Abstract
The formation of a single lumen during tubulogenesis is crucial for the development and function of many organs. Although 3D cell culture models have identified molecular mechanisms controlling lumen formation in vitro, their function during vertebrate organogenesis is poorly understood. Using light sheet microscopy and genetic approaches we have investigated single lumen formation in the zebrafish gut. Here we show that during gut development multiple lumens open and enlarge to generate a distinct intermediate, which consists of two adjacent unfused lumens separated by basolateral contacts. We observed that these lumens arise independently from each other along the length of the gut and do not share a continuous apical surface. Resolution of this intermediate into a single, continuous lumen requires the remodeling of contacts between adjacent lumens and subsequent lumen fusion. We show that lumen resolution, but not lumen opening, is impaired in smoothened (smo) mutants, indicating that fluid-driven lumen enlargement and resolution are two distinct processes. Furthermore, we show that smo mutants exhibit perturbations in the Rab11 trafficking pathway and demonstrate that Rab11-mediated trafficking is necessary for single lumen formation. Thus, lumen resolution is a distinct genetically controlled process crucial for single, continuous lumen formation in the zebrafish gut.
Keywords: Lumen, Remodeling, Tubulogenesis
INTRODUCTION
Tubulogenesis is a crucial process during the formation of many organs, including the pancreas, lungs, vasculature, mammary gland and gut. Tube formation mechanisms are diverse across organ systems, but they all result in a structure with a single lumen. Tubes arising from a polarized epithelium typically undergo a process of epithelial wrapping or budding that is driven primarily by changes in cell shape. By contrast, tubes originating from unpolarized cells form through a process of cord hollowing or cavitation that requires the establishment of cell polarity and de novo lumen formation (Lubarsky and Krasnow, 2003; Martin-Belmonte and Mostov, 2008). Lumen formation occurs through the coordinated effort of several distinct cellular processes, including intracellular trafficking to a prospective apical domain, de novo apical membrane biogenesis, lumen enlargement and, in many cases, epithelial remodeling.
De novo lumen formation is integral to the development of tubes that form from an unpolarized epithelium and has been extensively studied in vitro in 3D cysts. To initiate lumen formation, apical membrane proteins such as Podocalyxin accumulate in Rab11 and Rab8a-positive vesicles. These vesicles are then delivered to the plasma membrane where, together with the exocyst and the Par3 complex, they fuse to generate an apical surface (Bryant et al., 2010). Although these studies highlight the importance of apical membrane trafficking in lumen formation, such in vitro systems cannot fully recapitulate the complexity of a three-dimensional organ. For example, in most 3D cyst models the lumen typically forms between two differentiated epithelial cells by recycling of apical membrane components from the surface to the new lumen (Bryant et al., 2010). Thus, lumen formation in vitro does not require processes of epithelial transformation and remodeling that are necessary for tube formation in vivo in many organs. Because of these differences, the cellular mechanisms controlling lumen formation in large unbranched tubes, particularly in vertebrates, remain poorly understood.
The zebrafish intestine begins as a solid rod of endodermal cells that differentiate into epithelial cells and undergo a cord hollowing process to form a tube. Lumen formation initiates with the development of multiple actin-rich foci between cells and is followed by the localization of junctional proteins at multiple points within the intestine (Horne-Badovinac et al., 2001). Small lumens form at these points and expand, coalesce and eventually form a single continuous lumen (Bagnat et al., 2007). Interestingly, intestinal villus formation in the rat epithelium may also form through fusion of small secondary lumens (Madara et al., 1981). Previous work in zebrafish showed that paracellular ion transport regulated by Claudin15 and the Na+/K+-ATPase drives fluid accumulation, promoting lumen expansion and coalescence in to a single lumen (Bagnat et al., 2007). However, as the gut lumen forms without apoptosis (Ng et al., 2005), other cellular processes such as epithelial remodeling must occur to facilitate lumen coalescence.
Here we provide a high-resolution in vivo characterization of lumen formation in the zebrafish gut. Using this approach we identified a crucial stage in lumen coalescence that is regulated by smoothened (smo) signaling. We show that single, continuous lumen formation starts with rapid lumen expansion in an anterior to posterior (AP) manner. Expansion is followed by an intermediate structure characterized by adjacent unfused lumens. These lumens fuse through both luminal membrane expansion and the loss of adhesion at the fusion site. We also found that smo mutants are unable to undergo the crucial process of lumen fusion and thus fail to form a single continuous lumen in the gut. Furthermore, we show smo mutants exhibit altered localization of Rab11a and demonstrate that proper Rab11-mediated trafficking is important to the formation of a single continuous lumen in the zebrafish intestine.
RESULTS
Single lumen formation involves a distinct double lumen intermediate
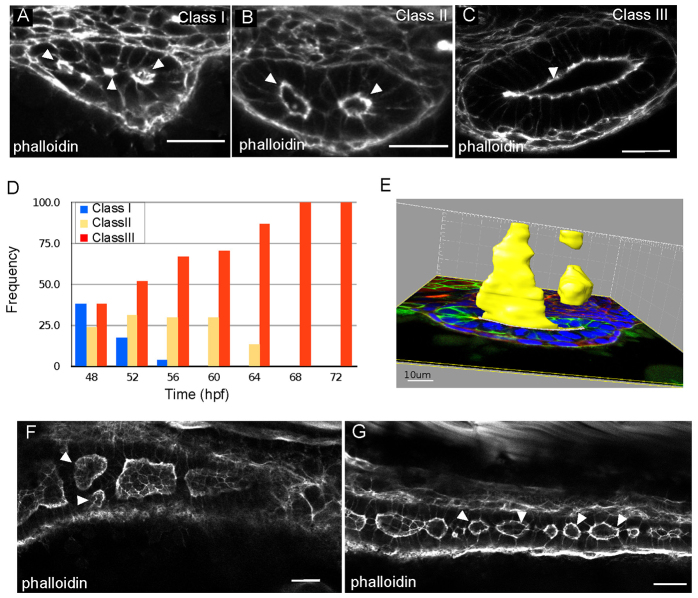
Lumen formation in the zebrafish gut begins with the appearance of multiple small lumens that enlarge through fluid accumulation and coalesce to form a single lumen (Bagnat et al., 2007). However, fluid accumulation alone cannot drive the cellular rearrangements necessary for lumen coalescence. The complexity of the tissue suggests that other processes are required. To elucidate the process of lumen formation in the zebrafish intestine we performed a timecourse analysis from 48 hours post fertilization (hpf) to 72 hpf and characterized the appearance of the lumen at 4-hour intervals. Analysis of fixed, thick transverse sections by confocal microscopy revealed a range of lumen morphologies. We classified the intestinal tubes into three categories: class I, containing multiple small lumens, in which two to four actin foci or small lumens span the intestine (Fig. 1A); class II, represented by enlarged, unfused lumens, in which a bridge of cells separate open lumens (Fig. 1B); class III, single lumens, characterized by one enlarged continuous lumen (Fig. 1C). In 48 hpf embryos, all three lumen types are apparent with relatively similar frequency. Class I and class III lumens were found in 38% of embryos, whereas class II lumens were found in 24% of gut sections. Over the next 12 hours the appearance of class I lumens decreased, whereas the frequency of class II lumens increased to 30% and class III lumens increased to 70% at 60 hpf. During the subsequent 12 hours, the number of embryos with class II lumens decreased and by 68 hpf only single lumen guts were observed (Fig. 1D). Thus, single lumen formation is preceded by two stereotypic luminal arrangements that include both multiple small lumens and enlarged, unfused lumens.
Fig. 1.
Lumens enlarge and fuse during single lumen formation in the zebrafish gut. (A-C) Confocal images of cross sections of wild-type embryos exhibiting class I (A), class II (B) and class III (C) lumens, stained with phalloidin. Arrowheads indicate the lumens. (D) Quantification of lumen phenotypes between 48 and 72 hpf: 48 hpf n=21, 52 hpf n=29, 56 hpf n=27, 60 hpf n=27, 64 hpf n=30, 68 hpf n=21, 72 hpf n=26. (E) Space-fill projection from a 200 μm confocal stack of an intestine section at the resolution stage. Yellow, lumen; green, GFP-CaaX; blue, DAPI. (F) Confocal whole-mount image of the anterior gut at 58 hpf stained with phalloidin (red). Arrowheads point to adjacent unfused lumens. (G) Confocal whole-mount image of the posterior gut at 58 hpf stained with phalloidin (red). Arrowheads point to lumens. Scale bars: 20 μm in A-C; 10 μm in E; 20 μm in F,G.
The most frequently observed luminal arrangement before single lumen formation is two enlarged lumens. However, this arrangement is not simply the result of two parallel lumens spanning the intestine. Using Imaris imaging software, we generated a 3D rendering of lumen size and shape from a 200-μm transverse confocal stack. Even within this small region, lumens are discontinuous and highly dynamic in shape and size (Fig. 1E). To gain a better understanding of how these discontinuous lumens are arranged along the AP axis we performed whole-mount confocal imaging. Analysis of the anterior intestinal bulb at 58 hpf revealed two enlarged lumens side by side (Fig. 1F), which is representative of the unfused lumens (class II) we observed in transverse cross section. These adjacently arranged lumens are most frequently observed in the anterior gut, probably owing to the larger diameter and number of cells in this region. Enlarged discontinuous lumens were found along the AP length of the intestine. Toward the posterior end of the intestine, discontinuous lumens were more abundant and of smaller size (Fig. 1G). The unfused lumen phenotype (class II) represents a previously uncharacterized stage in normal lumen formation, and we have termed this phase the ‘lumen resolution stage’. Together, these data suggest that lumen formation occurs through stages of multiple small, and expanded unfused lumens before resolving into a single continuous lumen.
Rapid lumen expansion and fusion during tubulogenesis
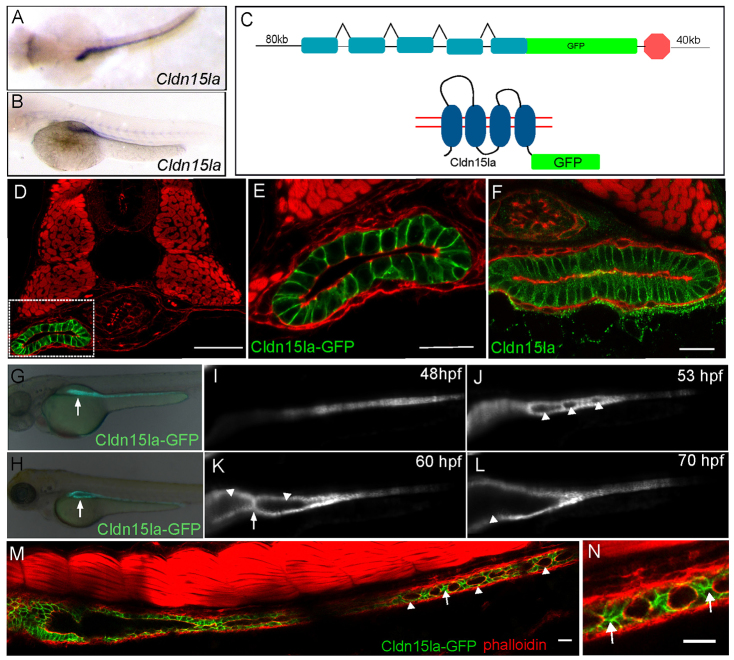
Analysis of fixed tissue sections suggested that initial lumen expansion and lumen resolution are distinct phases of intestinal lumen formation. As sectional analysis only offers a static snapshot of lumen formation, we wanted to monitor lumen formation in the intestine using live imaging. To image lumen coalescence in vivo, we required a new transgenic line that is intestine-specific and expresses before single lumen formation. To identify intestine-specific genes we isolated intestinal epithelial cells using a Tg(-1.0ifabp:GFP-CaaX) line which expresses membrane green fluorescent protein (GFP) in the intestinal cells starting around 120 hpf. Using RNA isolated from these cells we performed a microarray analysis and found that one of the most highly intestine-enriched genes was claudin 15-like a (cldn15la) (data not shown), a member of the Claudin family of tetraspanning membrane proteins (Furuse et al., 1998). By in situ hybridization, cldn15la was highly expressed and restricted to the intestine by 50 hpf (Fig. 2A,B). To generate a transgenic line expressing Cldn15la-GFP we used bacterial artificial chromosome (BAC) recombineering to create a C-terminal fusion protein (Fig. 2C).
Fig. 2.
Generation of an intestine-specific transgenic line. (A,B) Dorsal, top panel, and lateral view, bottom panel, in situ hybridization showing claudin 15-like a expressed specifically in the intestine at 56 hpf. (C) Schematic of TgBAC(cldn15la-GFP) generation. The recombination target is shown on top, and the expected protein structure is shown on the bottom. (D) Confocal cross-section of a 72 hpf TgBAC(cldn15la-GFP) embryo. (E) Magnification of box from D. (F) Immunolocalization of Cldn15la to the basolateral membranes of intestinal epithelial cells. (G,H) Whole-mount fluorescent images of 55 hpf and 75 hpf embryos expressing TgBAC(cldn15la-GFP). Arrows indicate the gut tube. (I-L) Live imaging of TgBAC(cldn15la-GFP) using SPIM. Snapshots from a single plane from 48-70 hpf. Arrowheads indicate lumens; arrows point to cell-cell contacts between lumens. (M) Stitched confocal whole-mount images of a TgBAC(cldn15la-GFP) embryo show unfused lumens (arrowhead) in the posterior intestine at 68 hpf that are separated by cell-cell contacts (arrow). (N) Magnification of cell-cell contacts from M. Arrows indicate contacts. Phalloidin (red). Scale bars: 50 μm in D-F; 20 μm in M,N.
Cldn15la-GFP expression was first observed at 48 hpf in the intestinal epithelium and remained expressed throughout the course of lumen formation (Fig. 2G,H). An analysis of transverse sections revealed that Cldn15la-GFP is restricted to the intestine and is not expressed in other endoderm-derived organs (Fig. 2D). Cldn15la-GFP was found localized to the lateral surface of the intestinal epithelium (Fig. 2E). Although Claudin proteins are components of tight junctions and typically localize to the subapical region (Furuse et al., 1998), studies have shown that several members of this protein family also localize to lateral membranes during morphogenesis (Gregory et al., 2001; Inai et al., 2007; Westmoreland et al., 2012), including the closely related zebrafish claudin Cldn15lb (Cheung et al., 2012). To determine whether Cldn15la-GFP lateral membrane localization represented the endogenous protein localization we generated an antibody against the C-terminus of Cldn15la. Similar to the BAC transgenic construct, Cldn15la localized to the lateral membrane in intestinal epithelial cells, indicating that the Cldn15la-GFP fusion recapitulates endogenous expression and localization (Fig. 2F). Furthermore, we generated additional transgenic lines with a different linker sequence between GFP and Cldn15la and observed a similar localization pattern (data not shown). The Cldn15la-GFP transgene allowed for improved examination of the cellular and luminal arrangements within the intestine. Whole-mount imaging of the entire intestine revealed that lumen fusion begins in the anterior region and proceeds in an anterior to posterior direction (Fig. 2M). In addition, we observed that unfused lumens were frequently separated by single cell-cell contacts (Fig. 2N).
To visualize the process of lumen formation live, we used selective plane illumination microscopy (SPIM) (Huisken and Stainier, 2009) to image TgBAC(cldn15la-GFP) embryos (supplementary material Movies 1, 2). Initially, several small lumens were seen opening along the AP length of the intestine (Fig. 2J). These lumens were often separated by a few cells, which is similar to those observed in fixed whole-mount embryos at the resolution stage. Initially, the expansion of these lumens was rapid and followed by local fusion events that resulted in two to three large luminal compartments. The larger lumens remained separated by a one- or two-cell-thick cellular bridge for an extended period of time, yielding a distinct intermediate (Fig. 2K). Ultimately, these large lumens resolve into one (Fig. 2L). Taken together, our morphological and live imaging studies reveal that single lumen formation in the zebrafish intestine involves two distinct morphological and kinetic phases and identify a previously unknown stage characterized by the presence of large, unfused lumens.
Basolateral adhesions localize to cell-cell contacts between lumens
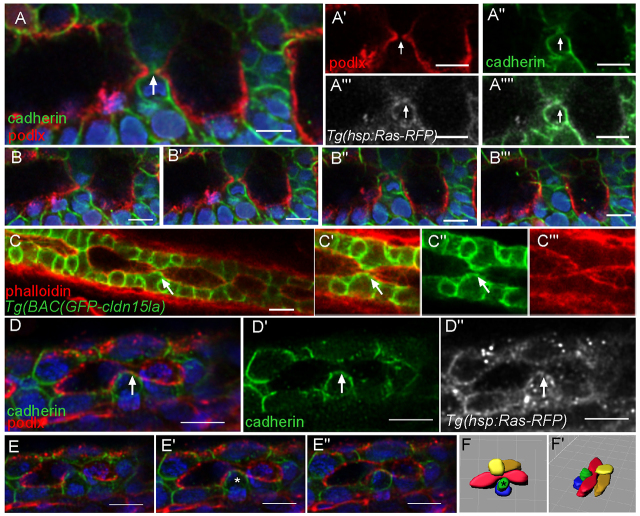
As lumen coalescence occurs in the absence of apoptosis (Ng et al., 2005), processes must be involved to facilitate tissue remodeling during lumen resolution. To address the process of lumen fusion, we further characterized the resolution stage. Analysis of the cellular architecture of transverse intestinal sections at the resolution stage using a membrane GFP marker, Tg(hsp70l:GFP-CaaX)pd1008, revealed that lateral lumens were often separated by a bridge of cells the contacts of which form a Y- or T-shaped arrangement between two adjacent lumens (Fig. 3A,A′). To determine the identity of the bridge contacts, we examined the localization of specific apical and basolateral proteins. Using a Tg(hsp70l:GFP-podxl)pd1080 line, we found that the apical membrane protein Podocalyxin localized to the apical surface surrounding the lumens and was absent from the connecting bridge (Fig. 3B). Similarly, the tight junction protein Zo-1 (Tjp1a - ZFIN) was restricted to the subluminal area and was not found at the membrane between lumens (Fig. 3C). By contrast, the adhesion proteins cadherin and β-catenin were localized to all basolateral membranes and were also located on the bridge membrane separating the two lumens (Fig. 3D,E). These data reveal that during the resolution stage, cells surrounding the lumens are polarized and adjacently arranged lumens within an intestinal cross section are separated by basolateral contacts that exclude apical proteins.
Fig. 3.
Cell-cell contacts are found between lumens. (A-E) Confocal cross sections of embryos at the lumen resolution stage. (A) Tg(hsp70l:GFP-CaaX) labels all cell membranes. (A′) Cartoon diagram of Fig. 3A depicting laterally arranged lumens in red and ‘bridge’ contacts in green. (B,B′) Apical protein, GFP-Podocalyxin surrounds the lumen but is not found at bridge contacts. Phalloidin (red). (C,C′) Antibody staining against Zo-1 labels tight junctions. Phalloidin (red). (D-E′) Antibody staining against cadherin and β-catenin labels basolateral contacts and ‘bridge’ contacts between lumens. Phalloidin (green). (F) Cartoon depicting two scenarios of lumen fusion along the AP axis. Apical membrane (red) can be deposited on membranes between lumens (top) or lumens may arise isolated and fuse directly without an apical membrane linker (bottom). (G-G′) Whole-mount confocal image of a lumen resolution stage embryo expressing GFP-Podocalyxin (red) and stained for cadherin in green. Cadherin localizes to basolateral contacts separating lumens. Arrows indicate lumens; arrowheads indicate bridge contact. Scale bars: 20 μm.
We next examined the process of lumen resolution along the length of the intestine. The generation of a single continuous lumen is a more complex process involving the coordination of several lumens along the gut. There are two possible scenarios in which a single continuous lumen can resolve from multiple discontinuous lumens (Fig. 3F). One possibility is that apical membrane can be deposited at bridge contacts between lumens, forming a continuous path to connect the enlarging lumens. Alternatively, each lumen may be an autonomous unit separated by basolateral contacts, similar to adjacently arranged lumens. In this case, single lumen formation would require the disengagement of the cell-cell contacts between adjacent lumens. To determine which scenario most accurately represents the process of lumen resolution along the intestine we performed whole-mount analysis of Tg(hsp70l:GFP-podxl)pd1080 embryos stained for cadherin. Consistent with the transverse section data, lumens along the AP axis were frequently separated by Y- and T-shaped cadherin-positive contacts and GFP-Podxl was restricted to the membrane surrounding the lumens (Fig. 3G-G′). Thus, the organization of adjacent lumens seen in transverse sections is analogous to the organization of adjacent lumens along the AP axis. Furthermore, we found no evidence of apical membrane deposition between two lumens before lumen fusion.
Lumen resolution may occur via the expansion and direct fusion of luminal membranes, or through the reduction and breaking of contacts between the lumens or both. Before lumen fusion adjacent lumens expand and the connecting bridge appears to shrink. We observed that in regions where the basolateral bridge contact was particularly narrow, GFP-podlx-positive membranes protruded from the adjacent luminal surfaces toward a central area with diffuse cadherin signal, probably originating from the internalization of the contact (Fig. 4A-A′; supplementary material Movie 3). We termed this type of resolution event ‘luminal fusion’. Further analysis of cadherin-stained and TgBAC(GFP-cldn15la) embryos revealed that in some instances during the fusion process, cadherin and GFP-Cldn15la can still be found at the fusion site. Although the basolateral proteins are not completely removed, cell-cell adhesion is lost. This localization at the cell surface probably originates from the separated bridge contact, suggesting that the adhesions between the cells had snapped before their complete internalization (Fig. 4C-F). Resolution through luminal fusion seems to be the predominant mode (65%), whereas snapping accounted for 35% of the events (n=20). These data indicate that lumen resolution involves remodeling of bridge contacts through both apical membrane expansion and the reduction of the adhesion contact.
Fig. 4.
Basolateral adhesions separate lumens along the AP axis. (A-A′′) Whole-mount confocal image of an embryo expressing GFP-Podocalyxin (false colored in red) and stained for cadherin (green) shows luminal expansion during a fusion event. Ras-RFP (white) marks cell membranes. Arrows mark fusion event. Blue, DAPI. (B-B′′) Optical sections from a z-stack surrounding a fusion event. (C-C′) Whole-mount confocal image of a TgBAC(GFP-cldn15la) embryo shows a putative adhesion snapping event during fusion. The arrow marks adhesion at the surface. (D-D′) Whole-mount confocal image of an embryo expressing GFP-Podocalyxin (red) and stained for cadherin (green) shows adhesion snapping during fusion. Ras-RFP (white) marks cell membranes. The arrow marks adhesion at the surface. (E-E′) Optical sections from z-stack surrounding a fusion event. The asterisk marks a cell with adhesion. (F,F′) Space-fill projection labeling cells surrounding the fusion event. Lumen, red. The asterisk marks a cell with adhesion. Scale bars: 10 μm.
smoothened mutants exhibit impaired lumen fusion
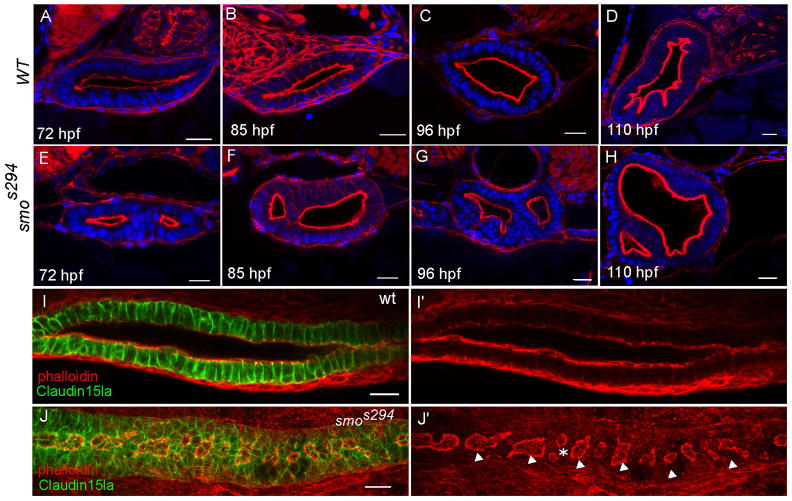
We next sought to identify a genetic model to investigate the resolution stage of lumen formation in the intestine. The hedgehog (Hh) pathway is known to be involved in gastrointestinal tract morphogenesis in mammals and cloaca formation in zebrafish (Parkin et al., 2009; Ramalho-Santos et al., 2000; Wallace and Pack, 2003). Therefore, we examined lumen formation in embryos mutant for smoothened (smo), the hedgehog co-receptor. We performed transverse sectional analyses of homozygous smos294 (Aanstad et al., 2009) mutant embryos at 72 hpf, a time point when a single continuous lumen is well established in wild-type embryos. The smos294 allele contains a mutation in a conserved cystine residue in the extracellular domain of the protein and is essential for full activation of the Hh pathway (Aanstad et al., 2009). At 72 hpf, ∼43% of smos294 mutant embryos (n=21 mutants) exhibit impaired intestinal lumen fusion (Fig. 5A,E). To confirm that the smoothened mutation is responsible for the lumen formation defect, we examined a null allele of smoothened, smohi1640 (Chen et al., 2001) and found the same phenotype in a similar proportion of embryos (44%, n=27) (data not shown). The smos294 phenotype was similar to the class II wild-type intermediate, which indicates a failure at the resolution stage. To determine if unfused lumens resolve at a later time in development, we also examined embryos beyond 72 hpf. The unfused lumen phenotype was observed at 85 hpf, 96 hpf and 110 hpf (Fig. 5B-D,F-H). At 96 hpf 27% of smos294 embryos (n=26 mutants) continued to exhibit unfused lumens, indicating that impaired lumen fusion in mutants is not due to a developmental delay. In addition to transverse sectional analysis, we also examined smos294 in whole mount to determine if impaired fusion was displayed along the entire intestine or was restricted to the anterior intestinal bulb. At 72 hpf, smos294 mutants exhibited several unfused lumens along the intestine (Fig. 5I-J), which is consistent with the results observed in wild-type embryos at the resolution stage. It is important to note that in smos294 mutants, unfused lumens are fully open and continue to expand as cells divide (Fig. 5E-H). These results indicate that the smos294 phenotype results from a failure in lumen resolution and not from impaired fluid accumulation. Together, these data support the idea that fluid alone cannot drive single lumen formation and reveal that lumen opening and lumen fusion are two distinct events required for single lumen formation that can be genetically uncoupled.
Fig. 5.
smos294 mutants exhibit a lumen fusion defect. (A-H) Confocal cross sections of wild-type (top) and smos294 (bottom) intestines at 72 hpf, 85 hpf, 96 hpf and 110 hpf. Phalloidin (red). (I,J) Confocal whole-mount image of wild-type and smos294 embryos expressing TgBAC(cldn15la-GFP) to highlight the cellular and luminal architecture of the intestine at 72 hpf. (I) Wild-type intestine, (J) smos294 intestine. Arrowheads, lumens; asterisk, adjacent lumens. Scale bars: 20 μm.
To examine the spatiotemporal expression of smo signaling we used a transgenic reporter line for the Hh pathway target gene patched (Choi et al., 2013). At 48 and 72 hpf signaling was observed in the mesenchyme surrounding the gut (supplementary material Fig. S1A,B). Hh signaling is known to play an important role in the differentiation of mesodermal precursors into smooth muscle. In smos294 the mesenchymal layer contains fewer, more elongated cells compared with wild type (Fig. 5E-H). Therefore, we examined the differentiation of the mesenchymal layer in smo mutants. In situ hybridization revealed that expression of the smooth muscle marker αSMA is lacking in mutant embryos at 72 hpf, indicating an absence of differentiated smooth muscle surrounding the gut (supplementary material Fig. S1D,E).
To determine if impaired lumen fusion in smo mutants is due to an early endoderm migration defect, we examined expression of the endoderm marker, foxa3. At 30 hpf smo mutants show a single, midline localized endodermal rod that is overall similar in shape to that of wild-type embryos (supplementary material Fig. S2A,B). Examination of TgBAC(cldn15la-GFP) embryos at 48 hpf revealed that the intestinal epithelium is also similar in size and shape in wild type and smo mutants (supplementary material Fig. S2C,D,G). Furthermore, we determined that there is no significant difference in cell number, or cell proliferation between wild type and smo mutants (supplementary material Fig. S2E-F′,H,I). There was also no observable apoptosis in the gut of wild-type or smo embryos (data not shown). Therefore, the lumen fusion phenotype observed in smo mutants does not result from defects in early endoderm migration, or impaired regulation of epithelial cell numbers.
Rab11-mediated recycling is misregulated in smos294 mutants
Because lumen formation in the zebrafish gut occurs without apoptosis (Ng et al., 2005), lumen resolution must involve epithelial remodeling and the rearrangement of cellular contacts between lumens. This remodeling can be achieved by changing the identity of bridge contacts, from basolateral to apical, or by breaking adhesions. To undergo remodeling, cellular contacts and adhesions can be internalized and trafficked to lysosomes for degradation or they can be recycled back to the cell surface (Le et al., 1999; Palacios et al., 2005). To determine if lysosomal degradation is important for lumen fusion, we inhibited the degradation pathway by expressing a dominant-negative form of Rab7, a small GTPase that regulates late endosomal trafficking (Bucci et al., 2000). We also inhibited lysosomal acidification using bafilomycin, and examined embryos mutant for members of the vacuolar H+-ATPase complex (Nuckels et al., 2009). However, no lumen fusion defects were observed (supplementary material Fig. S3A-F).
Several studies have found that endocytic recycling and trafficking are important for epithelial remodeling during morphogenesis. The Rab11 family of small GTPases as well as the Rab11 effector proteins Rab11-FIP and MyoVb are well-known regulators of the recycling pathway (Hales et al., 2001; Lapierre et al., 2001; Ullrich et al., 1996) and play a key role in apical trafficking and basolateral recycling during epithelial morphogenesis (Kerman et al., 2008; Satoh et al., 2005; Shaye et al., 2008). To determine if recycling is involved in single lumen formation, we utilized a dominant-negative construct to disrupt endogenous Rab11a function. We crossed Tg(UAS:mcherry-rab11a-S25N)mw35 (Rab11aDN) (Clark et al., 2011) to a Tg(hsp70l:gal4) line to temporally control expression of Rab11aDN. Mosaic expression of Rab11aDN before the resolution stage of lumen formation resulted in an unfused lumen phenotype, similar to that of smos294 mutants and class II wild-type embryos, in 45% of embryos (n=20) at 72 hpf (Fig. 6A). These embryos contained the same number of epithelial cells in the gut as wild-type embryos, indicating that failed lumen resolution is not due to differences in cell numbers (supplementary material Fig. S5A). Upon expression of Rab11aDN, cadherin accumulated intracellularly, indicating that it is recycled in a Rab11a-dependent manner (supplementary material Fig. S4A-E). In addition, the apical protein 4e8 was also found to colocalize to Rab11aDN compartments (supplementary material Fig. S4F-J). We also tested the function of Rab11b, which is highly similar to Rab11a, yet resides in distinct apical vesicles in epithelial cells and colocalizes with different cargo proteins (Lai et al., 1994; Lapierre et al., 2003). Unlike DN-Rab11a, expression of DN-Rab11b did not cause a lumen formation phenotype (Fig. 6B). Thus, Rab11a-mediated recycling of basolateral and apical membrane proteins is necessary for lumen fusion during single lumen formation.
Fig. 6.
Rab11 is abnormally localized in smos294 mutants. (A,A′) Confocal cross section of a Tg(hsp70l:gal4); Tg(UAS:rab11aS25N) embryo. Phalloidin (green). (B,B′) Confocal cross section of a Tg(hsp70l:rab11bS25N) embryo. Phalloidin (red) (C,D) Confocal cross sections of smos294 and wild-type clutchmates expressing Tg(hsp70l:GFP-RAB11a). Arrowheads point to abnormal enlarged Rab11 positive vesicles in smos294. Phalloidin (red) (E-H) Confocal cross section of wild-type and smos294 embryos expressing Tg(hsp70l:GFP-RAB11a) stained for the apical marker 4e8 (E,F) or cadherin (G,H). Arrowheads point to areas of colocalization. (I,J) Confocal cross sections of uninjected and RFP-Rab11fipa1-injected Tg(hsp70l:GFP-RAB11a) embryos. Arrowheads point to dispersed compartments. (K) Expression levels of Rab11 family members in the intestinal epithelium of smos294 mutants relative to wild-type clutchmates. Rab11fip1a P<0.011, n=3. All embryos are 72 hpf. Scale bars: 20 μm.
To determine if Rab11aDN embryos exhibit a mesenchymal differentiation defect similar to that of smo mutants, we examined the expression of αSMA. Expression of αSMA in Rab11aDN embryos revealed proper differentiation of the mesenchymal layer (supplementary material Fig S5B,C). Furthermore, staining for smooth muscle myosin, Myh11, showed that mesenchymal cells expressing Rab11aDN differentiated as well as the controls (supplementary material Fig. S5D,E), suggesting that Rab11aDN expression does not affect the differentiation of the gut mesenchymal layer and that the lumen resolution phenotype was not due to mesenchymal defects.
The similar lumen phenotype shared by smos294 mutants and Rab11aDN-expressing embryos next led us to investigate whether defects in the recycling pathway contribute to the smos294 phenotype. To this end we generated a GFP-Rab11a transgenic line, Tg(hsp70l:GFP-RAB11a)pd1031, to mark recycling endosomes. In wild-type embryos, GFP-Rab11a was localized to small subapical compartments surrounding the lumen (Fig. 6C). By contrast, smos294 mutants exhibited abnormally enlarged GFP-Rab11a compartments that were dispersed from the apical surface (Fig. 6D). These enlarged Rab11a compartments in smo mutants contained the apical protein 4e8, indicating a defect in trafficking of apical membrane proteins (Fig. 6E,F). Colocalization with cadherin was not as apparent (Fig. 6G,H); however, owing to the transient nature of internalized cadherin, cadherin colocalization with recycling endosomes is often limited (Desclozeaux et al., 2008). This probably accounts for the minimal amount of colocalization with Rab11a compartments we observe. These data, together with the Rab11DN data, suggest that Rab11 trafficking of both apical and basolateral proteins is important in lumen fusion.
Previous studies in the Drosophila trachea have shown a similar accumulation of Rab11 in enlarged compartments upon overexpression of the Rab11 effector protein Rip11, an ortholog of Rab11Fip1a (Shaye et al., 2008). Rab11Fip1a and MyoVb interact with Rab11 family members and regulate plasma membrane recycling (Hales et al., 2001; Lapierre et al., 2001). To investigate Rab11 effectors in smos294 mutants we examined the expression levels of Rab11a, Rab11b, Rab11Fip1a and MyoVb in wild-type and smos294 embryos. We used fluorescence-activated cell sorting (FACS) to isolate intestinal cells from mutant TgBAC(cldn15la-GFP)pd1034; smos294 embryos and wild-type clutchmates, isolated RNA and performed quantitative polymerase chain reaction (qPCR) to evaluate differential gene expression. In smos294 cells, expression of rab11fip1a was increased threefold. However, the expression of rab11a, rab11b and myo5b was not significantly different from that in wild-type cells (Fig. 6K). To assess how an increase in Rab11fip1a levels may contribute to the smo gut phenotype, we overexpressed Rab11fipa1 in Tg(hsp70l:GFP-RAB11a) embryos through RNA injection. Upon mild overexpression, GFP-Rab11a compartments became enlarged and disorganized compared with non-injected embryos, similar to that observed in smo mutants (Fig. 6I,J). The data suggest that increased levels of Rab11fip1a are likely to be responsible for the abnormally enlarged GFP-Rab11a compartments observed in smos294 mutants.
Altogether, these studies identify an intermediate stage in the process of single lumen formation and reveal that lumen resolution is a genetically regulated process crucial for continuous lumen formation in the zebrafish gut. Our data also highlight the role of smoothened signaling from the mesenchyme in the regulation of lumen morphogenesis in the gut epithelium.
DISCUSSION
In this study we identify lumen resolution as a crucial process during single lumen formation. Single lumen formation begins with multiple small lumens that enlarge through fluid accumulation driven by Cldn15 and Na+/K+ ATPase (Fig. 7A). Before lumen coalescence, enlarged lumens are found along the length of the gut, separated by basolateral bridge contacts. Our studies reveal that cell-cell bridge contacts lack apical and tight junction markers between lumens, indicating that these bridge contacts do not change identity before lumen fusion. Instead, we observed that lumen fusion occurs through both apical membrane expansion and the shrinking and breaking of basolateral bridge contacts. The most common bridge cell arrangement involves cells that have one apical surface; however, occasionally bridge cells exhibit two apical surfaces. Bipolar cells have also been observed during tubulogenesis in the Ciona notochord (Denker and Jiang, 2012). Although the mechanism by which a cell acquires two apical membranes is unknown, it is possible that the cells between lumens are unable to receive proper polarizing cues from the basement membrane.
Fig. 7.
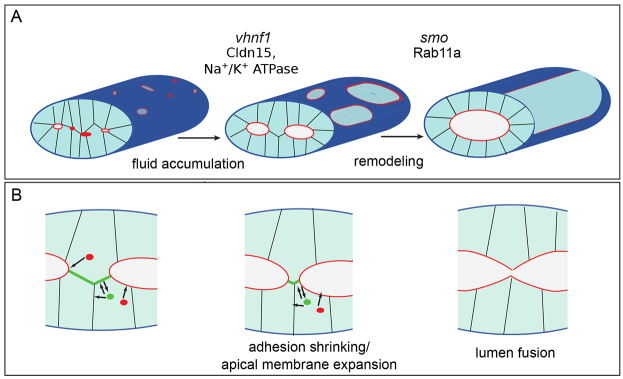
Lumens enlarge and fuse during single lumen formation. (A) During single lumen formation, vhnf1 drives lumen enlargement through Cldn15 and Na+/K+ ATPase regulated fluid accumulation. Next, smoothened regulates remodeling through Rab11a-mediated trafficking to facilitate lumen fusion. Red indicates the luminal surface. (B) During the fusion process, Rab11 traffics apical proteins (red) to the luminal surface and recycles basolateral proteins (green) from bridge contacts to lateral membranes. As the lumens expand, the bridge contacts between the lumens shrink and split, and the lumens fuse.
Based on our studies, we propose that Smo signaling facilitates the remodeling and weakening of bridge contacts and the enlargement of apical membrane via Rab11a-mediated trafficking and recycling to generate a single continuous lumen (Fig. 7A). In this model basolateral recycling relocalizes the adhesion from the bridge to lateral surfaces, thus shrinking and weakening the contacts between lumens. In addition, apical membrane is delivered to the luminal surface to facilitate membrane expansion. As adhesions shrink, the bridge contacts eventually break and lumens fuse (Fig. 7B). This may also be facilitated by the insertion of anti-adhesive apical proteins on the edge of the bridge contact, as shown in blood vessels (Strilić et al., 2010). Interestingly, zebrafish mutant for aPkcλ, which regulates adherens and tight junctions, also exhibit a single lumen formation defect (Horne-Badovinac et al., 2001), underscoring that proper regulation of adherens is critical in single lumen formation.
Work in 3D cysts has established the importance of functional Rab11 and recycling endosomes in E-cadherin trafficking, cyst morphogenesis and lumen formation (Bryant et al., 2010; Desclozeaux et al., 2008). In 3D cysts, Rab11 is crucial for lumen initiation by mediating the relocation of apical membrane from the outer surface of cells to a central patch where a lumen subsequently forms (Bryant et al., 2010). However, in most in vivo tubular systems, including the zebrafish intestine, mammalian pancreas and mammary gland, lumens initiate at several different sites within a rod of cells and must connect with each other to form a continuous luminal network. Our work shows that Rab11-mediated trafficking is needed to facilitate lumen resolution. Therefore, Rab11 may play a role in two distinct processes of lumen formation: lumen initiation and lumen resolution.
During the resolution stage lumens may merge through direct apical-apical membrane fusion or through fusion at cell junctions. Studies in the zebrafish vasculature and ascidian notochord provide insight into the possible mechanisms involved in membrane and junctional coalescence during lumen fusion. During ascidian notochord tubulogenesis, cellular remodeling involves a reduction of intracellular junctions between neighboring cells and the establishment of a new junction between two previously unconnected cells (Dong et al., 2009). Furthermore, work in the zebrafish dorsal longitudinal anastomotic vessel suggests that two apical membranes can merge directly (Herwig et al., 2011). However, a more detailed study will be needed to determine whether lumens merge through apical fusion in the zebrafish gut.
Examination of recycling pathway members revealed differential expression of the Rab11 effector protein, Rab11fip1a, and an accumulation of enlarged Rab11a compartments in smos294 intestinal epithelium. Overexpression of Rab11fip1a caused a similar accumulation of Rab11a compartments compared with wild-type embryos but did not produce a lumen fusion phenotype. This suggests that additional genes are also involved or that higher levels of Rab11fip1a expression are required to cause a lumen formation defect. In the Drosophila trachea, the overexpression of the Rab11 effector protein, Rip11, causes an accumulation of large Rab11 vesicles, similar to that observed in smos294 mutants, and results in impaired morphogenesis (Shaye et al., 2008). Therefore, our findings in smos294 mutants are consistent with previous studies and highlight the importance of effector protein levels in modulating endocytic recycling.
Although smos294 exhibits aberrant Rab11a localization and increased expression of Rab11 effector proteins, it is unclear how smo signaling regulates these recycling pathway members. We found that smo signaling acts in the surrounding mesenchyme but not in the intestinal epithelium. In the zebrafish esophagus and swimbladder, molecular interactions between epithelial Hh and mesenchymal Fgf10 regulate proliferation and differentiation (Korzh et al., 2011). In addition, Hh signaling from the endoderm is required for posterior gut development in zebrafish embryos (Parkin et al., 2009). Thus, smo probably regulates lumen fusion through epithelial-mesenchymal interactions and/or morphogen signaling such as the Bmp or Fgf pathways. Signaling from the mesenchyme through secreted factors and/or mechanical interactions are undoubtedly important for epithelial organization during tubulogenesis. Future studies should dissect the specific role both types of interactions play in regulating gut morphogenesis.
In the zebrafish intestine, lumens open at multiple sites within the gut tube, rather than at a single initiation point as seen in other models of tube formation such as 3D cysts and the C. elegans excretory cell (Bryant et al., 2010; Khan et al., 2013; Kolotuev et al., 2013). We observed that lumens form along the entire length of the gut tube and are typically separated from each other by one or two cells. This architecture allows lumens to fuse through localized cellular rearrangements, thus facilitating the generation of a continuous lumen within a long tube.
MATERIALS AND METHODS
Fish stocks
Zebrafish were maintained at 28°C and bred as previously described (Westerfield, 2000). Zebrafish lines used in this study include: EK, smos294 (Aanstad et al., 2009), Tg(UAS:mCherry-rab11a-S25N)mw35 (Clark et al., 2011), Tg(hsp70l:Gal4) (Scheer et al., 2001), Tg(hsp:GFP-podlx)pd1080 (Navis et al., 2013), atp6v1e1bhi577aTg/hi577aTg, atp6v1fhi1988Tg/hi1988Tg (Nuckels et al., 2009), Tg(GBS-ptch2:EGFP)umz23 (Choi et al., 2013), TgBAC(cldn15la-GFP)pd1034, Tg(-1.0ifabp:GFP-CaaX)pd1005, Tg(hsp70l:GFP-CaaX)pd1008, Tg(hsp70l:GFP-rab11bS25N)pd1090, Tg(hsp70l:GFP-RAB11a)pd1031, Tg(hsp70l:GFP-RAB7)pd1033, Tg(hsp70l:GFP-RAB7T22N)pd1032 (this study). To induce expression from the hsp70l promoter, embryos were heat-shocked for 40 minutes in a 40°C water bath.
Transgenics
Transgenic lines were generated using the Tol2kit (Kwan et al., 2007). Plasmids used include p5E-MCS, p5E-hsp70l, pME-MCS, pME-EGFP-CaaX, p3E-polyA, pDestTol2pA2 and pDestTol2CG2. pME-RAB11a was generated from Addgene plasmid 12674: GFP-RAB11aWT. pMe-GFP-Rab7 and pMe-GFP-Rab7DN were generated from Addgene plasmids 12605 and 12660, respectively. Rab11b was amplified from cDNA using primers: Rab11b_BamHI_F, GGATCCATGGGGACCCGTGACGAC; Rab11b_NotI_R, GCGGCCGCTCACAGGTCCTGACAGC and the Rab11bS25N mutation was created using the QuikChange kit (Agilent Technologies).
BAC recombineering
A BAC containing cldn15la was modified as previously described (Navis et al., 2013). For the C-terminal GFP fusion, a 20-aa spacer (DLPAEQKAASEEDLDPPVAT) was used. Recombination was performed with homology primers: cldn15la-spGFP_hom_F, CCATCTATACCACAGCTCAATCGAACGCAGAAACATCCAAAGCCTACGTCGATCTCCCCGCCGAACAGAAA, and cldn15la-spGFP_hom_R, TAAACAAACATCAACGTAACAACAGTTCAGCCTTGTTAAAATGGGAAATCATTGGAGCTCCACCGCGGTG. The cldn15la-GFP BAC was linearized using AsiSI (NEB), injected into one-cell stage embryos to generate TgBAC(cldn15la-GFP)pd1034.
RNA injection and BrdU labeling
pCS2-RFP-Rab11fip1a was linearized using NotI and RNA was transcribed using the mMESSAGE mMACHINE SP6 kit (Ambion). RFP-Rab11fip1a (294 pg) RNA was injected into embryos at the one-cell stage.
To label proliferating cells, 72 hpf embryos were incubated in 16 mM bromodeoxyuridine (BrdU) with 10% dimethyl sulfoxide (DMSO) in egg water for 1 hour at 28°C. Percentage of proliferating cells was calculated by comparing the number of BrdU-positive cells in a gut section versus total cells.
Histology and immunofluorescence
Cross sections were cut with a vibratome (VT 1000S; Leica) and stained as described previously (Bagnat et al., 2007). Primary antibodies: pan-cadherin (Sigma; 1:1200), ZO-1 (339100, Invitrogen; 1:500), 4e8 (73643, AbCam; 1:500), β-catenin (SC 7199, Santa Cruz Biotechnology; 1:500), caspase 3 (AB3623, Millipore; 1:500), Myh11 (BT-562, Biomedical Technologies; 1:150) and BrdU (033900, Invitrogen; 1:500). Secondary antibodies (Molecular Probes) were used at 1:300. A custom Cldn15la antibody was generated in rabbit using a peptide derived from the C-terminus of Cldn15la (YQRFSKSKEKGAYYPC) and used at 1:500. Cldn15la staining was performed as previously described (Dong et al., 2007). Imaging was carried out on an SP5 confocal microscope (Leica, Wetzlar, Germany) with 40×/1.25-0.75 HCX PL APO oil objective, For whole-mount imaging, fixed embryos were mounted in 0.8% agarose and imaged on a SP5 confocal with 20×/0.70 HC PL APO oil objective, Application Suite (Leica) and Huygens Essential deconvolution software were used for image processing.
Live imaging
Zebrafish embryos at 48 hpf were anesthetized and embedded in 1.5% agarose. SPIM was performed using three 10×/0.3 water dipping lenses (Leica), two for illumination and one for detection in an mSPIM configuration (Huisken and Stainier, 2007). A 488 nm laser (Coherent) was used for excitation. A stack of 100 planes (3 μm apart) was recorded with an EMCCD camera (Andor) every 10 minutes for a total duration of 24 hours at 24°C.
Embryo dissociation and FACS
Embryos were rinsed with calcium free Ringer’s solution for 10 minutes. Ringer’s solution was removed and embryos were incubated in 0.25% trypsin (Gibco) and 300 μg/ml collagenase (Sigma) for 45 minutes at 28°C with pipetting every 15 minutes until a single cell suspension was attained. Cells were washed with PBS plus 5% fetal calf serum (FCS) and passed through a 70 μm filter (BD Falcon). Cell suspensions were stained with propidium iodide (Invitrogen) and sorted on a BD FACS Diva sorter at the Flow Cytometry Shared Resource center (Duke University). Cells were collected in Buffer RLT (Qiagen) and stored at -80°C.
RNA isolation qPCR
RNA was extracted using the RNeasy Micro Kit (Qiagen) according to the manufacturer’s protocol. cDNA was synthesized using First Strand cDNA Synthesis Kit (Roche). Quantitative PCR was performed using a Bio-Rad CFX96 Real-Time System C1000 Thermocycler and Bio-Rad iQ SYBR Green Supermix. Reactions were performed in duplicate and data from three independent runs were obtained. Primers used include: elfa_F, CTTCTCAGGCTGACTGTGC; elfa_R, CCGCTAGGATTACCCTCC; myoVb_F, AGGACATGCTGGACCACTTC; myoVb_R, TCCAGCTCTTGCACTTTCTTC; rab11fip1a_F, TCAAACACGTTGGGACCATA; rab11fip1a_R, TTTGGGCCTTGTAAGGACAG; rab11a_F, GAAAGACCGTCAAGGCTCAG; rab11a_R, ACCTGGATGGACACCACATT; rab11b_F, GGACAGGAACGCTACAGAGC; rab11b_R, TGCCCTTTAACCCGTCAGTA. Expression levels were normalized to elfa for each cDNA set.
In situ hybridization
To make an in situ probe, cldn15la was amplified from cDNA using the following primers: cldn_probe_F, GGGGCTGGTTGGTTTAGTTT; cldn_probe_R, CCGCATCCATGAAAATTGA and ligated into pGEMT-Easy (Promega). The plasmid was linearized and DIG RNA Labeling Kit (Roche) was used to make digoxygenin-labeled RNA. In situ hybridization for cldn15la, foxa3 (Field et al., 2003) and αSMA (Georgijevic et al., 2007) was performed as described previously (Navis et al., 2013) and images were acquired on a Discovery V20 stereoscope (Zeiss).
Pharmacological treatment
Embryos were dechorionated at 48 hpf and placed in a 12-well dish with 1 μM bafilomycin (Sigma) or 1 μM DMSO (Sigma) in egg water. Embryos were incubated at 28°C and fixed at 72 hpf.
Statistical analysis
Gut diameter and perimeter was determined using ImageJ software. Number of cells in the gut were obtained by counting nuclei in AP-position-matched sections. Statistical significance for all measurements was determined using Student’s t-test.
Supplementary Material
Acknowledgments
We thank the Zebrafish Facility and the Flow Cytometry Shared Resource center at Duke University; Brian Link, Jeff Gross, Sarah Kucenas, Ken Poss and Didier Stainier for fish lines and Sarah Childs and Jim Patton for in situ probes; Didier Y. Stainier for generous support during the initial phase of this work; the M.B. lab for advice and discussion; and Ken Poss, Calga Eroglu and Terry Lechler for the critical reading of this manuscript.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
M.B. conceived the study. A.L.A. performed the majority of the experiments with help from S.R., and J.H. performed SPIM imaging of the gut. The gut lumen phenotype in smo mutants was initially observed by P.J.S. and M.B. in Didier Stainier’s lab at UCSF. A.L.A. and M.B. wrote the manuscript with input from other authors.
Funding
This work was funded by a National Institutes of Health Innovator Grant [1DP2OD006486 to M.B.]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.100313/-/DC1
References
- Aanstad P., Santos N., Corbit K. C., Scherz P. J., Trinh A., Salvenmoser W., Huisken J., Reiter J. F., Stainier D. Y. R. (2009). The extracellular domain of Smoothened regulates ciliary localization and is required for high-level Hh signaling. Curr. Biol. 19, 1034–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M., Cheung I. D., Mostov K. E., Stainier D. Y. R. (2007). Genetic control of single lumen formation in the zebrafish gut. Nat. Cell Biol. 9, 954–960 [DOI] [PubMed] [Google Scholar]
- Bryant D. M., Datta A., Rodríguez-Fraticelli A. E., Peränen J., Martín-Belmonte F., Mostov K. E. (2010). A molecular network for de novo generation of the apical surface and lumen. Nat. Cell Biol. 12, 1035–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci C., Thomsen P., Nicoziani P., McCarthy J., van Deurs B. (2000). Rab7: a key to lysosome biogenesis. Mol. Biol. Cell 11, 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Burgess S., Hopkins N. (2001). Analysis of the zebrafish smoothened mutant reveals conserved and divergent functions of hedgehog activity. Development 128, 2385–2396 [DOI] [PubMed] [Google Scholar]
- Cheung I. D., Bagnat M., Ma T. P., Datta A., Evason K., Moore J. C., Lawson N. D., Mostov K. E., Moens C. B., Stainier D. Y. R. (2012). Regulation of intrahepatic biliary duct morphogenesis by Claudin 15-like b. Dev. Biol. 361, 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W. Y., Gemberling M., Wang J., Holdway J. E., Shen M. C., Karlstrom R. O., Poss K. D. (2013). In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development 140, 660–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark B. S., Winter M., Cohen A. R., Link B. A. (2011). Generation of Rab-based transgenic lines for in vivo studies of endosome biology in zebrafish. Dev. Dyn. 240, 2452–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denker E., Jiang D. (2012). Ciona intestinalis notochord as a new model to investigate the cellular and molecular mechanisms of tubulogenesis. Semin. Cell Dev. Biol. 23, 308–319 [DOI] [PubMed] [Google Scholar]
- Desclozeaux M., Venturato J., Wylie F. G., Kay J. G., Joseph S. R., Le H. T., Stow J. L. (2008). Active Rab11 and functional recycling endosome are required for E-cadherin trafficking and lumen formation during epithelial morphogenesis. Am. J. Physiol. Cell Physiol. 295, C545–C556 [DOI] [PubMed] [Google Scholar]
- Dong P. D., Munson C. A., Norton W., Crosnier C., Pan X., Gong Z., Neumann C. J., Stainier D. Y. (2007). Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat. Genet. 39, 397–402 [DOI] [PubMed] [Google Scholar]
- Dong B., Horie T., Denker E., Kusakabe T., Tsuda M., Smith W. C., Jiang D. (2009). Tube formation by complex cellular processes in Ciona intestinalis notochord. Dev. Biol. 330, 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field H. A., Ober E. A., Roeser T., Stainier D. Y. (2003). Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev. Biol. 253, 279–290 [DOI] [PubMed] [Google Scholar]
- Furuse M., Fujita K., Hiiragi T., Fujimoto K., Tsukita S. (1998). Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 141, 1539–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgijevic S., Subramanian Y., Rollins E. L., Starovic-Subota O., Tang A. C., Childs S. J. (2007). Spatiotemporal expression of smooth muscle markers in developing zebrafish gut. Dev. Dyn. 236, 1623–1632 [DOI] [PubMed] [Google Scholar]
- Gregory M., Dufresne J., Hermo L., Cyr D. (2001). Claudin-1 is not restricted to tight junctions in the rat epididymis. Endocrinology 142, 854–863 [DOI] [PubMed] [Google Scholar]
- Hales C. M., Griner R., Hobdy-Henderson K. C., Dorn M. C., Hardy D., Kumar R., Navarre J., Chan E. K., Lapierre L. A., Goldenring J. R. (2001). Identification and characterization of a family of Rab11-interacting proteins. J. Biol. Chem. 276, 39067–39075 [DOI] [PubMed] [Google Scholar]
- Herwig L., Blum Y., Krudewig A., Ellertsdottir E., Lenard A., Belting H. G., Affolter M. (2011). Distinct cellular mechanisms of blood vessel fusion in the zebrafish embryo. Curr. Biol. 21, 1942–1948 [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S., Lin D., Waldron S., Schwarz M., Mbamalu G., Pawson T., Jan Y., Stainier D. Y., Abdelilah-Seyfried S. (2001). Positional cloning of heart and soul reveals multiple roles for PKC lambda in zebrafish organogenesis. Curr. Biol. 11, 1492–1502 [DOI] [PubMed] [Google Scholar]
- Huisken J., Stainier D. Y. (2007). Even fluorescence excitation by multidirectional selective plane illumination microscopy (mSPIM). Opt. Lett. 32, 2608–2610 [DOI] [PubMed] [Google Scholar]
- Huisken J., Stainier D. Y. (2009). Selective plane illumination microscopy techniques in developmental biology. Development 136, 1963–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inai T., Sengoku A., Hirose E., Iida H., Shibata Y. (2007). Claudin-7 expressed on lateral membrane of rat epididymal epithelium does not form aberrant tight junction strands. Anat. Rec. (Hoboken) 290, 1431–1438 [DOI] [PubMed] [Google Scholar]
- Kerman B. E., Cheshire A. M., Myat M. M., Andrew D. J. (2008). Ribbon modulates apical membrane during tube elongation through Crumbs and Moesin. Dev. Biol. 320, 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan L. A., Zhang H., Abraham N., Sun L., Fleming J. T., Buechner M., Hall D. H., Gobel V. (2013). Intracellular lumen extension requires ERM-1-dependent apical membrane expansion and AQP-8-mediated flux. Nat. Cell Biol. 15, 143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolotuev I., Hyenne V., Schwab Y., Rodriguez D., Labouesse M. (2013). A pathway for unicellular tube extension depending on the lymphatic vessel determinant Prox1 and on osmoregulation. Nat. Cell Biol. 15, 157–168 [DOI] [PubMed] [Google Scholar]
- Korzh S., Winata C. L., Zheng W., Yang S., Yin A., Ingham P., Korzh V., Gong Z. (2011). The interaction of epithelial Ihha and mesenchymal Fgf10 in zebrafish esophageal and swimbladder development. Dev. Biol. 359, 262–276 [DOI] [PubMed] [Google Scholar]
- Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P., Chien C. B. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 [DOI] [PubMed] [Google Scholar]
- Lai F., Stubbs L., Artzt K. (1994). Molecular analysis of mouse Rab11b: a new type of mammalian YPT/Rab protein. Genomics 22, 610–616 [DOI] [PubMed] [Google Scholar]
- Lapierre L. A., Kumar R., Hales C. M., Navarre J., Bhartur S. G., Burnette J. O., Provance D. W., Jr, Mercer J. A., Bähler M., Goldenring J. R. (2001). Myosin vb is associated with plasma membrane recycling systems. Mol. Biol. Cell 12, 1843–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre L. A., Dorn M. C., Zimmerman C. F., Navarre J., Burnette J. O., Goldenring J. R. (2003). Rab11b resides in a vesicular compartment distinct from Rab11a in parietal cells and other epithelial cells. Exp. Cell Res. 290, 322–331 [DOI] [PubMed] [Google Scholar]
- Le T. L., Yap A. S., Stow J. L. (1999). Recycling of E-cadherin: a potential mechanism for regulating cadherin dynamics. J. Cell Biol. 146, 219–232 [PMC free article] [PubMed] [Google Scholar]
- Lubarsky B., Krasnow M. A. (2003). Tube morphogenesis: making and shaping biological tubes. Cell 112, 19–28 [DOI] [PubMed] [Google Scholar]
- Madara J. L., Neutra M. R., Trier J. S. (1981). Junctional complexes in fetal rat small intestine during morphogenesis. Dev. Biol. 86, 170–178 [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte F., Mostov K. (2008). Regulation of cell polarity during epithelial morphogenesis. Curr. Opin. Cell Biol. 20, 227–234 [DOI] [PubMed] [Google Scholar]
- Navis A., Marjoram L., Bagnat M. (2013). Cftr controls lumen expansion and function of Kupffer’s vesicle in zebrafish. Development 140, 1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng A. N. Y., de Jong-Curtain T. A., Mawdsley D. J., White S. J., Shin J., Appel B., Dong P. D. S., Stainier D. Y. R., Heath J. K. (2005). Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Dev. Biol. 286, 114–135 [DOI] [PubMed] [Google Scholar]
- Nuckels R. J., Ng A., Darland T., Gross J. M. (2009). The vacuolar-ATPase complex regulates retinoblast proliferation and survival, photoreceptor morphogenesis, and pigmentation in the zebrafish eye. Invest. Ophthalmol. Vis. Sci. 50, 893–905 [DOI] [PubMed] [Google Scholar]
- Palacios F., Tushir J. S., Fujita Y., D’Souza-Schorey C. (2005). Lysosomal targeting of E-cadherin: a unique mechanism for the down-regulation of cell-cell adhesion during epithelial to mesenchymal transitions. Mol. Cell. Biol. 25, 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin C. A., Allen C. E., Ingham P. W. (2009). Hedgehog signalling is required for cloacal development in the zebrafish embryo. Int. J. Dev. Biol. 53, 45–57 [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M., Melton D. A., McMahon A. P. (2000). Hedgehog signals regulate multiple aspects of gastrointestinal development. Development 127, 2763–2772 [DOI] [PubMed] [Google Scholar]
- Satoh A. K., O’Tousa J. E., Ozaki K., Ready D. F. (2005). Rab11 mediates post-Golgi trafficking of rhodopsin to the photosensitive apical membrane of Drosophila photoreceptors. Development 132, 1487–1497 [DOI] [PubMed] [Google Scholar]
- Scheer N., Groth A., Hans S., Campos-Ortega J. A. (2001). An instructive function for Notch in promoting gliogenesis in the zebrafish retina. Development 128, 1099–1107 [DOI] [PubMed] [Google Scholar]
- Shaye D. D., Casanova J., Llimargas M. (2008). Modulation of intracellular trafficking regulates cell intercalation in the Drosophila trachea. Nat. Cell Biol. 10, 964–970 [DOI] [PubMed] [Google Scholar]
- Strilić B., Eglinger J., Krieg M., Zeeb M., Axnick J., Babál P., Müller D. J., Lammert E. (2010). Electrostatic cell-surface repulsion initiates lumen formation in developing blood vessels. Curr. Biol. 20, 2003–2009 [DOI] [PubMed] [Google Scholar]
- Ullrich O., Reinsch S., Urbé S., Zerial M., Parton R. G. (1996). Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 135, 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace K. N., Pack M. (2003). Unique and conserved aspects of gut development in zebrafish. Dev. Biol. 255, 12–29 [DOI] [PubMed] [Google Scholar]
- Wallace K. N., Dolan A. C., Seiler C., Smith E. M., Yusuff S., Chaille-Arnold L., Judson B., Sierk R., Yengo C., Sweeney H. L., et al. (2005). Mutation of smooth muscle myosin causes epithelial invasion and cystic expansion of the zebrafish intestine. Dev. Cell 8, 717–726 [DOI] [PubMed] [Google Scholar]
- Westerfield M. (2000). The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). Eugene, OR: University of Oregon Press; [Google Scholar]
- Westmoreland J. J., Drosos Y., Kelly J., Ye J., Means A. L., Washington M. K., Sosa-Pineda B. (2012). Dynamic distribution of claudin proteins in pancreatic epithelia undergoing morphogenesis or neoplastic transformation. Dev. Dyn. 241, 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.