Abstract
Epstein–Barr virus (EBV) is a tumorigenic human γ-herpesvirus, which produces several known structured RNAs with functional importance: two are implicated in latency maintenance and tumorigenic phenotypes, EBER1 and EBER2; a viral small nucleolar RNA (v-snoRNA1) that may generate a small regulatory RNA; and an internal ribosomal entry site in the EBNA1 mRNA. A recent bioinformatics and RNA-Seq study of EBV identified two novel EBV non-coding (nc)RNAs with evolutionary conservation in lymphocryptoviruses and likely functional importance. Both RNAs are transcribed from a repetitive region of the EBV genome (the W repeats) during a highly oncogenic type of viral latency. One novel ncRNA can form a massive (586 nt) hairpin, while the other RNA is generated from a short (81 nt) intron and is found in high abundance in EBV-infected cells.
Keywords: EBER, Epstein–Barr virus, latency, ncRNA, pseudoknot, stable intronic sequence RNA
Overview
Epstein–Barr virus (EBV), a human γ-herpesvirus (Fig. 1), is widely dispersed (infecting as many as 95% of humans)1 and is implicated in cancers such as Burkitt’s lymphomas (BL) and nasopharyngeal carcinomas,2,3 as well as in several serious autoimmune disorders.4 Like all herpesviruses, EBV infection results in lifelong latent infections, interrupted by intermittent lytic reactivation. EBV can infect epithelial cells or B cells; however, B cells, antibody-producing lymphocytes (white blood cells), are the ultimate targets of the virus. Establishment of latency upon EBV infection stimulates B cell immortalization in vitro and, in ways that are not yet fully understood, rewires the cell in such a manner as to increase the tumorigenic potential of EBV-infected cells.5,6 Latency proceeds primarily via three programs expressing various coding and non-coding (nc)RNAs (Fig. 2).7 Furthermore, various EBV-related malignancies are associated with each type of latency.8
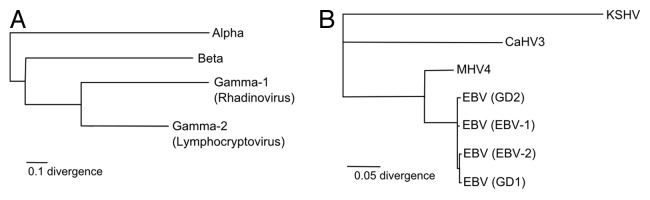
Figure 1. Phylogenetic relationships. (A) Herpesvirus phylogenetic tree. (B) Kaposi’s sarcoma-associated herpesvirus (KSHV), a gamma-1 herpesvirus, is included to demonstrate the relationships between four EBV strains (EBV-1, EBV-2, GD1, and GD2) and two other lymphocryptoviruses [Macacine herpesvirus 4 (MHV4) and Callitrichine herpesvirus 3 (CaHV3)].
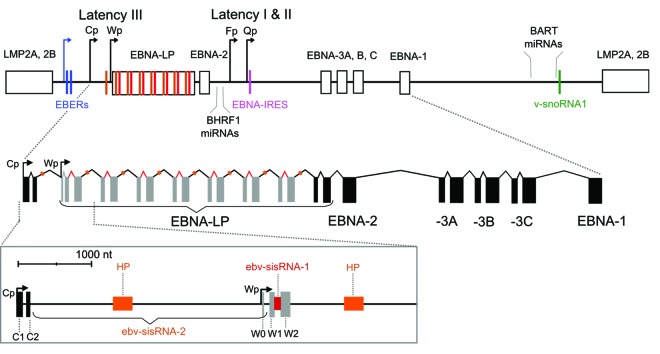
Figure 2. Top: Cartoon of the EBV genome with positions of ncRNAs indicated. Promoters are denoted with bent arrows: the blue arrow shows the promoter for the EBERs (used in all latency programs), while the black arrows show the Cp/Wp promoters used in latency III and Fp/Qp promoters used in latency I and II. Exons are indicated with boxes, introns with lines, and each ncRNA with a colored bar that matches the colored label (the orange bar shows the location of the hairpin (HP) in ebv-sisRNA-2 and the red bar shows ebv-sisRNA-1). Additional details on EBV latent gene expression are reviewed in reference 7. Bottom: Cartoon of EBNA primary transcript from latency III. Exons are indicated with solid boxes and introns with lines. The W repeat W1 and W2 coding exons are shown in gray. Orange and red colored regions indicate locations of the HP and ebv-sisRNA-1, respectively. The zoomed-in box below shows the 5′ end of the RNA. The intron that generates ebv-sisRNA-2 is indicated by the braces (and titled in orange). The HP within ebv-sisRNA-2 is indicated with the orange box. The red region indicates the intron that generates ebv-sisRNA-1.
In addition to nine expressed latent proteins [the six Epstein–Barr virus nuclear antigens (EBNAs) and three latent membrane proteins (LMPs)], EBV produces ncRNAs, including EBER1 and EBER2, a viral snoRNA, an internal ribosomal entry site (IRES) in the EBNA1 mRNA, and subsets of as many as 50 viral micro (mi)RNAs during latency. Additionally, EBV generates many lytic transcripts with likely non-coding functions.9,10 This review will provide an overview of the known EBV RNA families (suggesting a revision of the structure model of one family, which may form a conserved pseudoknot), as well as introduce two recently discovered EBV RNAs: both stable intronic sequence RNAs. A Wikipedia entry, to be linked to the Rfam database, has been created for these new RNAs. Additionally, the EBER page has been updated to reflect the most up-to-date research on EBER1, and to include info on EBER2 as well.
Known EBV RNA Families
EBER1 and EBER2 are highly structured, non-polyadenylated, and non-coding RNA-Pol III transcripts (Fig. 2) that are each ~170 nt long. High levels of EBER1 and EBER2 are typically found in EBV-infected cells throughout latency.11,12 The EBERs accumulate in the nucleus to approximately one million copies per cell, where they form stable ribonucleoprotein (RNP) complexes.13,14 The steady-state levels of EBER1 are higher (by a factor of approximately four) than those of EBER2 in EBV-infected cells,15 primarily due to EBER2 having a shorter half-life.16 The biological function of the EBERs is yet to be defined mechanistically. There are, however, a number of interesting reports regarding these RNAs. Two independent studies have shown that an EBER1/2-minus EBV recombinant strain is able to infect lymphocytes with no obvious phenotypic deficiency.17,18 Others have, however, shown that the deletion of EBER1 and EBER2 can compromise the ability of EBV to transform lymphocytes.19 In support of the latter finding, the expression of EBER1 in cell lines can induce a tumorigenic phenotype.20,21
EBER1
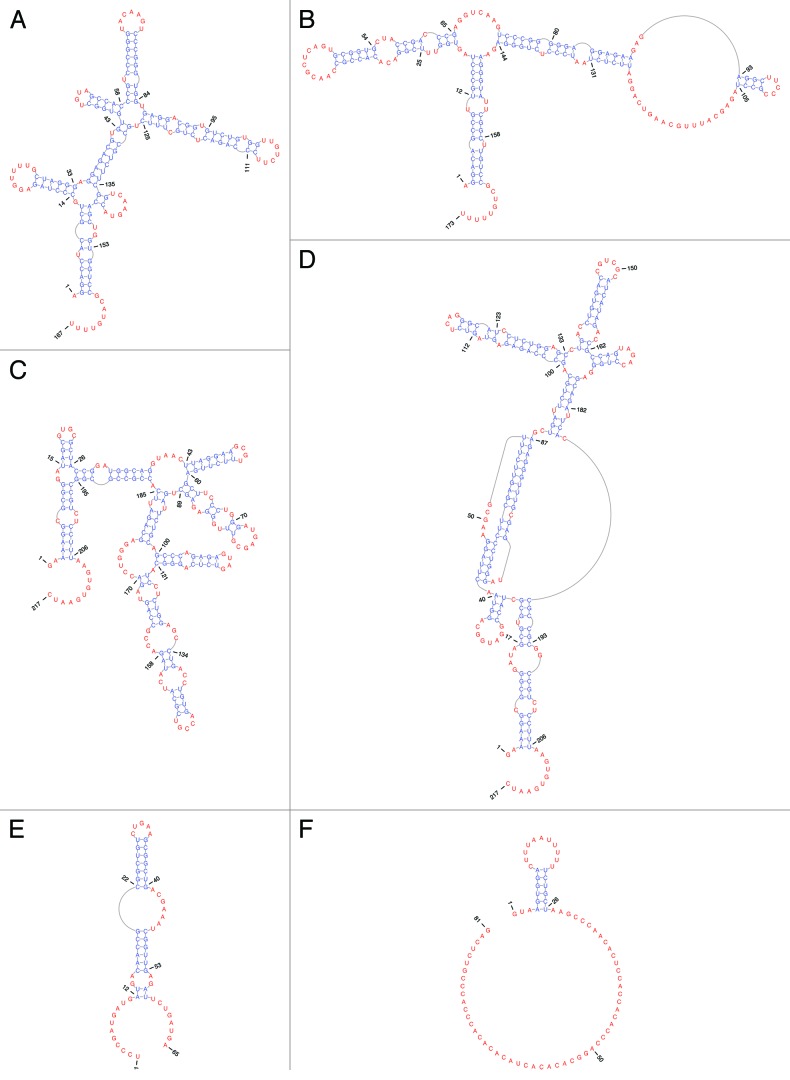
EBER1 (Rfam ID# RF01789) is highly conserved in sequence [~100% average pairwise sequence identity (APSI)] and structure (Fig. 3A) across EBV strains and is also found in two other lymphocryptoviruses: Macacine herpesvirus 4 (MHV4, a.k.a. rhesus lymphocryptovirus) and baboon herpesvirus (CeHV12), where sequence APSI (vs. EBV) is 57.3% and 68.9%, respectively. EBER1’s secondary structure is base pair rich, very thermodynamically stable,15 and is organized into five conserved hairpins (Fig. 3A) radiating from two multi-branch loop structures. In addition to its hairpins, EBER1 possesses a 9 nt single-stranded tail at its 3′ end. This RNA structure provides platforms for binding host proteins to form EBER1 RNPs.
Figure 3. EBV ncRNA secondary structures. Base-paired nt are in blue, while unpaired and non-canonically paired nt are in red (generated using PseudoViewer). (A) EBER1. (B) EBER2. (C) EBNA IRES minimum free energy structure.38 (D) EBNA IRES with pseudoknot. (E) v-snoRNA1. (F) ebv-sisRNA-1.
EBER1 (and EBER2) was initially discovered as a stoichiometric interactor of the host-cell RNA chaperone La/SSB (Sjögren's Syndrome type B antigen).13 Later, EBER1 was shown to also interact with the ribosomal protein L22.14 The tumorigenic potential of EBER1 has been associated with its ability to relocalize L22 from the nucleolus to the nucleoplasm,21 although the mechanistic details are still missing. Recently, it was determined that EBER1 also specifically interacts with at least three hnRNPs (A1, A2/B1, and D/AUF1); of these, there is biochemical evidence that the EBER1/AUF1 interaction is direct and occurs in vivo.22 Interestingly, EBNA1 (together with the EBERs) is highly expressed in the three primary EBV latency types,11,12 and co-purifies in an RNA-dependent manner with EBER1 and La, plus the hnRNPs AUF1 and A2/B1.23,24 This suggests that EBER1 and EBNA1 might associate with the same RNPs in the cells, and thus, have overlapping, if not complementary, functions.
EBER2
Like EBER1, EBER2 is highly abundant in latently infected cells.13,15 Whereas the sequence of EBER1 is well conserved among related lymphocryptoviruses, such as CeHV12 and MHV4, the conservation in the primary sequence of EBER2 in these viruses is less (only 50.3%).15 However, using the known secondary structure information of EBER225 as an in silico folding guide, its homologs in CeHV12 and MHV4 can be modeled to adopt homologous structures (Fig. 3B), suggesting that the yet unknown molecular mode of action of EBER2 and its homologs may be conserved despite divergent primary sequences.
To date, the only reported interacting partner of EBER2 is the cellular protein La, which binds EBER2 through the uridine stretch present at its 3′ end.13 As La binds all RNA polymerase III transcripts,26 no information regarding the function of EBER2 can be deduced from this interaction. Nevertheless, because EBER2 contains additional nucleotide stretches that could serve as potential protein binding sites, most notably the two major conserved stem-loop regions (Fig. 3B), future studies may reveal novel interacting partners that would point toward the molecular function of EBER2.
EBV strains that carry a deletion in EBER2 have been generated to study the phenotype upon EBV-infection in the absence of EBER2. Conflicting results have been reported that either do or do not substantiate a role of EBER2 in growth transformation of infected lymphoblastoid cell lines,18,27 and further studies are necessary to resolve this discrepancy. Even though the absence of EBERs does not appear to be detrimental to the life cycle of EBV in a laboratory setting, all EBV strains sequenced thus far have retained EBERs in their genome, indicating that EBERs do have a pivotal function outside the petri dish. Interestingly, an EBV strain from a nasopharyngeal carcinoma cell line has recently been sequenced that harbors a partial deletion in EBER2 from nucleotides 44 to 74 (Fig. S1).28 It is tempting to speculate that this partial deletion contributed to the carcinoma development. Ongoing functional studies addressing the importance of specific regions in EBER2 will help to further our understanding of the molecular mechanism of EBER2 action.
EBV miRNAs
EBV can generate as many as 50 different mature miRNAs (Table 1);29,30 many of which are also conserved in MHV4.31 EBV miRNAs are clustered in two regions on opposing sides of the virus’s genome: the BamHI fragment H rightward open reading frame 1 (BHRF1) and BamHI A rightward transcripts (BART) regions (Fig. 2). EBV miRNAs are differentially regulated during viral latency,32 and interestingly, in EBV-associated diseases.33-36 EBV miRNAs target a variety of viral and host genes for regulation30,37 and several EBV genes are regulated by both viral and host miRNAs.30
Table 1. EBV miRNAs.
| ID | Accession1 | Start2 | End2 |
|---|---|---|---|
| ebv-mir-BART1 | MI0001067 | 139346 | 139415 |
| ebv-mir-BART2 | MI0001068 | 152745 | 152806 |
| ebv-mir-BART3 | MI0003725 | 139076 | 139154 |
| ebv-mir-BART4 | MI0003726 | 139220 | 139295 |
| ebv-mir-BART5 | MI0003727 | 139661 | 139749 |
| ebv-mir-BART6 | MI0003728 | 140016 | 140107 |
| ebv-mir-BART7 | MI0003729 | 146425 | 146510 |
| ebv-mir-BART8 | MI0003730 | 146759 | 146840 |
| ebv-mir-BART9 | MI0003731 | 146946 | 147032 |
| ebv-mir-BART10 | MI0003732 | 147304 | 147393 |
| ebv-mir-BART11 | MI0003733 | 147524 | 147609 |
| ebv-mir-BART12 | MI0003734 | 147888 | 147970 |
| ebv-mir-BART13 | MI0003735 | 148512 | 148597 |
| ebv-mir-BART14 | MI0003736 | 148731 | 148815 |
| ebv-mir-BART15 | MI0004988 | 139507 | 139584 |
| ebv-mir-BART16 | MI0004989 | 139776 | 139874 |
| ebv-mir-BART17 | MI0004990 | 139894 | 139995 |
| ebv-mir-BART18 | MI0004991 | 145932 | 146050 |
| ebv-mir-BART19 | MI0004992 | 148198 | 148290 |
| ebv-mir-BART20 | MI0004993 | 148319 | 148417 |
| ebv-mir-BART21 | MI0010627 | 145503 | 145578 |
| ebv-mir-BART22 | MI0010628 | 147161 | 147231 |
| ebv-mir-BHRF1–1 | MI0001064 | 41471 | 41536 |
| ebv-mir-BHRF1–2 | MI0001065 | 42848 | 42912 |
| ebv-mir-BHRF1–3 | MI0001066 | 42966 | 43030 |
Notes: 1Accession numbers to miRNA entries in the miRBase database. 2Start and stop positions for pre-miRNA hairpins; nt numbering from EBV genome (GenBank # AJ507799.2).
The EBNA IRES
During Fp- and Qp-initiated transcription (Fig. 2; latency I and II), a segment of the 5′ UTR of EBNA1 mRNA, the U leader exon, was found to possess a sequence capable of stimulating cap-independent translation.38 This EBNA IRES (Rfam ID# RF00448) was found to be more active than the well-characterized encephalomyocarditis virus IRES,39 and remarkably, can increase EBNA1 expression by 4–14-fold (in various BL cell lines). The EBNA IRES allows cap-independent translation to occur under conditions of reduced canonical initiation (e.g., when strong RNA structure upstream of the canonical start site inhibits ribosomal scanning).40 It is also proposed to play roles in oncogenesis.41
RNA structure plays important roles in the activity of many known IRESs.42 The model secondary structure proposed for the EBNA IRES (Fig. 3C)38 was generated using a folding algorithm, mfold,43 which forbids pseudoknots. Using a folding algorithm based on statistical thermodynamics that allows pseudoknot prediction, DotKnot,44 we predict that a 46 nt portion of the EBNA IRES can form a pseudoknot (42–87 nt, Fig. 3D). The remainder of the EBNA IRES was refolded using RNAalifold.45,46 This program predicts RNA structure common to aligned sequences; the Rfam seed alignment was used as input, which resulted in the global refolding of the sequence shown in the model presented in Figure 3D. This alternative fold was used to search a database of known lymphocryptovirus sequences with the INFERNAL package;47,48 the structure formed by 42–185 nt is ~100% conserved throughout EBV strains (68–80 nt in Fig. 3D are deleted in sequence M125531, Fig. S2) and conservation extends outside EBV to include MHV4 (82.8% APSI). In the MHV4 pseudoknot region, conservation is even higher (91.3%): only four point mutations occur, which convert AU or GC pairs into GU, GA, or AC pairs (Fig. S2). Mutations that may form non-canonical pairs (GA and AC) are less disruptive to RNA structure than others,49,50 and GU pairs are almost as stable as canonical AU pairs.51 The presence of a possible, conserved, pseudoknot in the EBNA IRES is interesting, as these motifs play important roles in many known viral IRESs.52,53 Pseudoknot folding results in complex, thermodynamically stable54 3D shapes, which mimic interactions that can recruit elements of the translational machinery, including ribosomes.55,56 Further work is necessary to validate the proposed pseudoknot and to test its function in the EBNA IRES.
The viral snoRNA
A viral small nucleolar RNA (v-snoRNA1, Rfam ID# RF01516) is generated from an intronic sequence within the BART region. V-snoRNA1 occurs ~100 nt downstream of the BART2 miRNA (Fig. 2). This RNA was the first virally produced snoRNA to be discovered.57 It possesses sequence and structure similarities to eukaryotic C/D box snoRNAs (Fig. 3E), lacking only a terminal stem structure. During lytic reactivation, v-snoRNA1 may be further processed into a 24 nt small RNA (v-snoRNA124pp from 41–64 nt in Fig. 3E) that can target a sequence in the 3′ UTR of the BALF5 mRNA, encoding the viral DNA polymerase, and regulate BALF5 expression.57 Interestingly, a nearby BART2 miRNA also targets this gene. Conservation of v-snoRNA1 occurs throughout EBV strains and includes MHV4.
New EBV RNA Families
A recent survey of ncRNAs in EBV using bioinformatics and RNA-Seq identified multiple regions within its ~170 kbp genome likely to generate functional RNAs.15 These regions include EBER1, EBER2, v-snoRNA1, and most of the known viral miRNAs. In addition to these known EBV ncRNAs, this analysis also identified 249 novel regions likely to generate conserved structured RNAs. One repeat region of the EBV genome, the W repeats, is especially interesting as it has widespread predicted conserved and stable RNA structure (Fig. 2) that occurs within stable intronic sequence (sis)RNAs.58
The W repeats are transcribed as part of a long (~100 000 nt) EBNA pre-mRNA generated during lytic reactivation and also during latency program III: a highly oncogenic form of latency found in EBV-related B cell lymphomas.8 EBV W repeat number can vary, but the optimum number appears to be five to eight.59 The W repeats are transcribed from either an upstream promoter (Cp) or from one of the downstream (Wp) promoters; Wp is utilized early in latency program III and later there is a switch to Cp.60 W repeats are also transcribed during a latency program observed in ~15% of endemic Burkitt’s lymphomas, Wp-restricted latency.61 The six EBNA messages made in latency III are generated by alternative splicing, including one mRNA (for EBNA-LP), which is produced by the excision of a long and a short intron to join the W1 and W2 ORFs encoded within the W repeats (Fig. 2). It is within these two introns that two new EBV RNA families are found.
ebv-sisRNA-1
The short W repeat intron, rather than being excised and rapidly degraded, persists after splicing and is the third most abundant EBV-produced small ncRNA in latency III.15 Large stable introns (> 2 kbp) are found in other, distant, herpesviruses.62,63 In herpes simplex virus 1 (HSV1), for example, an abundant long (~2000 nt) intron, known as the latency-associated transcript (LAT), is able to suppress apoptosis by inhibiting cellular apoptotic pathways (e.g., caspase 8- and 9-induced apoptosis64). Unlike these other stable introns, ebv-sisRNA-1 is small (81 nt) and, where the functional form of the LAT is the spliced lariat structure,65 ebv-sisRNA-1 is a linear RNA molecule.15
The modeled structure of ebv-sisRNA-1 is shown in Figure 3F. Nucleotides 4–26 form a short hairpin that presents a U-rich sequence motif (a possible platform for protein interactions)66,67 into a loop. The remainder of the sequence includes a CA-rich region and is unlikely to form stable RNA structure. This unstructured stretch of sequence may be exposed to allow interactions with nucleic acids or other proteins. The ebv-sisRNA-1 sequence is ~100% conserved in EBV strains and homology extends to include other lymphocryptoviruses (Fig. S3): 91% conserved [vs. EBV in MHV4, 98% in Pongine herpesvirus 1, 94% in Pongine herpesvirus 3, 91% in CeHV12, and 60% in the more distant lypmphocryptovirus, Callitrichine herpesvirus 3 (CaHV3)]. The hairpin structure is also conserved and includes a structure-preserving compensatory mutation in CaHV3 that converts a GC to an AU pair.
ebv-sisRNA-2
Conserved and stable RNA structure includes approximately 40% of the long W repeat intron.15 RNA-Seq data from cultured latency III expressing human B cells (Jijoye cell line; data kindly provided by Prof. Erik Flemington, Tulane) provide evidence that the entire long intron is also a stable transcript (ebv-sisRNA-2). The presence of ebv-sisRNA-2 in latently infected human cells is very interesting as it more closely resembles the LATs in size than ebv-sisRNA-1. Perhaps ebv-sisRNA-2 also plays a role in the maintenance of latency in EBV.
Ebv-sisRNA-2 has one region that is modeled to fold into an unusually long (586 nt) and remarkably thermodynamically stable hairpin loop (Fig. 4). Canonically paired regions are interrupted by a number of internal and bulged “loops” that may have functionally relevant 2D and 3D structures. For example, 265–7 nt and 313–5 nt in Figure 4 are modeled to form a symmetric three-by-three nt internal loop (where three unpaired nt are arranged across from three unpaired nt). Comprised entirely of purines, this loop can form a stable and helical “3RRs” motif68 where opposing purines in the loop form sheared base pairs.
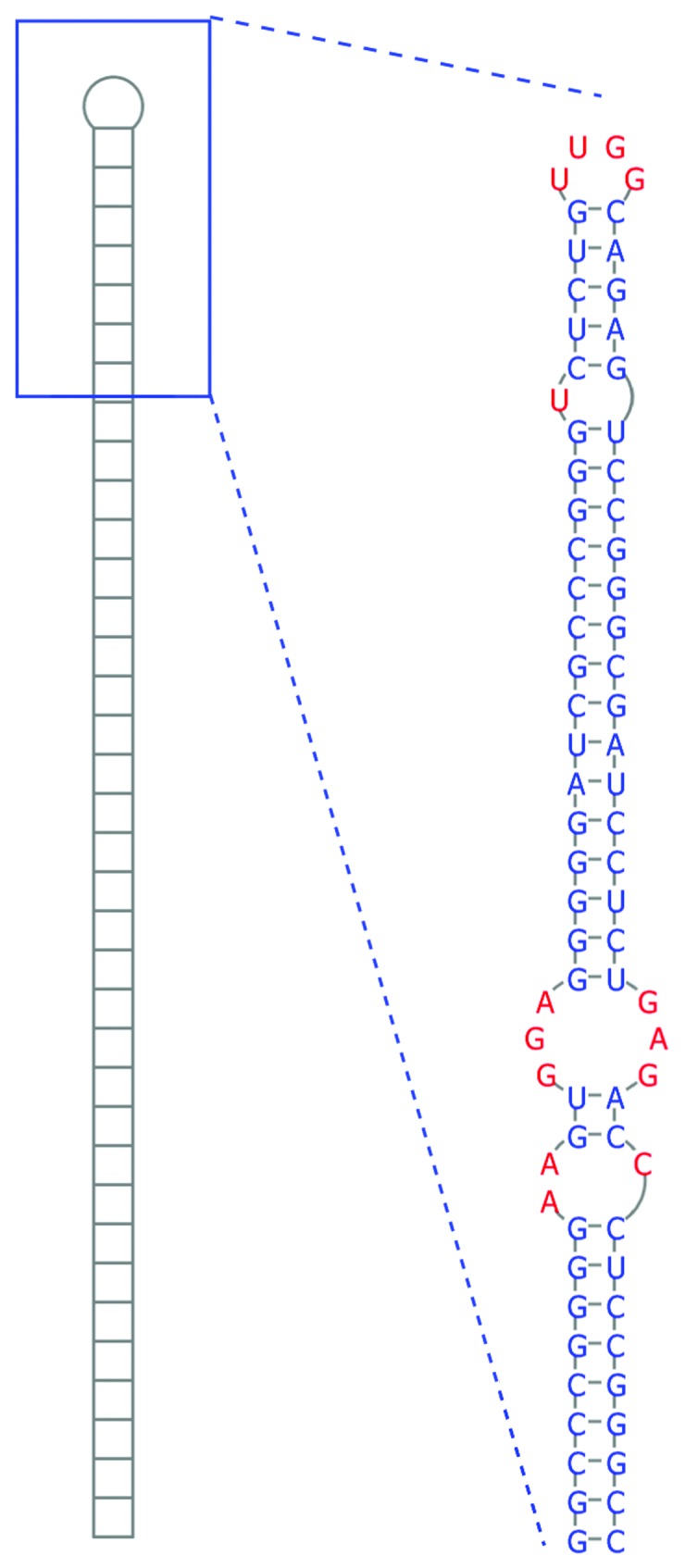
Figure 4. Secondary structure of the long hairpin RNA contained within ebv-sisRNA-2. At the left is a cartoon of the entire 586 nt hairpin and to the right is the structure of the terminal 76 nt of the hairpin, which includes the 3RRs motif (the three-by-three nt GGA/GAG internal loop).
In addition to EBV strains, where the hairpin is ~100% conserved in sequence, this structure is also found in MHV4, where it is multiplied six times in repeat introns, in (CeHV12) (repeated three times in a partial sequence), and in CaHV3 (repeated four times). The hairpin is 79% conserved in sequence between EBV and MHV4, while in in CeHV12 and CaHV3 the sequence is 74 and 38% conserved, respectively (Fig. S4). Despite the high divergence of sequence, long hairpins form in these homologous introns. Other examples of very long hairpins are found in the 3′ UTRs of humans and Caenorhabditis elegans. These hairpins are substrates for adenosine deaminase acting on RNA (ADAR), which converts adenosine to inosine (A-to-I) in double-stranded RNA. The long EBV hairpin was predicted to contain residues that could be strong ADAR editing sites15 and there is a previous report of an editing site in this region.69
Concluding Remarks
EBV-generated ncRNAs play important roles in mediating host-virus interactions and in regulating viral processes. A better understanding of the precise roles played by EBV ncRNAs may shed light on important aspects of EBV virology and offer general insights into herpesviruses. Conservation of ebv-sisRNA-1 and the ebv-sisRNA-2 hairpin structure in MHV4 and CeHV12 (as well as in other lymphocryptoviruses) suggests conserved functional significance. Future work will elucidate the functions of these interesting novel EBV ncRNAs and shed light on their roles in latency III. These new RNAs join the growing list of EBV ncRNA families. Studies to decipher the various functions of EBV-generated ncRNAs continue and the future surely holds many important discoveries.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We sincerely thank Prof Erik Flemington (Tulane) and his lab members for providing RNA-Seq data for the analysis of ebv-sisRNA-2. This work was supported by grant CA16038 from the NIH. J.A.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/27488
References
- 1.CDC. Epstein–Barr Virus and Infectious Mononucleosis, 2006. Available from: http://www.cdc.gov/ncidod/diseases/ebv.htm
- 2.Flavell KJ, Murray PG. Hodgkin’s disease and the Epstein-Barr virus. Mol Pathol. 2000;53:262–9. doi: 10.1136/mp.53.5.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burkitt D. A sarcoma involving the jaws in African children. Br J Surg. 1958;46:218–23. doi: 10.1002/bjs.18004619704. [DOI] [PubMed] [Google Scholar]
- 4.Niller HH, Wolf H, Minarovits J. Regulation and dysregulation of Epstein-Barr virus latency: implications for the development of autoimmune diseases. Autoimmunity. 2008;41:298–328. doi: 10.1080/08916930802024772. [DOI] [PubMed] [Google Scholar]
- 5.Nanbo A, Takada K. The role of Epstein-Barr virus-encoded small RNAs (EBERs) in oncogenesis. Rev Med Virol. 2002;12:321–6. doi: 10.1002/rmv.363. [DOI] [PubMed] [Google Scholar]
- 6.Nanbo A, Yoshiyama H, Takada K. Epstein-Barr virus-encoded poly(A)- RNA confers resistance to apoptosis mediated through Fas by blocking the PKR pathway in human epithelial intestine 407 cells. J Virol. 2005;79:12280–5. doi: 10.1128/JVI.79.19.12280-12285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowe DT. Epstein-Barr virus immortalization and latency. Front Biosci. 1999;4:D346–71. doi: 10.2741/Rowe. [DOI] [PubMed] [Google Scholar]
- 8.Rezk SA, Weiss LM. Epstein-Barr virus-associated lymphoproliferative disorders. Hum Pathol. 2007;38:1293–304. doi: 10.1016/j.humpath.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 9.Concha M, Wang X, Cao S, Baddoo M, Fewell C, Lin Z, Hulme W, Hedges D, McBride J, Flemington EK. Identification of new viral genes and transcript isoforms during Epstein-Barr virus reactivation using RNA-Seq. J Virol. 2012;86:1458–67. doi: 10.1128/JVI.06537-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Grady T, Cao S, Strong MJ, Concha M, Wang X, Bondurant SS, Adams M, Baddoo M, Srivastav SK, Lin Z, et al. Global Bidirectional Transcription of the Epstein-Barr Virus Genome During Reactivation. J Virol. 2013 doi: 10.1128/JVI.02989-13. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–68. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 12.Thorley-Lawson DA, Allday MJ. The curious case of the tumour virus: 50 years of Burkitt’s lymphoma. Nat Rev Microbiol. 2008;6:913–24. doi: 10.1038/nrmicro2015. [DOI] [PubMed] [Google Scholar]
- 13.Lerner MR, Andrews NC, Miller G, Steitz JA. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1981;78:805–9. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fok V, Friend K, Steitz JA. Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. J Cell Biol. 2006;173:319–25. doi: 10.1083/jcb.200601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss WN, Steitz JA. Genome-wide analyses of Epstein-Barr virus reveal conserved RNA structures and a novel stable intronic sequence RNA. BMC Genomics. 2013;14:543. doi: 10.1186/1471-2164-14-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke PA, Sharp NA, Clemens MJ. Expression of genes for the Epstein-Barr virus small RNAs EBER-1 and EBER-2 in Daudi Burkitt’s lymphoma cells: effects of interferon treatment. J Gen Virol. 1992;73:3169–75. doi: 10.1099/0022-1317-73-12-3169. [DOI] [PubMed] [Google Scholar]
- 17.Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci U S A. 1991;88:1546–50. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregorovic G, Bosshard R, Karstegl CE, White RE, Pattle S, Chiang AK, Dittrich-Breiholz O, Kracht M, Russ R, Farrell PJ. Cellular gene expression that correlates with EBER expression in Epstein-Barr Virus-infected lymphoblastoid cell lines. J Virol. 2011;85:3535–45. doi: 10.1128/JVI.02086-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yajima M, Kanda T, Takada K. Critical role of Epstein-Barr Virus (EBV)-encoded RNA in efficient EBV-induced B-lymphocyte growth transformation. J Virol. 2005;79:4298–307. doi: 10.1128/JVI.79.7.4298-4307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto N, Takizawa T, Iwanaga Y, Shimizu N, Yamamoto N. Malignant transformation of B lymphoma cell line BJAB by Epstein-Barr virus-encoded small RNAs. FEBS Lett. 2000;484:153–8. doi: 10.1016/S0014-5793(00)02145-1. [DOI] [PubMed] [Google Scholar]
- 21.Houmani JL, Davis CI, Ruf IK. Growth-promoting properties of Epstein-Barr virus EBER-1 RNA correlate with ribosomal protein L22 binding. J Virol. 2009;83:9844–53. doi: 10.1128/JVI.01014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee N, Pimienta G, Steitz JA. AUF1/hnRNP D is a novel protein partner of the EBER1 noncoding RNA of Epstein-Barr virus. RNA. 2012;18:2073–82. doi: 10.1261/rna.034900.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu CC, Wu CW, Chang SC, Chen TY, Hu CR, Yeh MY, Chen JY, Chen MR. Epstein-Barr virus nuclear antigen 1 is a DNA-binding protein with strong RNA-binding activity. J Gen Virol. 2004;85:2755–65. doi: 10.1099/vir.0.80239-0. [DOI] [PubMed] [Google Scholar]
- 24.Malik-Soni N, Frappier L. Proteomic profiling of EBNA1-host protein interactions in latent and lytic Epstein-Barr virus infections. J Virol. 2012;86:6999–7002. doi: 10.1128/JVI.00194-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glickman JN, Howe JG, Steitz JA. Structural analyses of EBER1 and EBER2 ribonucleoprotein particles present in Epstein-Barr virus-infected cells. J Virol. 1988;62:902–11. doi: 10.1128/jvi.62.3.902-911.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolin SL, Cedervall T. The La protein. Annu Rev Biochem. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- 27.Wu Y, Maruo S, Yajima M, Kanda T, Takada K. Epstein-Barr virus (EBV)-encoded RNA 2 (EBER2) but not EBER1 plays a critical role in EBV-induced B-cell growth transformation. J Virol. 2007;81:11236–45. doi: 10.1128/JVI.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu P, Fang X, Feng Z, Guo YM, Peng RJ, Liu T, Huang Z, Feng Y, Sun X, Xiong Z, et al. Direct sequencing and characterization of a clinical isolate of Epstein-Barr virus from nasopharyngeal carcinoma tissue by using next-generation sequencing technology. J Virol. 2011;85:11291–9. doi: 10.1128/JVI.00823-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barth S, Meister G, Grässer FA. EBV-encoded miRNAs. Biochim Biophys Acta. 2011;1809:631–40. doi: 10.1016/j.bbagrm.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Riley KJ, Rabinowitz GS, Yario TA, Luna JM, Darnell RB, Steitz JA. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 2012;31:2207–21. doi: 10.1038/emboj.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riley KJ, Rabinowitz GS, Steitz JA. Comprehensive analysis of Rhesus lymphocryptovirus microRNA expression. J Virol. 2010;84:5148–57. doi: 10.1128/JVI.00110-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cameron JE, Fewell C, Yin Q, McBride J, Wang X, Lin Z, Flemington EK. Epstein-Barr virus growth/latency III program alters cellular microRNA expression. Virology. 2008;382:257–66. doi: 10.1016/j.virol.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen SJ, Chen GH, Chen YH, Liu CY, Chang KP, Chang YS, Chen HC. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS One. 2010;5:5. doi: 10.1371/journal.pone.0012745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu J, Cosmopoulos K, Pegtel M, Hopmans E, Murray P, Middeldorp J, Shapiro M, Thorley-Lawson DA. A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog. 2011;7:e1002193. doi: 10.1371/journal.ppat.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu JY, Pfuhl T, Motsch N, Barth S, Nicholls J, Grässer F, Meister G. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J Virol. 2009;83:3333–41. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lo AK, Dawson CW, Jin DY, Lo KW. The pathological roles of BART miRNAs in nasopharyngeal carcinoma. J Pathol. 2012;227:392–403. doi: 10.1002/path.4025. [DOI] [PubMed] [Google Scholar]
- 37.Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, Nusbaum JD, Feederle R, Delecluse HJ, Luftig MA, et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012;8:e1002484. doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isaksson A, Berggren M, Ricksten A. Epstein-Barr virus U leader exon contains an internal ribosome entry site. Oncogene. 2003;22:572–81. doi: 10.1038/sj.onc.1206149. [DOI] [PubMed] [Google Scholar]
- 39.Jang SK, Kräusslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–43. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 41.Endo R, Kikuta H, Ebihara T, Ishiguro N, Kobayashi K. Possible involvement in oncogenesis of a single base mutation in an internal ribosome entry site of Epstein-Barr nuclear antigen 1 mRNA. J Med Virol. 2004;72:630–4. doi: 10.1002/jmv.20022. [DOI] [PubMed] [Google Scholar]
- 42.Kieft JS. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci. 2008;33:274–83. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 44.Sperschneider J, Datta A. DotKnot: pseudoknot prediction using the probability dot plot under a refined energy model. Nucleic Acids Res. 2010;38:e103. doi: 10.1093/nar/gkq021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J Mol Biol. 2002;319:1059–66. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 47.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–5. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–7. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jang SB, Hung LW, Chi YI, Holbrook EL, Carter RJ, Holbrook SR. Structure of an RNA internal loop consisting of tandem C-A+ base pairs. Biochemistry. 1998;37:11726–31. doi: 10.1021/bi980758j. [DOI] [PubMed] [Google Scholar]
- 50.Pace NR, Smith DK, Olsen GJ, James BD. Phylogenetic comparative analysis and the secondary structure of ribonuclease P RNA--a review. Gene. 1989;82:65–75. doi: 10.1016/0378-1119(89)90031-0. [DOI] [PubMed] [Google Scholar]
- 51.Chen JL, Dishler AL, Kennedy SD, Yildirim I, Liu B, Turner DH, Serra MJ. Testing the nearest neighbor model for canonical RNA base pairs: revision of GU parameters. Biochemistry. 2012;51:3508–22. doi: 10.1021/bi3002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berry KE, Waghray S, Doudna JA. The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA. 2010;16:1559–69. doi: 10.1261/rna.2197210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brierley I, Pennell S, Gilbert RJ. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat Rev Microbiol. 2007;5:598–610. doi: 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu B, Mathews DH, Turner DH. RNA pseudoknots: folding and finding. F1000 Biol Rep. 2010;2:8. doi: 10.3410/B2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson SR. Tricks an IRES uses to enslave ribosomes. Trends Microbiol. 2012;20:558–66. doi: 10.1016/j.tim.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathews DH, Moss WN, Turner DH. Folding and finding RNA secondary structure. Cold Spring Harb Perspect Biol. 2010;2:a003665. doi: 10.1101/cshperspect.a003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hutzinger R, Feederle R, Mrazek J, Schiefermeier N, Balwierz PJ, Zavolan M, Polacek N, Delecluse HJ, Hüttenhofer A. Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome. PLoS Pathog. 2009;5:e1000547. doi: 10.1371/journal.ppat.1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gardner EJ, Nizami ZF, Talbot CC, Jr., Gall JG. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev. 2012;26:2550–9. doi: 10.1101/gad.202184.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tierney RJ, Kao KY, Nagra JK, Rickinson AB. Epstein-Barr virus BamHI W repeat number limits EBNA2/EBNA-LP coexpression in newly infected B cells and the efficiency of B-cell transformation: a rationale for the multiple W repeats in wild-type virus strains. J Virol. 2011;85:12362–75. doi: 10.1128/JVI.06059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woisetschlaeger M, Yandava CN, Furmanski LA, Strominger JL, Speck SH. Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc Natl Acad Sci U S A. 1990;87:1725–9. doi: 10.1073/pnas.87.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kelly GL, Long HM, Stylianou J, Thomas WA, Leese A, Bell AI, Bornkamm GW, Mautner J, Rickinson AB, Rowe M. An Epstein-Barr virus anti-apoptotic protein constitutively expressed in transformed cells and implicated in burkitt lymphomagenesis: the Wp/BHRF1 link. PLoS Pathog. 2009;5:e1000341. doi: 10.1371/journal.ppat.1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen W, Sa e Silva M, Jaber T, Vitvitskaia O, Li S, Henderson G, Jones C. Two small RNAs encoded within the first 1.5 kilobases of the herpes simplex virus type 1 latency-associated transcript can inhibit productive infection and cooperate to inhibit apoptosis. J Virol. 2009;83:9131–9. doi: 10.1128/JVI.00871-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang S, Patel A, Krause PR. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J Virol. 2009;83:1433–42. doi: 10.1128/JVI.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Henderson G, Peng W, Jin L, Perng GC, Nesburn AB, Wechsler SL, Jones C. Regulation of caspase 8- and caspase 9-induced apoptosis by the herpes simplex virus type 1 latency-associated transcript. J Neurovirol. 2002;8(Suppl 2):103–11. doi: 10.1080/13550280290101085. [DOI] [PubMed] [Google Scholar]
- 65.Hesselberth JR. Lives that introns lead after splicing. Wiley Interdiscip Rev RNA. 2013;4:677–91. doi: 10.1002/wrna.1187. Forthcoming. [DOI] [PubMed] [Google Scholar]
- 66.Alian A, DeGiovanni A, Griner SL, Finer-Moore JS, Stroud RM. Crystal structure of an RluF-RNA complex: a base-pair rearrangement is the key to selectivity of RluF for U2604 of the ribosome. J Mol Biol. 2009;388:785–800. doi: 10.1016/j.jmb.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.You Y, Chen CY, Shyu AB. U-rich sequence-binding proteins (URBPs) interacting with a 20-nucleotide U-rich sequence in the 3′ untranslated region of c-fos mRNA may be involved in the first step of c-fos mRNA degradation. Mol Cell Biol. 1992;12:2931–40. doi: 10.1128/mcb.12.7.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lerman YV, Kennedy SD, Shankar N, Parisien M, Major F, Turner DH. NMR structure of a 4 x 4 nucleotide RNA internal loop from an R2 retrotransposon: identification of a three purine-purine sheared pair motif and comparison to MC-SYM predictions. RNA. 2011;17:1664–77. doi: 10.1261/rna.2641911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arvey A, Tempera I, Tsai K, Chen HS, Tikhmyanova N, Klichinsky M, Leslie C, Lieberman PM. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe. 2012;12:233–45. doi: 10.1016/j.chom.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.