1]. PinX1 knockout in mice can result in embryonic lethality in the PinX1 null mice (PinX1−/−) and the spontaneous development of a variety of malignant tumors in the PinX1 knockout heterozygous mice (PinX1+/−) [3], indicating that PinX1 is a potent telomerase inhibitor and a putative tumor suppressor [1, 4]. The functions of PinX1 are mainly attributable to the maintenance of telomerase activity and chromosomal stability [3, 4]. Although decreased expression of PinX1 was observed in breast cancer cell lines, and knockout of PinX1 in mice could cause breast cancer [3], the role of PinX1 in growth control of breast cancer cells and its molecular mechanism remains unclear. Therefore, in this study, we generated MCF-7, MDA-MB-231, and SK-BR-3 breast cancer cells stably overexpressing PinX1 and MCF-10A nontumorigenic breast cell knocking down PinX1 and assessed the role of PinX1 in growth control of the cells by MTT assay, focus formation, and flow cytometry. The localization of PinX1 in different phases in the cell cycle was observed. In addition, we also performed a genome wide screen of the mRNA and lncRNA expression profile alterations.
2. Materials and Methods
2.1. Cell Lines and Culture
Breast cancer cell lines MCF-7, MDA-MB-231, SK-BR-3, and nontumorigenic breast cell line MCF-10A were obtained from laboratory preservation. Three breast cancer cell lines were cultured in DMEM high glucose (Hyclone, Beijing) medium supplemented with 10% fetal bovine serum (Hyclone, South America) and 1% penicillin-streptomycin (10000 U/mL Penicillin and 10000 μg/mL Streptomycin, SolarBio, Beijing). MCF-10A was grown in DMEM/F12 medium (15 mM hepes buffer, Hyclone, USA) containing 5% donor equine serum (Sijiqing, Hangzhou), 20 ng/mL epidermal growth factor (Gibco, USA), 100 ng/mL cholera toxin (Merck Millipore, Germany), 500 ng/mL hydrocortisone (SolarBio, Beijing), 10 μg/mL insulin (Sigma, USA), and 1% penicillin-streptomycin (10000 U/mL Penicillin and 10000 μg/mL Streptomycin, SolarBio, Beijing). All the cells were cultured in a humidified atmosphere at 37°C with 95% air and 5% CO2.
2.2. Overexpression and Knockdown of PinX1 in Breast Cell Lines
PinX1 was cloned into the pcDNA3.1 (+) vector (Invitrogen, USA); the plasmid pcDNA3.1-PinX1 and empty vector were prepared using the QIAGEN Plasmid Midi Kit (QIAGEN, Germany). Then reconstructed and empty vector were transfected separately into the breast cancer cells by Lipofectamine 2000 (Invitrogen, USA) and a fresh cell culture medium containing 700 μg/mL, 1200 μg/mL, and 600 μg/mL of G418 (Amresco, USA) was applied 24 hours after transfection to MCF-7, MDA-MB-231, and SK-BR-3 accordingly. 3 weeks after the G418 screening, the cell clones were harvested using an Eclipse Ti-s (Nikon, Japan) microscope. The cell clones of pcDNA3.1-PinX1 group and empty vector group were maintained in the culture medium with 300 μg/mL of G418, and those with a high expression level of PinX1 were selected for later uses. Three different PinX1 siRNA fragments and siRNA NC were designed and synthesized by Ribbio (Guangzhou, China); the sequences were as follows: siRNA1, sense 5′-GGAGCCACAGAUCAUAUUA dTdT-3′, antisense 3′-dTdT CCUCGGUGUCUAGUAUAAU-5′; siRNA2, sense 5′-GGAGUAAUGACGAUUCCAA dTdT-3′, antisense 3′-dTdT CCUCAUUACUGCUAAGGUU-5′; siRNA3, sense 5′-GGACGCUACACUAGAAGAA dTdT-3′, antisense 3′-dTdT CCUGCGAUGUGAUCUUCUU-5′. MCF-10A cells were transfected separately with the three siRNAs and siRNA NC by Lipofectamine 2000 (Invitrogen, USA) to knockdown the PinX1.
2.3. Isolation of Total RNA and qRT-PCR Analysis
Total RNA was extracted from the cells mentioned above using a RNAiso plus kit (Takara, Dalian, China), according to the manufacturer's protocol. Qualified total RNA was reversely transcribed into 1st strand cDNA using the PrimeScript RT reagent Kit (Takara, Dalian, China). The PCR reaction included 25 μL of the 2× SYBR Premix Ex Taq II (Takara, Dalian, China), 80 ng of the cDNA template, 0.4 μM of the forward and reverse primers (Table 1) each, and ddH2O in a total volume of 50 μL. The PCR amplification was performed in a 7500 real-time PCR amplifier (ABI, USA) under the following conditions: 95°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C for 34 s. 7500 system SDS software (ABI, USA) was applied for acquiring the Ct values with manual thresholds. Expression of PinX1 was normalized by using GADPH as internal control; 2−ΔΔCt values were calculated to analyze the fold changes between different sample groups.
Table 1.
Primers used for real-time RT-PCR.
| mRNAs/lncRNAs | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| PinX1 | CCAGAGGAGAACGAAACCACG | ACCTGCGTCTCAGAAATGTCA |
| CUL2 | CATGTTCGGCATTTGCATAAGAG | GCACCCTTGCTGTATTCTTCC |
| HIF1A | CACCACAGGACAGTACAGGAT | CGTGCTGAATAATACCACTCACA |
| RET | ACACGGCTGCATGAGAACAA | GCCCTCACGAAGGGATGTG |
| JAK1 | CTTTGCCCTGTATGACGAGAAC | ACCTCATCCGGTAGTGGAGC |
| STAT1 | CGGCTGAATTTCGGCACCT | CAGTAACGATGAGAGGACCCT |
| BCL-2 | GGTGGGGTCATGTGTGTGG | CGGTTCAGGTACTCAGTCATCC |
| RP11-124O11.2 | TGCACCCATGATGAGGAAAT | CTGAAGAGGTAAGCCCTTTGT |
| lincRNA-TMEM30B-1 | CCGACTTGGTATCGACAACTT | CAGCATAGAGGTCTCCTGTTTC |
| GAPDH | CTGGGCTACACTGAGCACC | AAGTGGTCGTTGAGGGCAATG |
2.4. Western Blotting Analysis
Total proteins from all the cell samples were extracted using RIPA buffer containing 1 mM PMSF (Beyotime, Haimen, China). 50 μg of each protein was loaded on 10% SDS polyacrylamide gels for electrophoresis, and then the proteins were transferred to PVDF membranes (Millipore, USA) by a Transblot SD Cell semidry transfer machine (Bio Rad, USA). After being blocked by 5% nonfat milk, the membranes were incubated with goat anti-PinX1 antibody (1 : 500, Santa Cruz Biotechnology, Santa Cruz, USA) and HRP conjugated rabbit anti-goat IgG (1 : 5000, Multisciences, Hangzhou, China) for the detection of PinX1 protein. β-actin was detected by using the mouse anti-β-actin antibody (1 : 2000, Multisciences, Hangzhou, China) and HRP conjugated goat anti-mouse IgG antibody (1 : 5000, Multisciences, Hangzhou, China). HRPs on the immune complex were visualized by the BeyoECL Plus Kit (Beyotime, Haimen, China) and images were taken by the Image Station 4000R PRO scanner (Carestream Health, USA).
2.5. MTT Assay and Colorimetric Focus-Formation Assay
The role of PinX1 in growth control of MCF-7 and MCF-10A cells was determined by the MTT assay and focus-formation analysis. For MTT assay, cells were plated in 96-well plates at a density of 4,000 cells per well. The cells were incubated with 0.5% of MTT (Sigma, USA) for 4 h before assay. Then cells were lysed by DMSO (Sigma, USA) and the absorbance was determined by the TECAN Infinite 200 microplate reader (TECAN, Austria) at 490 nm. For colorimetric focus-formation assay analysis, MCF-7 cells stably transfected with pcDNA3.1-PinX1 and empty vector were plated in 6-well plates at a density of 10,000 cells per well, fixed by 100% of cold EtOH after 2 weeks of culture, and dyed by 5% crystal violate (Amresco, USA) in 30% of EtOH.
2.6. Cell Cycle Analysis by Flow Cytometry
For each of the MCF-7 and MCF-10A cell lines, 2 × 106 cells were harvested and fixed with 1.5 mL of 75% EtOH at 4°C overnight. Cells were resuspended in 0.5 mL of PBS and incubated with 50 μg/mL of RNaseA at 37°C for 30 min. Then the cells were stained by 50 μg/mL of PI at 4°C for 30 min and analyzed by a flow cytometry (BD Corporation, USA).
2.7. cRNA Labeling and Microarray Hybridization
mRNA from MCF-7 cells transfected with pcDNA3.1-PinX1 and empty vector was purified from the total RNA using a mRNA-ONLY Eukaryotic mRNA Isolation Kit (Epicentre, USA); subsequently, Cyanine-3-CTP (NEB, USA) was incorporated and the fluorescent cRNA of each sample was linearly amplified and transcribed using a Quick Amp Labeling Kit, One-Color (Agilent, USA). The labeled cRNA was then purified using a RNeasy Mini Kit (Qiagen, Germany). The hybridization solution for each sample was prepared using a Gene Expression Hybridization Kit (Agilent, USA) and 1.5 μg of the labeled cRNA. The hybridization solution was applied on the Human LncRNA Array V2.0 (Arraystar, USA) in a SureHyb chamber (Agilent, USA), and the hybridization was conducted in a hybridization oven (Agilent, USA) at 65°C for 17 hours.
2.8. Data Extraction and Analysis
Following the wash steps, the slide was scanned using a G2505C scanner (Agilent, USA). The raw data of both the array images were then extracted using the Feature Extraction software version 11.0.1.1 (Agilent, USA), and a quantile normalization of the raw data was performed using the GeneSpring GX v11.5.1 software (Agilent, USA). Boxplots and scatterplots were generated for the log2 ratios of mRNAs and lncRNAs between the two groups. Hierarchical clustering was performed to highlight the expression pattern of the distinguishable mRNAs and lncRNAs between the two samples. mRNAs and lncRNAs that had flags in the present or marginal (“All Targets Value”) in both samples were chosen for fold-change comparisons, and the differentially expressed mRNAs and lncRNAs with a fold change of ≥2 were identified for further analysis.
2.9. Bioinformatics Analysis
The differentially expressed mRNAs were submitted to the Gene Ontology database for GO category analysis and were then submitted to the Kegg database for pathway analysis. lncRNAs with enhancer-like functions were identified using a GENCODE annotation [5] of the human genes [6]. Rinn lincRNAs [7, 8] profiling and HOX cluster [9] were analyzed based on papers published by the John Rinn laboratory. The differentially expressed lncRNAs, especially the enhancer-like lncRNAs and Rinn lincRNAs, were remapped on the genome and their nearby coding gene pairs (distance <300 kb) were identified for lncRNA-mRNA coexpression analysis.
2.10. Validation of the Differentially Expressed mRNAs and lncRNAs by Real-Time RT-PCR
The expression of 6 cancer pathway genes and two of their nearby lncRNA pairs was validated by qRT-PCR in all the cell lines. The methods were the same as mentioned in Section 2.3. The forward and reverse primers for validation were listed in Table 1.
3. Results
3.1. Expression of PinX1 mRNA and Protein in the Transfected Cell Lines
The qRT-PCR and western blotting results indicated that pcDNA3.1-PinX1 transfected breast cancer cell lines MCF-7, MDA-MB-231 and SK-BR-3 showed a higher PinX1 expression level than their counterpart untransfected cells and empty vector transfected control cells (Figures 1(a) and 1(b)). For the three different PinX1 siRNA fragments transfected MCF-10A cells, all of them showed a lower PinX1 expression level than their counterpart untransfected cells and siRNA NC transfected control cells. PinX1 siRNA3 showed the highest interference rate (Figures 1(c) and 1(d)) and was applied to investigate the effects of PinX1 knockdown in the MCF-10A cells.
Figure 1.
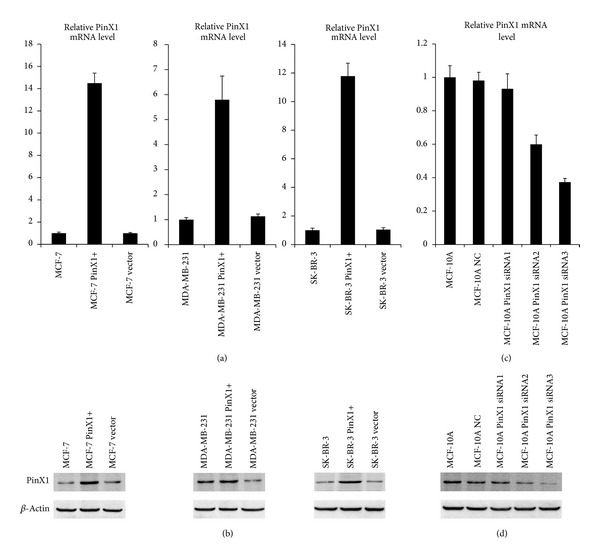
qRT-PCR and western blotting analysis of PinX1 expression in PinX1 overexpressed and knocked-down breast cell lines. (a) Fold changes (2−ΔΔCt values) by qRT-PCR showed increased expression of PinX1 mRNA in the pcDNA3.1-PinX1 transfected breast cancer cell lines MCF-7, MDA-MB-231, and SK-BR-3 when compared with their counterpart untransfected cells and empty vector transfected control cells. Expression levels were normalized for GAPDH. (b) Western blotting indicated upregulation of PinX1 protein in the pcDNA3.1-PinX1 transfected breast cancer cell lines MCF-7, MDA-MB-231, and SK-BR-3 in comparison with untransfected cells and empty vector transfected control cells. (c) Fold changes (2−ΔΔCt values) by qRT-PCR showed decreased expression of PinX1 mRNA in the PinX1 siRNA fragments transfected MCF-10A cells, when compared with the untransfected cells and siRNA NC transfected control cells. (d) Western blotting indicated downregulation of PinX1 protein in the PinX1 siRNA fragments transfected MCF-10A cells in comparison with untransfected cells and siRNA NC transfected control cells.
3.2. Growth Control of Breast Cell Lines by PinX1 Overexpression and Knockdown
MTT assay and colorimetric focus-formation assay were utilized to evaluate the growth control of PinX1 overexpression and knockdown in breast cell lines. Cell growth curves were plotted according to the data by MTT assay (Figures 2(a) and 2(b)). The results showed a lower growth rate in the pcDNA3.1-PinX1 transfected MCF-7 breast cancer cell line than the untransfected and vector transfected control cells and a higher growth rate in the PinX1 knockdown MCF-10A cell line than the untransfected and siRNA NC transfected control cells. The focus-formation assay indicated that pcDNA3.1-PinX1 stably transfected MCF-7 cells had a lower focus counting than the empty vector stably tranfected control cells (Figure 2(c)).
Figure 2.
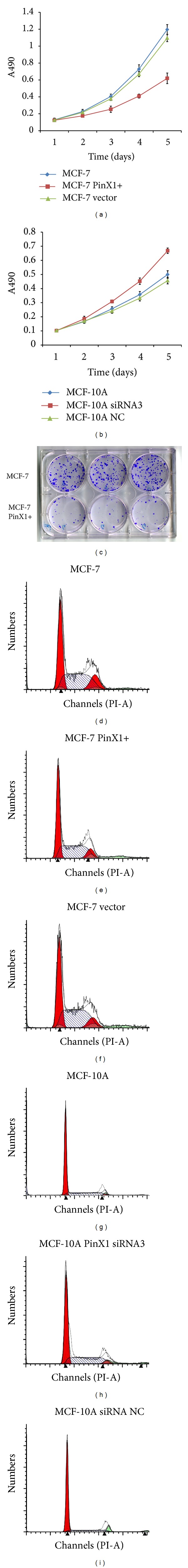
Growth control of breast cell lines by PinX1 overexpression and knockdown. (a) MTT assay showed a lower growth rate in the pcDNA3.1-PinX1 transfected MCF-7 cells than the untransfected and vector transfected control cells. (b) MTT assay showed a higher growth rate in the PinX1 siRNA3 tranfected MCF-10A cell line than the untransfected and siRNA NC transfected control cells. (c) Colorimetric focus-formation assay showed pcDNA3.1-PinX1 stable transfected MCF-7 cells had a lower focus counting than the empty vector stably tranfected control cells. (d), (e), and (f) Flow cytometry analysis indicated a G0/G1 phase arrest and S phase inhibition in the pcDNA3.1-PinX1 transfected MCF-7 cells compared to the untransfected and vector transfected control cells. (g), (h), and (i) Flow cytometry analysis indicated a decreased G0/G1 phase and increased S phase rate in the PinX1 knockdown MCF-10A cells compared to the untransfected and siRNA NC transfected control cells.
The effect of PinX1 overexpression and knockdown on the cell cycle was examined by the flow cytometry analysis. The results indicated a G0/G1 phase arrest and S phase inhibition in the pcDNA3.1-PinX1 transfected MCF-7 breast cancer cell line compared to the untransfected and vector transfected control cells (Figures 2(d)–2(f)) and a decreased G0/G1 phase and increased S phase rate in the PinX1 knockdown MCF-10A cells compared to the untransfected and siRNA NC transfected control cells (Figures 2(e)–2(i)).
3.3. Microarray Hybridization Data
The microarray data was deposited in the Gene Expression Omnibus (GEO) database (GEO accession GSE46756). After the quantile normalization and data filtering steps, 15,728 mRNAs and 14164 lncRNAs (Tables S1 and S2 in Supplementary Material available online at http://dx.doi.org/10.1155/2014/978984) out of the 33,000 probes of lncRNAs and 30,200 probes of coding genes were identified from the pcDNA3.1-PinX1 group and the empty vector group for fold-change comparison. The heat map of the hierarchical clustering results showed a distinguishable mRNA and lncRNA expression profiling between the two groups (Figures 3(a) and 3(b)). The scatterplot results showed that the distribution and expression variation of the log2 ratios of lncRNAs and mRNAs between the two groups were nearly the same (Figures 3(c) and 3(d)).
Figure 3.
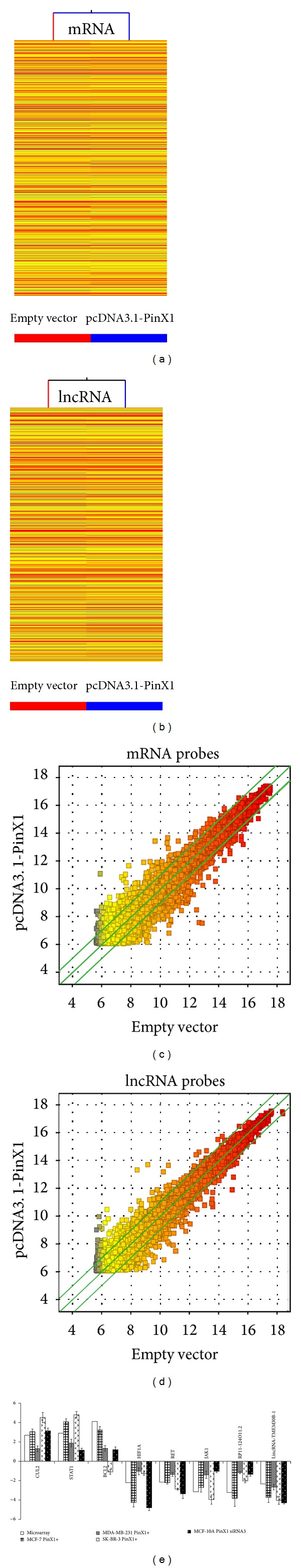
Microarray screening of the mRNA and lncRNA expression profile alterations in PinX1 overexpressed MCF-7 cells and qRT-PCR validation. (a), (b), (c), and (d) Heat maps and scatterplots of the distinguishable mRNA and lncRNA expression profiles between the pcDNA3.1-PinX1 group and the empty vector group of MCF-7 cells. Hierarchical clustering was performed and the results were displayed as a heat map, in which red denotes high relative expression levels and blue denotes low relative expression levels. (e) qRT-PCR validation of the microarray data in different breast cell lines. The expression fold change of the pcDNA3.1-PinX1 group versus the empty vector group of MCF-7 cells was verified by calculating the 2−ΔΔCt of real-time RT-PCR results. The result showed that the fold change in expression by qRT-PCR was mainly consistent with the microarray data.
3.4. Differentially Expressed mRNAs and lncRNAs
The differentially expressed mRNAs (Tables S3) and lncRNAs (Tables S4) were identified by the fold change in filtering, in which 366 mRNAs and 328 lncRNAs were upregulated, whereas 611 mRNAs and 303 LncRNAs were downregulated in the pcDNA3.1-PinX1 group. Microarray analysis found that PinX1 expression of the pcDNA3.1-PinX1 group was elevated by 8.49-fold.
3.5. GO Analysis and Pathway Analysis of the Differentially Expressed mRNAs
The results of the GO analysis of the differentially expressed mRNAs by biological processes, cellular components, and molecular function are presented in Table S5. The Kegg pathway analysis indicated that the differentially expressed mRNAs were involved in 203 pathways, among which 52 were cancer related pathways that had more than two differentially expressed mRNAs as regulating modules (Table 2).
Table 2.
Cancer related pathways of the differentially expressed mRNAs by Kegg pathway analysis.
| Pathway list | Upregulated genes | Downregulated genes |
|---|---|---|
| Metabolic pathways (19) | ALOX15B; FPGT; GAA; GART; HPSE; KYNU; PTGES; SQLE | ALDH9A1; FUT4; GAMT; GCLC; GOT2; NDUFV1; PCCB; POLD3; QARS; RPA1; RRM1; UMPS |
| Pathways in cancer (14) | BCL2; BRCA2; CUL2; FGFR1; FOS; STAT1; WNT2 | E2F2; FZD4; HIF1A; JAK1; LAMC1; MLH1; RET |
| MAPK signaling pathway (11) | FGFR1; FOS; NF1; NTRK2; PAK2; RASGRP1; RASGRP4 | CACNG1; MAP2K6; MAP3K5; NTF4 |
| Proteoglycans in cancer (11) | CAV2; FGFR1; HPSE; PXN; WNT2 | CD44; DDX5; ERBB3; FZD4; HIF1A; TIAM1 |
| Spliceosome (11) | DDX42; SNRPB2 | CDC5L; DDX23; DDX5; HNRNPM; ISY1; SF3B1; SF3B2; SFRS3; SR140 |
| Protein processing in endoplasmic reticulum (10) | BAG2; BCL2; CALR; EIF2AK2 | EIF2AK1; MAP3K5; OS9; TXNDC5; UBE2G2; XBP1 |
| Cell cycle (9) | CDKN2D; PCNA | BUB1; BUB1B; CDC45; CDC6; E2F2; MCM3; SMC3 |
| RNA transport (7) | EIF2B4; RNPS1 | EIF2B1; EIF2B5; NMD3; NUP133; NUP85 |
| Cytokine-cytokine receptor interaction (7) | CCL5; CXCL16; IL1RAP | BMP7; CXCL12; EDA; IL4R |
| Regulation of actin cytoskeleton (7) | FGFR1; ITGAL; MYL9; PAK2; PXN | CHRM1; TIAM1 |
| Chemokine signaling pathway (7) | CCL5; CXCL16; PXN; STAT1 | CXCL12; HCK; TIAM1 |
| Ubiquitin mediated proteolysis (7) | CUL2; NEDD4L; UBE2W; WWP2 | PIAS3; TRIP12; UBE2G2 |
| PI3K-Akt signaling pathway (7) | BCL2; FGFR1 | CHRM1; IL4R; JAK1; LAMC1; SGK3 |
| Leukocyte transendothelial migration (6) | ITGAL; MYL9; PXN; SIPA1 | CXCL12; F11R |
| Calcium signaling pathway (6) | ORAI2 | CHRM1; ERBB3; GNA14; GNAS; PPIF |
| Endocytosis (6) | CAV2; NEDD4L | ERBB3; RAB7A; RABEP1; RET |
| Focal adhesion (6) | BCL2; CAV2; MYL9; PAK2; PXN | LAMC1 |
| Cell adhesion molecules (CAMs) (6) | CDH15; ITGAL | ALCAM; CDH3; F11R; LRRC4B |
| Jak-STAT signaling pathway (5) | IRF9; STAT1 | IL4R; JAK1; PIAS3 |
| PPAR signaling pathway (5) | CD36; DBI; SCD; SORBS1 | SLC27A2 |
| RNA degradation (5) | XRN1 | CNOT4; DCP1A; DCP1B; DHX36 |
| Hippo signaling pathway (5) | AREG; BMP5; WNT2 | BMP7; FZD4 |
| ECM-receptor interaction (5) | CD36; CD47 | CD44; HMMR; LAMC1 |
| Purine metabolism (4) | GART | POLD3; RPA1; RRM1 |
| Toll-like receptor signaling pathway (4) | CCL5; FOS; STAT1 | MAP2K6 |
| mRNA surveillance pathway (4) | RNPS1; SMG7 | CSTF3; SMG5 |
| DNA replication (4) | PCNA | MCM3; POLD3; RFC1 |
| Biosynthesis of secondary metabolites (4) | GART; SQLE | ALDH9A1; GOT2 |
| Insulin secretion (4) | PCLO; RIMS2 | GNAS; KCNMB4 |
| Phagosome (4) | CALR; CD36 | CLEC7A; RAB7A |
| Transcriptional misregulation in cancer (4) | TMPRSS2 | DDX5; SIX1; WHSC1 |
| ErbB signaling pathway (4) | AREG; PAK2 | ERBB3; NRG1 |
| Lysosome (4) | GAA | AP4S1; CTSL2; GLA |
| Mismatch repair (4) | PCNA | MLH1; POLD3; RFC1 |
| Nucleotide excision repair (4) | PCNA | ERCC5; POLD3; RFC1 |
| Pyrimidine metabolism (4) | POLD3; RPA1; RRM1; UMPS | |
| Tight junction (3) | CASK; MYL9 | F11R |
| p53 signaling pathway (3) | RPRM | MDM4; PMAIP1 |
| HIF-1 signaling pathway (3) | BCL2; CUL2 | HIF1A |
| NF-kappa B signaling pathway (3) | BCL2; BLNK | CXCL12 |
| Cytosolic DNA-sensing pathway (3) | ADAR; CASP1; CCL5 | |
| T-cell receptor signaling pathway (3) | FOS; PAK2; RASGRP1 | |
| Arginine and proline metabolism (3) | ALDH9A1; GAMT; GOT2 | |
| ABC transporters (3) | ABCG2 | ABCA10; ABCC4 |
| GnRH signaling pathway (3) | CGA | GNAS; MAP2K6 |
| NOD-like receptor signaling pathway (3) | CASP1; CCL5; SUGT1 | |
| Homologous recombination (2) | BRCA2 | POLD3 |
| TGF-beta signaling pathway (2) | BMP5 | BMP7 |
| Adherens junction (2) | FGFR1; SORBS1 | |
| Apoptosis (2) | BCL2; IL1RAP | |
| Wnt signaling pathway (2) | WNT2 | FZD4 |
3.6. lncRNA Classification and Subgroup Analysis
Of the 849 enhancer-like lncRNAs (Table S6) that were detected by the GENCODE annotation, 15 were identified to be upregulated and 9 as downregulated lncRNAs. The adjacent coding genes that were differentially expressed (distance <300 kb) were detected among 11 out of the 24 differentially expressed enhancer-like lncRNAs; 4 of the lncRNA-mRNA pairs were regulated in the same direction (down-down) and 7 pairs in the opposite direction (up-down), as shown in Table S7.
Of all the 3,019 lincRNAs listed in the studies of the John Rinn laboratory [7, 8], 1,828 lincRNAs were identified by the present study (the profiling data of all the probes are provided in Table S8), including 50 upregulated and 24 downregulated lincRNAs. Further analysis indicated that 25 differentially expressed lincRNAs had adjacent coding gene pairs, of which 20 lincRNA-mRNA pairs were regulated in the same direction (up-up or down-down) and 5 pairs in the opposite direction (up-down). The results are shown in Table S9.
The profiling data of all the probes targeting the four HOX loci listed in Rinn's paper are presented in Table S10. Of all the 407 targeted discrete transcribed regions, lncRNAs and coding transcripts, 125 coding transcripts, and 241 noncoding transcripts were detected, with 6 of the coding transcripts being upregulated and 2 downregulated and 4 of the noncoding transcripts being upregulated and 4 downregulated.
3.7. Real-Time Quantitative RT-PCR Validation
The expression level of six differentially expressed mRNAs (CUL2, HIF1A, RET, JAK1, STAT1, and BCL2) in the cancer pathway and two of their nearby lncRNA pairs (RP11-124O11.2 and lincRNA-TMEM30B-1) was verified by real-time quantitative RT-PCR (qRT-PCR). The relative change in expression, as detected by the 2−ΔΔCt method, was mainly consistent with the microarray data (Figure 3(e)).
4. Discussion
PinX1 has been identified as an endogenous telomerase inhibitor and a major haploinsufficient tumor suppressor gene. Increasing evidence suggests that decreased expression of PinX1 plays a key role in different human cancers [1, 4]. Initially, when PinX1 was isolated, Zhou and Lu [1] reported that the gene was located on human chromosome 8p23.1, which is a region that frequently exhibits LOH in many human cancers, and that the depletion of endogenous PinX1 increased the tumorigenicity of HT1080 cells in nude mice. In subsequent follow-up studies, Kondo et al. [10] and Ma et al. [11] reported that LOH played a major role in the negative expression of PinX1 in gastric carcinoma and its level might be associated with the TNM stage of the cancer specimens. Wang et al. [12] suggested that PinX1 inhibited the telomerase activity in gastric cancer cells through the induction of the Mad1/c-Myc pathway and overexpression of PinX1 in MKN28 gastric carcinoma cells could enhance its sensitivity to 5-fluorouracil [13]. Park et al. [14] concluded that LOH of PinX1 might occur as an early event in the development of HCC. Cai et al. [15] suggested that the low expression level of PinX1 was correlated with ovarian carcinoma and could be used as an independent factor of poor prognosis. Moreover, Lai et al. [16], Chen et al. [17], and Zhang et al. [18] also reported that the overexpression of PinX1 could inhibit the tumorigenicity of nasopharyngeal carcinoma, hepatoma, and Burkitt's lymphoma cells.
The role of PinX1 in breast cancer was demonstrated by Zhou [3, 4] which decreased expression of PinX1 was observed in breast cancer cell lines, and knockout of PinX1 in mice could cause different epithelial cancers including breast cancer. However, the role of PinX1 in growth control of breast cancer cells and its molecular mechanism remains unclear. Therefore, in this study, overexpression and knockdown of PinX1 were generated in breast cell line to validate the role of growth control in carcinogenesis by PinX1. In addition, a microarray-based lncRNA and mRNA expression profile screening was also performed to evaluate the potential molecular pathways PinX1 may involved.
Our study suggested the role of PinX1 as a major tumor suppressor gene in breast cancer cell lines. Overexpression of PinX1 in breast cancer cell lines caused lower growth rate, G0/G1 phase arrest, and S phase inhibition, whereas knockdown of PinX1 in nontumorigenic breast cell line resulted in higher growth rate, decreased G0/G1 phase, and increased S phase rate. PinX1 might exert its tumor suppressor function in breast cancer cell lines by inhibiting cell proliferation through the Jak/STAT pathway and the HIF-1 signaling pathway; by resisting the protooncogene RET, transcription factor E2F2, focal adhesion related LAMC1, and DNA mismatch repair related MLH1; and by activating the tumor suppressor BRAC2.
lncRNAs are nonprotein coding transcripts, and they are more than 200 nucleotides in length. Because lncRNAs are generally expressed at low levels and are not strongly conserved, they had simply been disregarded as transcriptional noise over the past several years [19], and only a few were functionally annotated. However, recent studies showed that lncRNAs can regulate not only basal transcription but also the posttranscriptional [20] processes, including pre-mRNA processing, splicing, transport, translation, and siRNA-directed gene regulation. Furthermore, lncRNAs were also involved in epigenetic modifications, including DNA methylation [21] and histone modification [22], followed by chromatin remodeling. Some lncRNAs could directly bind proteins and regulate protein function [23]. Several association studies had recognized that lncRNAs may function on various aspects of cell biology and identified a large number of lncRNAs that were differentially expressed in disease states, including oncogenesis [24]. In our study, we also found that the overexpression of PinX1 could alter the lncRNA expression profile in MCF-7 breast cancer cells. Although the function of most aberrantly expressed lncRNAs was yet unknown, we determined that lncRNA-mRNA pairs like RP11-124O11.2 and RET as well as lincRNA-TMEM30B-1 and HIF1A were coexpressed and these pairs may function in the PinX1 regulated network.
5. Conclusions
In summary, our study confirmed the role of PinX1 as a major tumor suppressor gene in breast cancer cell lines, and identified the differentially expressed mRNAs and lncRNAs in PinX1 overexpressed MCF-7 breast cancer cells, which provides information for further research on the molecular mechanisms of PinX1 in tumorigenesis.
Supplementary Material
Human LncRNA Array V2.0 was applied for screening the alterations of lncRNA and mRNA expression profile between PinX1 overexpressed and control MCF-7 cells. After the quantile normalization and data filtering steps, the mRNA expression profile data qualified for fold-change comparison was listed in Tables S1, and lncRNA expression profile data was listed in Table S2. The differentially expressed mRNAs was listed in Tables S3 and differentially expressed lncRNAs was listed in Tables S4. GO analysis of the differentially expressed mRNAs by biological processes, cellular components and molecular functions was showed in Table S5. Profiling data of enhancer-like lncRNAs was listed in Table S6. The enhancer-like lncRNAs and their adjacent coding gene pairs was listed in Table S7. Profiling data of the Rinn lincRNAs was listed in Table S8. The Rinn lincRNAs and their adjacent coding gene pairs was listed in Table S9. Profiling data of the Hox clusters was listed in Table S10.
Acknowledgments
The work was supported by the National Natural Science Foundation of China (Grant nos. 81101536 and 81201565), the Program of the Pearl River Young Talents of Science and Technology in Guangzhou, China (2011J2200070), the Specialized Research Fund for the Doctoral Program of Higher Education (20104433120001 and 20124433120001), and the Guangdong Natural Science Foundation (Grant nos. S2012010009294 and S2012010009404).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contribution
Rong Shi and Jue-Yu Zhou contributed equally to this work.
References
- 1.Zhou XZ, Lu KP. The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell. 2001;107(3):347–359. doi: 10.1016/s0092-8674(01)00538-4. [DOI] [PubMed] [Google Scholar]
- 2.Soohoo CY, Shi R, Lee TH, Huang P, Lu KP, Zhou XZ. Telomerase inhibitor PinX1 provides a link between TRF1 and telomerase to prevent telomere elongation. Journal of Biological Chemistry. 2011;286(5):3894–3906. doi: 10.1074/jbc.M110.180174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou XZ, Huang P, Shi R, et al. The telomerase inhibitor PinX1 is a major haploinsufficient tumor suppressor essential for chromosome stability in mice. Journal of Clinical Investigation. 2011;121(4):1266–1282. doi: 10.1172/JCI43452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou XZ. PinX1: a sought-after major tumor suppressor at human chromosome 8p23. Oncotarget. 2011;2(10):810–819. doi: 10.18632/oncotarget.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrow J, Denoeud F, Frankish A, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biology. 2006;7(supplement 1, article S4) doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ørom UA, Derrien T, Beringer M, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondo T, Oue N, Mitani Y, et al. Loss of heterozygosity and histone hypoacetylation of the PINX1 gene are associated with reduced expression in gastric carcinoma. Oncogene. 2005;24(1):157–164. doi: 10.1038/sj.onc.1207832. [DOI] [PubMed] [Google Scholar]
- 11.Ma Y, Wu L, Liu C, Xu L, Li D, Li J-C. The correlation of genetic instability of PINX1 gene to clinico-pathological features of gastric cancer in the Chinese population. Journal of Cancer Research and Clinical Oncology. 2009;135(3):431–437. doi: 10.1007/s00432-008-0471-6. [DOI] [PubMed] [Google Scholar]
- 12.Wang H-B, Wang X-W, Zhou G, et al. PinX1 inhibits telomerase activity in gastric cancer cells through Mad1/c-Myc pathway. Journal of Gastrointestinal Surgery. 2010;14(8):1227–1234. doi: 10.1007/s11605-010-1253-4. [DOI] [PubMed] [Google Scholar]
- 13.Wang H-B, Wang W-Q, Wang X-W, et al. PinX1 gene transfection enhances the sensitivity of gastric carcinoma cell line to 5-fluorouracil. Hepato-Gastroenterology. 2011;58(106):682–686. [PubMed] [Google Scholar]
- 14.Park WS, Lee JH, Park JY, et al. Genetic analysis of the liver putative tumor suppressor (LPTS) gene in hepatocellular carcinomas. Cancer Letters. 2002;178(2):199–207. doi: 10.1016/s0304-3835(01)00841-2. [DOI] [PubMed] [Google Scholar]
- 15.Cai M-Y, Zhang B, He W-P, et al. Decreased expression of PinX1 protein is correlated with tumor development and is a new independent poor prognostic factor in ovarian carcinoma. Cancer Science. 2010;101(6):1543–1549. doi: 10.1111/j.1349-7006.2010.01560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai X-F, Shen C-X, Wen Z, Qian Y-H, Yu C-S, Wang J-Q. PinX1 regulation of telomerase activity and apoptosis in nasopharyngeal carcinoma cells. Journal of Experimental & Clinical Cancer Research. 2012;31(12) doi: 10.1186/1756-9966-31-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen G, Da L, Wang H, et al. HIV-tatmediated delivery of an LPTS functional fragment inhibits telomerase activity and tumorigenicity of hepatoma cells. Gastroenterology. 2011;140(1):332–343. doi: 10.1053/j.gastro.2010.08.046. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Jiang Y, Zheng Y, et al. Selective killing of Burkitt’s lymphoma cells by mBAFF-targeted delivery of PinX1. Leukemia. 2011;25(2):331–340. doi: 10.1038/leu.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu S, Zhang X-O, Yang L. Panning for long noncoding RNAs. Biomolecules. 2013;3(1):226–241. doi: 10.3390/biom3010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. Journal of Molecular Biology. 2012;425(19):3723–3730. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohammad F, Pandey GK, Mondal T, et al. Long noncoding RNA-mediated maintenance of DNA methylation and transcriptional gene silencing. Development. 2012;139(15):2792–2803. doi: 10.1242/dev.079566. [DOI] [PubMed] [Google Scholar]
- 22.Chu C, Qu K, Zhong F, Artandi S, Chang H. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Molecular Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arun G, Akhade VS, Donakonda S, Rao MR. mrhl RNA, a long noncoding RNA, negatively regulates Wnt signaling through its protein partner Ddx5/p68 in mouse spermatogonial cells. Molecular and Cellular Biology. 2012;32(15):3140–3152. doi: 10.1128/MCB.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maruyama R, Suzuki H. Long noncoding RNA involvement in cancer. Biochemistry and Molecular Biology Reports. 2012;45(11):604–611. doi: 10.5483/BMBRep.2012.45.11.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Human LncRNA Array V2.0 was applied for screening the alterations of lncRNA and mRNA expression profile between PinX1 overexpressed and control MCF-7 cells. After the quantile normalization and data filtering steps, the mRNA expression profile data qualified for fold-change comparison was listed in Tables S1, and lncRNA expression profile data was listed in Table S2. The differentially expressed mRNAs was listed in Tables S3 and differentially expressed lncRNAs was listed in Tables S4. GO analysis of the differentially expressed mRNAs by biological processes, cellular components and molecular functions was showed in Table S5. Profiling data of enhancer-like lncRNAs was listed in Table S6. The enhancer-like lncRNAs and their adjacent coding gene pairs was listed in Table S7. Profiling data of the Rinn lincRNAs was listed in Table S8. The Rinn lincRNAs and their adjacent coding gene pairs was listed in Table S9. Profiling data of the Hox clusters was listed in Table S10.


