Abstract
The Argonaute family of proteins is highly evolutionarily conserved and plays essential roles in small RNA-mediated gene regulatory pathways and in a wide variety of cellular processes. They were initially discovered by genetics studies in plants and have been well characterized as key components of gene silencing pathways guided by small RNAs, a phenomenon known as RNA interference. Conventionally, guided by different classes of small RNAs, Argonautes bind to and silence homologous target sequences at the post-transcriptional level. Increasing lines of evidence support their multi-functional roles in the nucleus. Advances in high-throughput genome-wide methodologies have greatly facilitated our understanding of their functions in post-transcriptional gene silencing as well as in other nuclear events. In this point-of-view, we will summarize key findings from genome-wide analyses of the Ago subfamily of proteins in mammals and Drosophila, discuss their nuclear functions in the regulation of transcription and alternative splicing identified in recent years, and briefly touch upon their potential implications in cancer.
Keywords: Argonaute proteins, histone modifications, transcription regulation, alternative splicing, cancer
Introduction
Argonaute proteins comprise a family of evolutionarily conserved proteins that are central to the RNA interference (RNAi) platform and microRNA (miRNA) function and biogenesis.1 They are best known as core components of the RNA-induced silencing complex (RISC) required for small RNA-mediated gene regulatory mechanisms. In post-transcriptional gene silencing (PTGS), Argonautes guided by the small RNAs (e.g., siRNAs, miRNAs, piRNAs, etc.) bind to the complementary transcripts via base-pairing and serve as platforms for recruiting proteins to facilitate gene silencing. Most of the molecular functions of Argonautes have been well studied in the cytoplasmic compartment of eukaryotic cells in which they regulate gene transcripts via PTGS mechanisms. However, some unique nuclear functions and RNAi-related processes have also been well characterized in other model organisms (i.e., Schizosaccharomyces pombe, Drosophila melanogaster, Arabidopsis thaliana, and C. elegans) in which the Ago protein guided by small RNAs assists in heterochromatin formation at centromeric regions and/or facilitates transcriptional gene silencing (TGS) associated with chromosome segregation, suppression of antisense RNA, maintenance of epigenetic inheritance,2-8 and transcriptome surveillence.9
Mammals have eight Argonaute proteins, which are divided into two subfamilies: the Piwi clade and the Ago clade. This review will focus on the Ago clade members (which will be referred to as Ago proteins or Agos in short). While Piwi proteins are exclusively expressed in the germline where they bind to piRNAs to facilitate silencing of transposable elements, Ago proteins (Ago1–4 or EIF2C1–4) are ubiquitously expressed in all tissues. Human Ago1, Ago3, and Ago4 genes are clustered on chromosome 1, whereas Ago2 is uniquely located on chromosome 8. Biologically, Ago2 is essential for embryonic development as constitutive knockout of Ago2 causes embryonic lethality in mice while single knockout of other family members appears to be dispensable.10-13 Ago2 is the primary mediator of RNAi given that it is the only family member possessing endonuclease activity.10 Crystal structures of full-length human Ago2 have been solved,14,15 which provide key mechanistic insights into their interactions with small RNAs. Similar to the bacteria counterpart, human Ago2 is a bilobular structure comprising the N-terminal (N), PAZ, MID, and PIWI domains. The PAZ domain anchors the 3′end of the small RNAs and is dispensable for the catalytic activity of Ago2. However, PAZ domain deletion disrupts the ability of the non-catalytic Agos to unwind small RNA duplex and to form functional RISC.16
Biochemical studies strongly support that Ago members can bind common sets of miRNAs with no functional selectivity.17 Specifically, human Ago proteins have been shown to be functionally redundant in miRNA-mediated gene silencing as subsequent re-introduction of single Ago (Ago1–4) into Ago-knockout mouse embryonic fibroblasts (MEFs) can rescue defect in miRNA-mediated gene silencing.18 However, it has been reported that many non-miRNA sequences are specifically enriched in human Ago1 immunoprecipitates in small RNA deep sequencing experiments,19 suggesting that Agos may have differential selectivity for non-miRNA RNA species, including promoter-associated transcripts.
Ago proteins are predominantly located in the cytoplasmic compartment known as the cytoplasmic processing bodies (P-bodies) and can be readily detected by immunofluorescence techniques.20 By biochemical fractionation and confocal immunofluorescence microscopy, endogenous Ago proteins have also been detected in the nuclear compartment of mouse and human cells in a number of studies.21-24 Sophisticated optical techniques such as fluorescence correlation spectroscopy (FCS) and fluorescence cross-correlation spectroscopy (FCCS) have been applied to directly study the nucleocytoplasmic shuttling of Ago-small RNA complexes.25 It is not until 2 years ago, several molecular studies investigating the nuclear activities of exogenous miRNAs26 and/or synthetic RNA duplexes in mammalian cells have implicated Ago proteins in several noncanonical pathways unique to the nucleus, including transcriptional silencing, transcriptional activation, alternative splicing, and quiescence-associated translational upregulation.21,23,27-32 Yet, despite such studies, the nuclear functions of Ago proteins remain mysterious in mammalian cells. Here, in addition to the cytoplasmic functions in PTGS, we will discuss novel findings of the nuclear functions of the Ago subfamily specifically in transcriptional regulation and alternative splicing obtained from high-throughput genome-wide studies.
Genome-Wide Characterization of Ago-RNA Interactions
Transcriptome-wide mapping of Ago-mRNA interactions by crosslinking immunoprecipitation (CLIP)-based high-throughput methods such as CLIP-seq (crosslinking immunoprecipitation high-throughput sequencing) or HITS-CLIP (high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation),33-35 PAR-CLIP (photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation),36 and CLASH (crosslinking, ligation, and sequencing of hybrids)37 allow systematic identification of Ago-bound target mRNA sequences in mammalian cells. The use of photocrosslinking and photoactivatable nucleoside in PAR-CLIP has greatly improved the efficiency of nucleic acid crosslinking and signal to noise ratio with single nucleotide resolution compared with the traditional RNA immunoprecipitation methods (reviewed in Ascano et al.38). These studies reveal that Ago proteins mediate binding of miRNAs to diverse RNA species via both canonical and noncanonical base pairings and have generated a wealth of information on ternary map of Ago-miRNA-mRNA interactions (Table 1). Furthermore, these studies confirm that miRNAs can regulate gene expression by binding to various regions of the transcripts not restricted to 3′ UTR regions. Surprisingly, many of these interactions can occur through noncanonical seed base pairings;39 for example, ~10% of the mRNA tags mapped to introns, suggesting that miRNA can target precursor mRNAs in the nucleus.33
Table 1. Summary of high-throughput Ago-mRNA/DNA mapping in mammalian systems.
| Experimental approach | Experimental system | Ago member(s) studied | Antibody used | Key findings | Reference |
|---|---|---|---|---|---|
| HITS-CLIP | Mouse brain | Endogenous Ago | anti-pan-Ago (2A8) | Ago-bound tags fell in 3′UTR (40%), coding sequences (25%), introns (12%), and non-coding RNAs (5%). | Chi et al.33 |
| PAR-CLIP | Human embryonic fibroblasts (HEK293) | Tagged Ago1–4 |
anti-HA | Identified seed-dependent miRNA target sites and nearly 50% of the binding sites located in the CDS. | Hafner et al.36 |
| CLIP-seq | Mouse embryonic stem cells (wt vs. Dicer −/−) | Endogenous Ago2 | anti-Ago2 (2D5) | miRNA dependent and GC rich binding sites were identified in 3′ untranslated and coding regions. | Leung et al.35 |
| CLASH | HEK293 (Doxycycline inducible) | Tagged Ago1 | anti-FLAG | Widespread (> 63%) noncanonical miRNA-mRNA seed interactions via noncanonical Watson-Crick base pairing. | Helwak et al.37 |
| CLIP-seq | HEK293S | Endogenous Ago2 | anti-Ago2(11A9), anti-Ago2 (4F9), anti-Ago2(2E12–1C9) | Cellular stress induced by arsenite strengthens Ago2-mRNA interactions in 3′UTR and CDS of genes repressed during translation. | Karginov et al.34 |
| ChIP-chip | Primary human diploid fibroblasts (WI38) | Endogenous Ago2 | anti-pan-Ago, anti-Ago2(9E8.2) | Ago2, RB1 and miRNAs functionally interact to repress E2F regulated genes during senescence. | Benhamed et al.28 |
| ChIP-seq | Human prostate cancer cells (PC-3) | Endogenous Ago1 | anti-Ago1(2A7) | Ago1 bound sites are associated with transcriptionally active loci marked by H3K4me3. | Huang et al.22 |
Noncanonical Functions of the Ago Proteins
Transcriptional regulation
The chromatin association with Ago proteins and other RNAi-related components in Drosophila suggests their fundamental roles in chromatin regulation.40,41 There has been a limited number of reports on the role of mammalian Ago proteins in transcription regulation (summarized in Fig. 1A and B) despite it has been well-documented in fission yeast and other lower eukaryotes.2,40,41 A series of studies in Drosophila published in the last couple of years has truly laid groundwork for the investigation of nuclear Ago proteins in mammalian cells. Cernilogar et al.40 found that Drosophila Ago2 (dAgo2) interacts with euchromatin and dAgo2-associated small RNAs are enriched at promoter regions of heat shock-responsive genes. Moshkovich et al.41 demonstrated that dAgo2 is associated with transcriptionally active sites which correspond to regions of low endo-siRNA productions. However, the effect of RNA polymerase II (RNAP II) activity on dAgo2-bound sites was not examined in this study. A more recent study by Taliaferro et al.42 obtained an almost identical dAgo2 binding profile as Moshkovich et al. and speculated that, in additional to its role in alternative splicing, nuclear dAgo2 regulates developmentally regulated genes by binding to their respective promoters and negatively regulates transcription. This group of promoters was also bound by Polycomb group (PcG) transcriptional repressor proteins, suggesting that dAgo2 may mediate gene transcription via recruitment of PcG. Consistently it has been previously shown that dAgo2 and components of the RNAi machinery are required for chromatin and nuclear organization of PcG chromatin targets.43
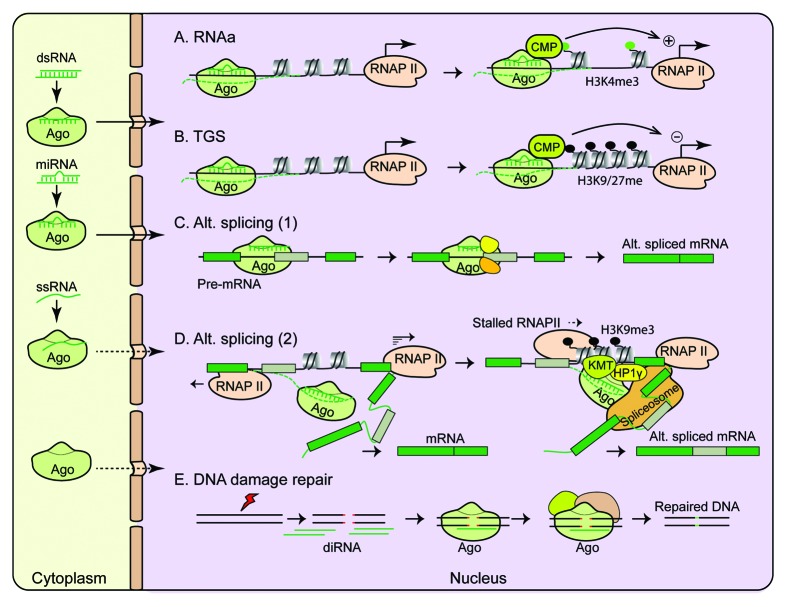
Figure 1. Multi-functional roles of nuclear Ago proteins. Cytoplasmic Agos enter the nucleus through nuclear import pathways (e.g., importin 8) via one of the four possible routes as depicted in the left panel of the figure: in complex with a guide dsRNA, miRNA, ssRNA, or as a free form. In the nucleus, they are guided by the bound RNA to its homologous target sites or recruited by nuclear-localized RNAs to chromatin sites to exert multi-functional roles. (A and B) Ago-mediated chromatin modification processes. (A) RNAa: Recruitment of CMPs by Ago-dsRNA or miRNA complex leads to increased active chromatin marks such as H3K4me3 methylation, allowing active transcription to occur at the targeted promoter. (B) TGS: Recruitment of CMPs by Ago-dsRNA or miRNA complex leads to increased repressive chromatin marks such as H3K9/K27 methylation thereby inhibits transcription at the targeted promoter. (C and D) RNA-mediated alteration of splicing. (C) Exogenous dsRNA-guided alternative splicing: dsRNA-Ago complex forces exon exclusion or inclusion by targeting intronic or exonic sequences involved in splicing. (D) Endogenous small RNA-mediated alternative splicing: Agos, possibly guided by unstructured single stranded small RNA (ssRNA), are recruited to chromatin at the 3′ ends of the variant regions possibly by an intragenic antisense transcript. Interactions with other chromatin components (such as HP1γ or SR proteins) result in H3K9me3 mark deposition by Suv39h1 and EHMT2 (KMT lysine methyl transferase). Consequently, RNAP II elongation rate is slowed, which in turn facilitates spliceosome recruitment and allows alternative splicing of the variant exons. (E) DNA damage repair process. diRNAs generated from DSBs are incorporated into Ago proteins which facilitate DNA damage repair by directly interacting with repair proteins or by affecting local chromatin structures via recruitment of CMPs. CMPs, chromatin modifying proteins; diRNAs, double-strand-break-induced small RNAs; DSBs, double strand breaks; dsRNAs, double stranded RNAs; miRNA, microRNA; RNAa, RNA activation; RNAP II, RNA polymerase II; ssRNA, single stranded small RNA; TGS, transcriptional gene silencing.
By using ChIP-chip (chromatin immunoprecipitation coupled with microarray) analysis, Benhamed et al.28 showed that nuclear Ago2 is involved in senescence-associated transcriptional gene silencing (SA-TGS) by directly interacting with retinoblastoma 1 (RB1) to repress E2F1 target genes in human fibroblasts. By deep-sequencing small RNAs co-immunoprecipitated with Agos, they further showed that the enrichment of heterochromatin-associated miRNAs, including the let-7 family, correlated with transcriptional repression and epigenetic changes at Ago-bound promoters (e.g., CDC2 and CDCA8). Based on these data, the authors proposed two non-mutually exclusive models for Ago2-mediated SA-TGS. Ago2-miRNP interacts with histone deacetylases (HDACs) or histone methyltransferases (HMTs) to block RNAP II at target promoters leading to transcriptional gene silencing (TGS). Alternatively, Ago2-miRNP complex may enhance the stability of RB1-associated repressor complexes to promote TGS.
Very recently, we provided the first unbiased mapping of endogenous Ago1-DNA interactions in human cancer cells by using ChIP-seq (chromatin immunoprecipitation coupled with massively parallel sequencing).22 We showed that Ago1 is pervasively associated with euchromatic regions defined by H3K4me3 marks. Gene expression profiling in Ago 1 depleted cells revealed a strong positive correlation between Ago1 occupancy and levels of gene expression, suggesting that Ago1 may play a positive role in gene regulation. Biochemically, Ago1 is associated with chromatin and physically interacts with RNAP II. Furthermore, Ago1 and RNAP II are both enriched in the promoters of many genes known to be involved in cancer pathways. Bioinformatics analysis also revealed that Ago1-bound sequences are enriched for oncomiR targets, suggesting that Ago1-chromatin interactions may partly depend on miRNAs. Although the underlying mechanism by which Ago1 is involved in transcriptional gene regulation remains to be determined, this study will spur general interests in nuclear Ago1-chromatin regulation and potential cross-talk between small RNA-mediated gene regulation and oncogenic pathways in cancer cells.
Alternative splicing
Several studies have reported that synthetic siRNAs targeting exonic or intronic sequences proximal to the splicing junction can regulate alternative splicing through TGS23,32 (Fig. 1C). It is thought that intronic siRNAs trigger heterochromatin formation by inducing histone H3K9 dimethylation, and subsequently, inhibiting transcriptional elongation resulting in changes in alternative splicing.32 A recent study by Ameyar-Zazoua et al.29 uncovered an unexpected role for Ago proteins in alternative splicing. The authors identified novel interacting proteins of chromatin-bound Agos in human cells using tandem affinity purification. Surprisingly, a large fraction of interacting partners of chromatin-bound Ago1 and Ago2 were part of spliceosome, including components of the U2 and U5 snRNP complexes, SR proteins, and heterogeneous nuclear ribonucleoproteins (hnRNPs). Other proteins involved in chromatin regulation include HP1γ and HMTs were also identified. By using the CD44 gene as a model, the authors demonstrated that Ago1 and Ago2 regulate alternative splicing by coupling RNAP II elongation and histone modifications. The authors proposed that Ago1/2, possibly guided by an intragenic antisense transcript, is recruited to the transcribed regions in CD44 where it interacts with other chromatin components (such as HP1γ or SR proteins), which in turn, induces H3K9 methylation on variant exons and generates heterochromatin-like environment through lysine methyl transferase (KMT) activity resulting in the slowdown of RNAP II. Consequently, this gives splicing factors more time to recognize splice sites and influences splicing decisions (Fig. 1D). In line with this finding, another study found that dAgo2 is also involved in alternative pre-mRNA splicing,42 suggesting that the splicing-related function of Ago proteins is evolutionarily conserved.
Double strand break repair
Recent studies have revealed an unexpected and conserved role for small RNAs and Ago2 in the double strand break (DSB) repair pathway. Wei et al.44 reported that in A. Thaliana and human cells, small RNAs are produced in vicinity of the DSBs and are referred to as diRNAs (double strand break induced small RNAs), which then serve as guiding molecules to recruit effector proteins to facilitate DNA repair. Intriguingly, the authors proposed that Ago2/diRNAs can either directly recruit repair proteins to DSB or by recruiting chromatin modifying complexes to modify local chromatin to facilitate DSB repair (Fig. 1E). How exactly Ago proteins are involved in DSB repair is a fascinating question that remains to be addressed.
Biological Implications of Ago Proteins in Cancer
Components of the small RNA-mediated pathways have been implicated in various cancers;45,46 however, the role of Ago proteins in cancer has never been examined closely. Several expression studies have found that Ago2 is highly expressed in breast cancer,47 high-risk myeloma,48 colon cancer,49 liver cancer,50 prostate, and esophageal cancer.45,51 Ago2 overexpression has been implicated as a result of increase in Ago2 DNA copy number on chromosome 8q2450 (Huang et al., unpublished data).
The link between miRNA maturation and epidermal growth factor receptor (EGFR) signaling has been recently established. Shen et al.52 reported that EGFR can directly phosphorylate Ago2 at tyrosine 393, which blocks the binding of Dicer to Ago2 and inhibits the maturation of a subset of miRNAs, and enhances cell invasiveness under hypoxia. Supported by clinical data, higher expression of p-Y393-Ago2 also correlated with poorer overall survival in breast cancer patients. Ago1 seems to be also involved in angiogenesis during tumorigenesis as it has been identified to be a miRNA target of hypoxia-induced translational de-repression.53 In support, our integrated analysis of ChIP-seq and gene expression profiling demonstrated that Ago1-bound and Ago1-responsive genes are enriched in major cancer-related signaling pathways.22 How the nuclear activities of Ago proteins are directly related to cancer signaling pathways await further investigation.
Conclusions and Future Perspective
Recent high-throughput genomic approaches allowed the mapping of Ago footprints on either mRNA or DNA sequences in vivo under different physiological conditions and demystifying the multi-functional roles of Ago proteins, particularly in the nucleus. The topic of nuclear RNAi remains controversial and there have been disparate observations as to which Ago family members plays a dominant role in a particular nuclear event. The results from these studies were highly dependent on the cell type and cell context and the experimental approaches employed (Table 1). Technical-wise, differences in the specificity and affinity of the antibodies used in ChIP- and CLIP-based studies can be a major contributing factor (summarized in Table 1).
Some of the outstanding questions remain to be addressed include (1) How do Ago proteins interact with chromatin? Due to the lack of a DNA binding domain in Ago proteins, it is conceivable that nuclear Ago-small RNA complex mediates gene regulation by localizing regulatory proteins (i.e., chromatin-modifying enzymes) to specific genomic DNA regions to modulate gene expression (reviewed by Li54). Although more direct evidence needs to be provided, our findings showed that overexpression of mutant Ago1 without the PAZ domain failed to induce the expression of Ago1-bound genes,22 implying a role for small RNAs in mediating Ago-chromatin interactions. (2) What types of RNA species might be involved in mediating Ago1-chromatin interactions? Ago1-RNAP II interaction was reduced in Dicer-knockdown cells,22 implying that Dicer-processed RNA species are responsible for Ago-chromatin interactions. Identification of RNAs associated with chromatin-bound Agos will provide clues to these questions. (3) How Ago1 and Ago2 are functionally distinct in the nucleus? Subcellular localization studies showed that these two family members appear to have differential nuclear distributions.22,24 While the nuclear localization of Ago1 is generally scattered throughout the nuclear interior, Ago2 predominantly localizes to the inner nuclear periphery, which may partly account for the lack of Ago2-chromatin association at steady-state although technical reasons cannot be completely excluded. As further supported by biochemical evidence, Ago1 and Ago2 have been reported to exhibit intrinsic preferences when selecting and/or loading RNA molecules.55,56 Quantitative proteomic analysis has revealed differential abundance of Ago proteins in human and mouse melanoma cells.17 It is plausible to hypothesize that Ago1 and Ago2 have differential nuclear abundance, which may influence their target specificity. Post-translational modifications of Ago proteins have been shown to affect Ago protein stability and homeostasis of miRNA-guided gene silencing.1,57 Another yet-to-be explored area is how post-translational modification(s) affect the nuclear functions of Ago proteins.
Ago proteins have attracted considerable attention for their involvement in gene regulation and other novel functions, which will continue to expand in the years to come. Coupling high-throughput genomic and computational analyses with biochemical characterization of nuclear Ago proteins will enhance our understanding of the complexity of small RNA-mediated gene regulatory mechanisms in various biological contexts. It would be of future interests to understand the crosstalk between Ago-mediated gene regulatory networks and major signaling pathways altered in pathological conditions. Ultimately, modulating nuclear Ago protein activities may potentially be a novel point of therapeutic intervention for cancer and others diseases.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by grants from the National Institutes of Health (1R01GM090293-0109 to Li L-C). Huang V was supported by a Prostate Cancer Postdoctoral Training Award from the Department of Defense (W81XWH-10-1-0505).
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/27604
References
- 1.Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14:447–59. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 2.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–7. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 3.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–9. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 4.Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–27. doi: 10.1016/S0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 5.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–6. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zofall M, Fischer T, Zhang K, Zhou M, Cui B, Veenstra TD, Grewal SI. Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature. 2009;461:419–22. doi: 10.1038/nature08321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–34. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–51. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marasovic M, Zocco M, Halic M. Argonaute and Triman generate dicer-independent priRNAs and mature siRNAs to initiate heterochromatin formation. Mol Cell. 2013;52:173–83. doi: 10.1016/j.molcel.2013.08.046. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 11.Morita S, Horii T, Kimura M, Goto Y, Ochiya T, Hatada I. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics. 2007;89:687–96. doi: 10.1016/j.ygeno.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Modzelewski AJ, Holmes RJ, Hilz S, Grimson A, Cohen PE. AGO4 regulates entry into meiosis and influences silencing of sex chromosomes in the male mouse germline. Dev Cell. 2012;23:251–64. doi: 10.1016/j.devcel.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–42. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150:100–10. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–40. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu S, Jin L, Huang Y, Zhang F, Kay MA. Slicing-independent RISC activation requires the argonaute PAZ domain. Curr Biol. 2012;22:1536–42. doi: 10.1016/j.cub.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D, Zhang Z, O’Loughlin E, Lee T, Houel S, O’Carroll D, Tarakhovsky A, Ahn NG, Yi R. Quantitative functions of Argonaute proteins in mammalian development. Genes Dev. 2012;26:693–704. doi: 10.1101/gad.182758.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–17. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Suzuki H, Hayashizaki Y, Daub CO. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8:158–77. doi: 10.4161/rna.8.1.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rüdel S, Flatley A, Weinmann L, Kremmer E, Meister G. A multifunctional human Argonaute2-specific monoclonal antibody. RNA. 2008;14:1244–53. doi: 10.1261/rna.973808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, Yu A, Shuman M, Yu J, Li LC. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 2012;40:1695–707. doi: 10.1093/nar/gkr934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang V, Zheng J, Qi Z, Wang J, Place RF, Yu J, Li H, Li LC. Ago1 Interacts with RNA polymerase II and binds to the promoters of actively transcribed genes in human cancer cells. PLoS Genet. 2013;9:e1003821. doi: 10.1371/journal.pgen.1003821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Hu J, Corey DR. Expanding the action of duplex RNAs into the nucleus: redirecting alternative splicing. Nucleic Acids Res. 2012;40:1240–50. doi: 10.1093/nar/gkr780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahlenstiel CL, Lim HG, Cooper DA, Ishida T, Kelleher AD, Suzuki K. Direct evidence of nuclear Argonaute distribution during transcriptional silencing links the actin cytoskeleton to nuclear RNAi machinery in human cells. Nucleic Acids Res. 2012;40:1579–95. doi: 10.1093/nar/gkr891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohrt T, Mütze J, Staroske W, Weinmann L, Höck J, Crell K, Meister G, Schwille P. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 2008;36:6439–49. doi: 10.1093/nar/gkn693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang V, Li LC. miRNA goes nuclear. RNA Biol. 2012;9:269–73. doi: 10.4161/rna.19354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Younger ST, Corey DR. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res. 2011;39:5682–91. doi: 10.1093/nar/gkr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat Cell Biol. 2012;14:266–75. doi: 10.1038/ncb2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol. 2012;19:998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- 30.Truesdell SS, Mortensen RD, Seo M, Schroeder JC, Lee JH, LeTonqueze O, Vasudevan S. MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci Rep. 2012;2:842. doi: 10.1038/srep00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsui M, Chu Y, Zhang H, Gagnon KT, Shaikh S, Kuchimanchi S, Manoharan M, Corey DR, Janowski BA. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res. 2013;41:10086–109. doi: 10.1093/nar/gkt777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alló M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16:717–24. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- 33.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karginov FV, Hannon GJ. Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev. 2013;27:1624–32. doi: 10.1101/gad.215939.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol. 2011;18:237–44. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr., Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–65. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip Rev RNA. 2012;3:159–77. doi: 10.1002/wrna.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chi SW, Hannon GJ, Darnell RB. An alternative mode of microRNA target recognition. Nat Struct Mol Biol. 2012;19:321–7. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Lo Sardo F, Saxena A, Miyoshi K, Siomi H, et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–5. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 2011;25:1686–701. doi: 10.1101/gad.16651211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taliaferro JM, Aspden JL, Bradley T, Marwha D, Blanchette M, Rio DC. Two new and distinct roles for Drosophila Argonaute-2 in the nucleus: alternative pre-mRNA splicing and transcriptional repression. Genes Dev. 2013;27:378–89. doi: 10.1101/gad.210708.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–71. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 44.Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang YG, Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–12. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 45.Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, Dhir R. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–20. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papachristou DJ, Korpetinou A, Giannopoulou E, Antonacopoulou AG, Papadaki H, Grivas P, Scopa CD, Kalofonos HP. Expression of the ribonucleases Drosha, Dicer, and Ago2 in colorectal carcinomas. Virchows Arch. 2011;459:431–40. doi: 10.1007/s00428-011-1119-5. [DOI] [PubMed] [Google Scholar]
- 47.Adams BD, Claffey KP, White BA. Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology. 2009;150:14–23. doi: 10.1210/en.2008-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Y, Chen L, Barlogie B, Stephens O, Wu X, Williams DR, Cartron MA, van Rhee F, Nair B, Waheed S, et al. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc Natl Acad Sci U S A. 2010;107:7904–9. doi: 10.1073/pnas.0908441107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li L, Yu C, Gao H, Li Y. Argonaute proteins: potential biomarkers for human colon cancer. BMC Cancer. 2010;10:38. doi: 10.1186/1471-2407-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng N, Li Y, Han ZG. Argonaute2 promotes tumor metastasis by way of up-regulating focal adhesion kinase expression in hepatocellular carcinoma. Hepatology. 2013;57:1906–18. doi: 10.1002/hep.26202. [DOI] [PubMed] [Google Scholar]
- 51.Yoo NJ, Hur SY, Kim MS, Lee JY, Lee SH. Immunohistochemical analysis of RNA-induced silencing complex-related proteins AGO2 and TNRC6A in prostate and esophageal cancers. APMIS. 2010;118:271–6. doi: 10.1111/j.1600-0463.2010.02588.x. [DOI] [PubMed] [Google Scholar]
- 52.Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim SO, Du Y, Wang Y, Chang WC, Chen CH, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–7. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Z, Lai TC, Jan YH, Lin FM, Wang WC, Xiao H, Wang YT, Sun W, Cui X, Li YS, et al. Hypoxia-responsive miRNAs target argonaute 1 to promote angiogenesis. J Clin Invest. 2013;123:1057–67. doi: 10.1172/JCI65344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li LC. Chromatin remodeling by the small RNA machinery in mammalian cells. Epigenetics. 2013;9:9. doi: 10.4161/epi.26830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mescalchin A, Detzer A, Weirauch U, Hahnel MJ, Engel C, Sczakiel G. Antisense tools for functional studies of human Argonaute proteins. RNA. 2010;16:2529–36. doi: 10.1261/rna.2204610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang B, Li S, Qi HH, Chowdhury D, Shi Y, Novina CD. Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat Struct Mol Biol. 2009;16:1259–66. doi: 10.1038/nsmb.1712. [DOI] [PubMed] [Google Scholar]
- 57.Johnston M, Hutvagner G. Posttranslational modification of Argonautes and their role in small RNA-mediated gene regulation. Silence. 2011;2:5. doi: 10.1186/1758-907X-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]