Abstract
Ribosomal proteins (RPs) have been shown to be able to impart selectivity on the translating ribosome implicating them in gene expression control. Many ribosomal proteins are highly conserved and recently a number of ribosomal protein paralogs have been described in mammals. We examined the expression pattern of RPs in differentiating mouse Embryonic Stem Cells (ESCs), paying particular attention to the RP paralogs. We find the RP paralog Rpl39l is highly expressed in ESC and its expression strongly correlates with hepatocellular carcinoma tumor (HCC) samples with high tumor grading and alpha-fetoprotein level giving it diagnostic potential. We further screen the expression pattern of all RPs and their paralogs across 22 different tissues. We find that the more recently evolved RP paralogs show a much greater level of tissue-specific expression. We propose that these RP paralogs evolved more recently to provide a greater level of gene expression control to higher eukaryotes.
Keywords: ribosomal protein, RPL39L, hepatocellular carcinoma, ribosomal protein paralogs
Introduction
Regulation of gene expression is central to the existence of all organisms and the complexity of the regulation increases with the complexity of the organism. Tissue- and context-specific control of gene expression is critical in all cellular processes and involves a dramatic network of regulators to ensure proteins are expressed at the appropriate times and places. Control is levied at the level of transcription, RNA stability, translation, and protein stability and function. While dramatic changes in transcription can drastically alter the RNA content of a cell, it is the translational controls that really dictate the protein repertoire of a cell. Translational regulation of specific mRNAs occurs to a large extent through the binding of target-specific miRNAs and RNA binding proteins. These regulate the translation machinery to control protein production. Recently, the ribosome itself has been shown to have the ability to regulate translation of specific mRNAs, adding a new level of complexity to the cells’ gene regulatory network.1-4
The ribosome is composed of four rRNAs and over 80 ribosomal proteins (RPs) assembled into two major subunits: the small 40S and large 60S subunits in eukaryotes. The ribosome is assembled in the nucleolus before being transported to the cytoplasm. The crystal structure of the yeast ribosome identified the location of the majority of RPs but the specific function of many of these proteins remains unknown.5,6 RPs are required for the assembly and function of the ribosome and in addition, many have additional non-ribosomal functions.7
Haploinsufficiency of RPs has been shown to cause a range of different phenotypes. For example the minute phenotype in Drosophila, characterized by developmental defects and impaired fertility, is caused by haploinsufficiency of one of a number of different ribosomal proteins.8 In humans, mutations in ribosomal proteins have been shown to be the cause of a number of different inherited disorders known as ribosomopathies.9 Over 50% of Diamond-Blackfan Anemia cases have mutations in a ribosomal protein, the most prevalent being RPS19.10 Reduced levels of RPS19 result in deregulated translation of specific mRNAs that correlate with the disease phenotype.11
Cancer is often associated with increased translation and aberrantly expressed RPs.12 RPL19 is upregulated in many prostate cancers and its knockdown results in a less severe malignant phenotype in vivo suggesting it has a functional role in promoting tumorogenesis.13 Haploinsufficiency of ribosomal proteins can also lead to aberrant ribosome assembly and nucleolar stress. Under such conditions certain ribosomal proteins bind to MDM2 preventing it from targeting p53 for degradation resulting in a p53-dependant cell cycle arrest. This pathway directly links ribosome biogenesis to the cell cycle.12 Ribosomal proteins are also important for p53 translation. Haploinsufficiency of a selection of RPs in zebrafish results in peripheral nerve sheath tumors that fail to translate p53 protein.14 During the DNA damage response, RPL26 has been shown to bind the 5′UTR of p53 and promote its translation in human cancer cell lines.15 While it is clear that ribosomal proteins are mis-regulated in many cancers, very little is currently known about what their role may be in tumor progression. RPL38 in mice is required for the translation of a subset of homeobox mRNAs and mutations result in homeotic transformations.3 These data point toward the existence of specialized ribosomes containing specific subsets of ribosomal proteins. These and other studies have shown that altering the levels of specific RPs can affect the translation of subsets of mRNAs demonstrating that the RPs can impart selectivity on the ribosome.2,11
In yeast, the majority of ribosomal proteins have a paralog that arose from a genome duplication event. While almost identical in sequence, these protein paralogs harbor different functions leading to the first suggestion of specialized ribosomes. Studies by Komili et al. showed the specific translation of Ash1 mRNA is dependent on a selection of RP paralogs.1 In Zebrafish RPL22 and its paralog RPL22L1 play distinct, antagonistic roles during hematopoietic development.16 A significant number of mammalian ribosomal protein paralogs have recently been identified though only a few have been studied.4,17-21
Ribosomal proteins have the potential to yield an enormous amount of control on the translation state of individual mRNAs in different tissues. Studies to date investigating the expression profiles of RPs suggest they can exhibit tissue-specific expression at the transcript and protein level.19,22,23 A recent study describes a highly diverse expression pattern among RPs during mouse embryogenesis.3 At the protein level, Sugihara and colleagues similarly detected differential expression of RPs in rodent livers by coupling techniques of 2D gel electrophoresis and mass spectrometry.19 The current notion is that RP genes are subject to tight tissue-specific control and the resulting differential RP expression may contribute to precise adjustment of the ribosome heterogeneity.
In this study, we examined the cell type-specific expression pattern of the human ribosomal protein paralogs. These recently annotated RPs are largely uncharacterized. We identify two RP paralogs, Rpl39l and Rpl10l, which are enriched in mouse embryonic stem cells. We go on to show that Rpl39l, the paralog of Rpl39, is abundantly expressed in cancer cell lines and its expression strongly correlates with vascular invasion in HCC tumor samples. On examination of the expression of the RP paralogs across different tissues, we find that the more recently evolved paralogs show a more tissue-specific expression pattern. We propose these paralogs evolved more recently to allow a greater level of gene expression control that is required by more complex organisms.
Results
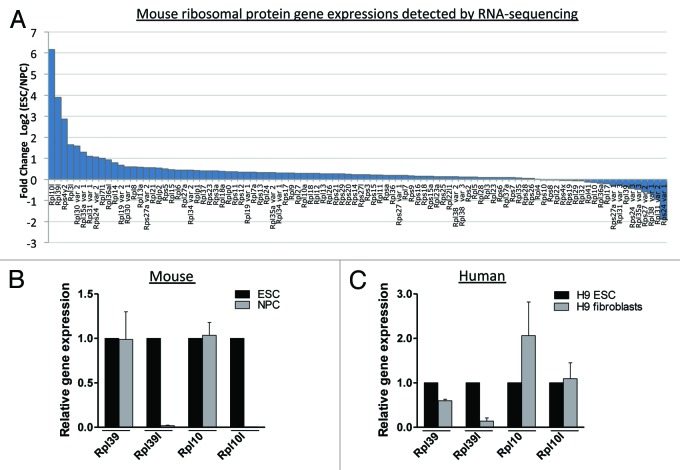
To identify RPs, specifically RP paralogs that are differentially regulated upon mouse Embryonic Stem Cell (ESC) differentiation, we interrogated RNA-seq data from ESCs and ESCs that had been differentiated into Neural Precursor Cells (NPCs) (Wong and Vardy unpublished data) (Table S1). Of the 98 RP mRNAs identified to be expressed, the majority showed no significant change in expression levels on differentiation. We identified 10 RPs with a greater than 2-fold difference in expression levels in NPCs compared with ESCs (Fig. 1A). Two of these, RPL39L and RPL10L, are paralogs of RPs RPL39 and RPL10, respectively. Gene expression changes were confirmed by qRT-PCR in mouse and human ESCs for RPl39l and for RPl10l in mouse only (Fig. 1B and C). Rpl39l and Rpl10l have previously been demonstrated to be expressed at very high levels in the testis and Rpl39l is upregulated in many cancer cell lines.21,24 Rpl39l and its paralog Rpl39 encode proteins that share 92% amino acid sequence similarity. In mammals, RPL39L has a highly conserved Arginine (R) to Glutamine (Q) amino acid change resulting in the loss of a positive charge when compared with RPL39 (Fig. S1). This conserved change suggests functional significance.
Figure 1. Ribosomal protein gene expression in mouse and human Embryonic Stem Cells (ESCs). (A) Relative levels of 98 RP mRNAs detected in mouse ESCs and NPCs by RNA-sequencing. In total, 10 RPs exhibited ≥ 2-fold change in expression on ESC differentiation. The values are presented as fold change in ESCs relative to NPCs. (B) qRT-PCR validation of Rpl39l and Rpl10l in mouse ESCs and cells differentiated to NPCs. (C) qRT-PCR analysis of Rpl39l and Rpl10l in H9 human ESC and its derived fibroblasts.
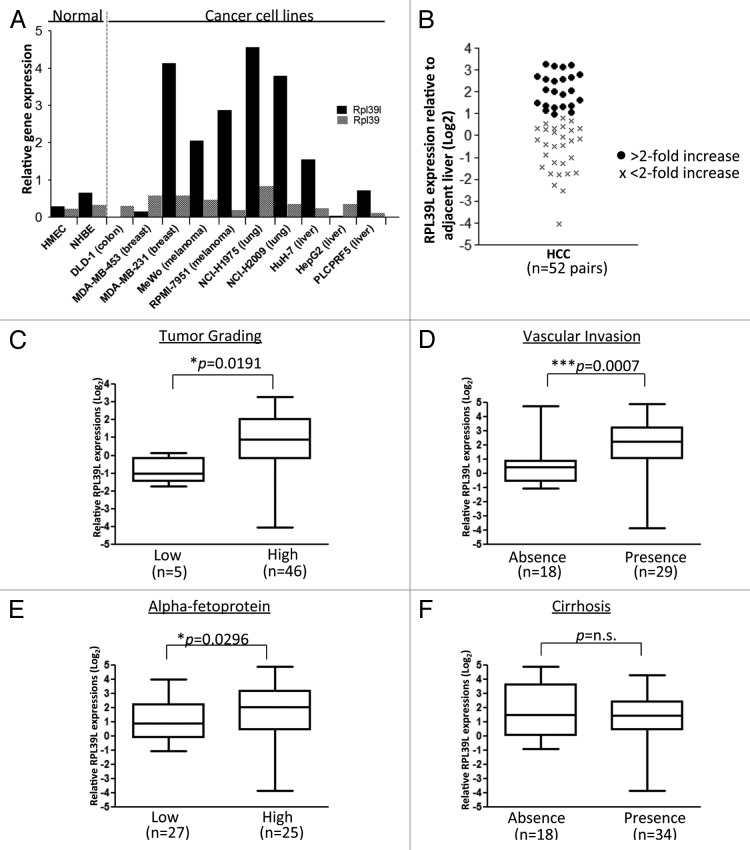
The high levels of RPl39l expression in ESCs and previously reported increased levels in cancer cell lines prompted us to further examine its levels in cancer cells and primary tissue. We examined the expression of Rpl39l in 10 cancer cell lines, including melanoma, lung, and liver cancers by qRT-PCR (Fig. 2A). Rpl39 was expressed at similar levels in both normal epithelial and cancer cell lines while Rpl39l was significantly upregulated in 60% of the cancer cell lines tested compared with normal epithelial cells. In an attempt to evaluate the potential clinical relevance of Rpl39l in cancer development, we extended the Rpl39l expression analysis to a cohort of 52 pairs of primary HCC and corresponding adjacent non-malignant liver tissues (Fig. 2B; Table 1). A significant upregulation of Rpl39l of ≥ 2-fold was observed in HCC tumors compared with non-malignant adjacent liver tissues (P = 0.0016). In total, 23 out of 52 cases (44.23%) displayed an upregulation of Rpl39l. Rpl39l induction in HCC was significantly associated with the presence of vascular invasion (P = 0.0007), high tumor grading (P = 0.0191) as well as high levels of α-fetoprotein (P = 0.0296) in HCC patients (Fig. 2C–E). There was no significant association with increased levels of Rpl39l with liver cirrhosis (Fig. 2F). These data suggest Rpl39l may be specifically upregulated in more advanced HCC tumors giving it diagnostic potential.
Figure 2. High Rpl39l mRNA expressions in human cancer cell lines and HCC tumor samples. (A) Rpl39l and Rpl39 mRNA expression levels were examined in two human epithelial cell lines and 10 cancer cell lines using qRT-PCR analysis. Data shown was normalized to Gapdh. (B) qRT-PCR analysis of Rpl39l in a panel of 52 HCC tumors and adjacent non-tumoral liver tissue. Expression levels were normalized to Gapdh. (C–F) Statistical correlative analyses were conducted between Rpl39l expression and clinico-pathological features of HCC patients. Boxplots show the second and third quartiles, and the whiskers show the maximum and minimum values.
Table 1. Clinico-pathological features of HCC patients.
| Number of HCC patients (n=52) |
||||
|---|---|---|---|---|
| Gender | ||||
| Male | 41 | |||
| Female | 11 | |||
| Median age (y; range) | 59 (49-67.75) | |||
| Underlying liver disease | ||||
| Cirrhosis | 34 | |||
| Chronic hepatitis | 18 | |||
| Tumor gradinga | ||||
| Low | 5 | |||
| High | 46 | |||
| Alpha-fetoproteinb | ||||
| Low | 25 | |||
| High | 27 | |||
| Vascular invasionc | ||||
| Presence | 18 | |||
| Absence | 29 | |||
a One patient case is the fibrolamellar variant HCC. bAn alpha-fetoprotein value of ≥ 400mg/ml detected was considered as high level. cReport for vascular invasion status is not available for five patient cases.
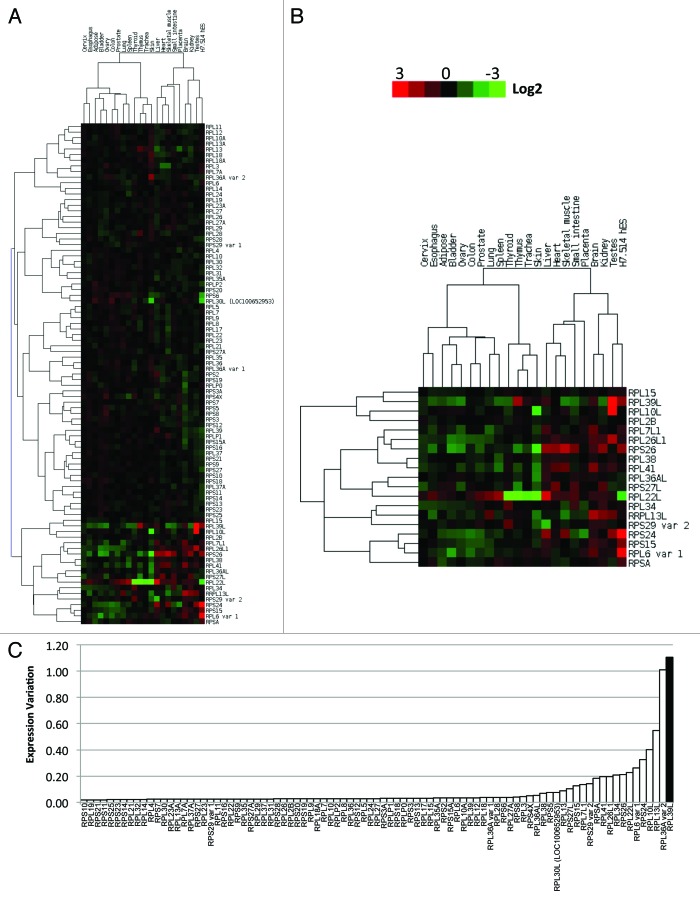
RPL39L is a recently evolved paralog of RPL39 and is only present in mammals. To determine the tissue specificity of all the RPs, we determined their expression levels in a cDNA panel from 22 normal human tissues (Table S2). Our RP targets included RP paralogs Rpl7l1, Rpl26l1, Rpl39l, Rpl36al, Rps27l, Rpl22l, Rpl30l, and Rpl10l, in addition to a select number of splice variants. These proteins have highly similar protein sequences to their original paralog (Figs. S1 and S3). Among the 22 human samples, the most abundant RP mRNA levels were detected in the ovary with a mean expression of 10.07 ± 3.27, whereas only a minimal average level was found in skeletal muscle (5.45 ± 2.60). 98.88% (88/89) of RP mRNAs were highly expressed in the ovary. In order to categorize RP genes based on their expression levels, hierarchical clustering was employed. As shown in the heatmap in Figure 3A, changes in expression levels for most RPs was below 4-fold across different tissues, suggesting that the majority of RP genes are not dramatically regulated between tissues. We identified a subset of 19 RPs that exhibit a distinctively heterogeneous expression pattern across samples (Fig. 3B). These include Rpl15, Rpl39l, Rpl10l, Rpl2b, Rpl7l1, Rpl26l1, Rps26, Rpl38, Rpl41, Rpl36al, Rps27l, Rpl22l, Rpl34, Rrpl13l, Rps29var2, Rps24, Rps15, Rpl6var1, and Rpsa. To determine the variability in expression pattern across different tissues for each RP, we calculated the expression variation for each mRNA. Comparison between the two gene clusters showed that the 19 RP genes have a higher expression variation relative to the remaining RPs (Fig. S2). Rpl39l was found to demonstrate the largest expression variation across tissues compared with other RP mRNAs, whereas Rps10 was the most stably expressed mRNA (Fig. 3C).
Figure 3. qRT-PCR analysis of 89 ribosomal protein mRNAs in 22 human tissues and a cell line. (A) Hierarchical clustering of the qPCR-derived expression levels segregates the RP genes into two clusters, with low or high expression levels across tissues. Red depicts high expression levels, whereas green and black correspond to low and unchanged expression levels, respectively. The color bar on the top shows the gene expression range from +3 to -3 in log2 scale. (B) A distinct cluster of 19 heterogeneously expressed RP genes. (C) Dynamic expressions of the RP genes across 22 human tissues and cell line were calculated in terms of expression variation of RP gene expression. As highlighted by the black bar, Rpl39l exhibits the greatest expression heterogeneity across different tissues and cells.
52.38% (11/21) of the clustered heterogeneously expressed mRNAs are variants or paralogs of other RPs. We asked if there was a relationship between the gene expression variation and evolutionary status of these genes. We determined the level of conservation for all the RP genes and assigned them to different E-stages (Evolution-stage) representing their relative evolutionary status. More recently evolved genes have a higher E-stage. We plotted the gene expression variation against the E-stage for each RP gene (Fig. 4A; Table S3). The majority of RP genes have an expression variation of < 0.3 and reside on the left bottom quadrant of low E-stage. In contrast, the heterogeneously expressed RP genes with a > 0.3 value of expression variation are distributed in the top right quadrant representing a high E-stage (Fig. 4A). Rpl39l, Rpl22l, Rpl10l, Rpl26l1, Rpl7l, Rpl36al, Rps27l, and Rpl30l are all recently evolved genes that show a high level of tissue expression variation (Fig. S2). These recently evolved genes are paralogs or variants of more conserved RPs. As further illustrated by the phylogenetic tree of eight RPs, in terms of protein sequence similarity, the corresponding paralogs are mostly found on the terminal node of branches, implying that these paralogs are more recently evolved (Fig. 4B). On analysis of the conservation of promoter sequences across the RPs and the paralogs, we find that the RP paralogs show a similar level of conservation to the older ribosomal proteins (Fig. S4). Our data suggests the RP paralogs may have evolved a greater level of tissue expression specificity and may play a significant role in gene expression control.
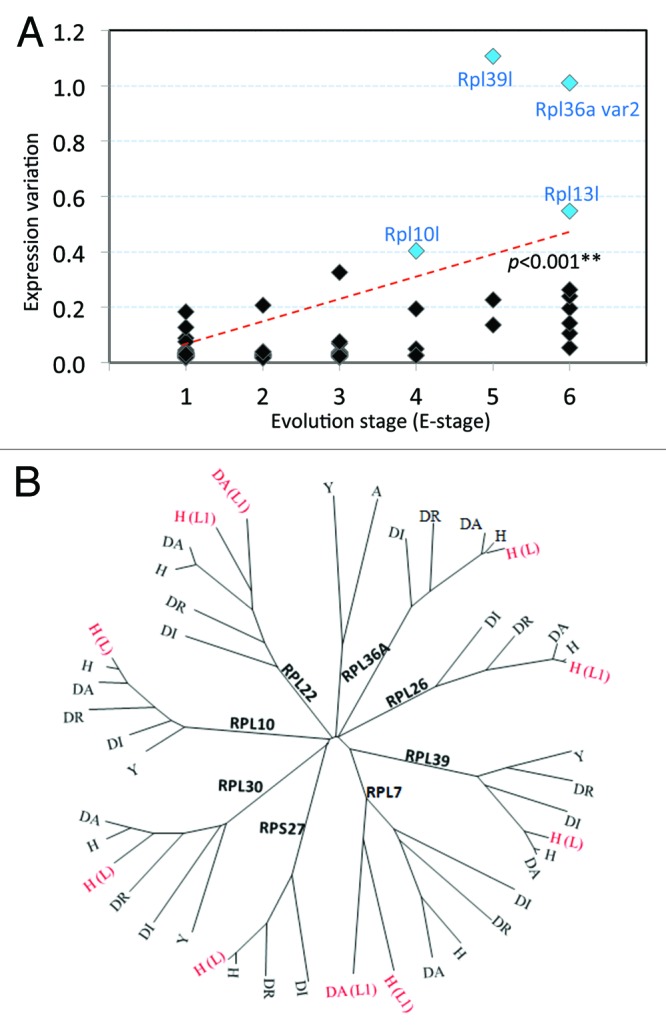
Figure 4. Expression variation and evolutionary stage of 89 RP mRNAs in human tissues. (A) Correlation between gene expression variations of ribosomal proteins in human tissues with their corresponding E-stage. Four RP paralogs with high expression variation were highlighted in blue. The positive correlation of expression variation and E-stage was indicated with a dotted trend line. (B) Phylogenetic tree based on sequence similarity for ribosomal protein orthologs across different species. H, Homo sapiens; A, Arabidopsis thaliana; DR, Drosophila melanogaster; DA, Danio rerio; DI, Dictyostelium discoideum; and Y, yeast species. L or L1 in red indicates a ribosomal protein-like paralog.
Discussion
Here we demonstrate the diverse expression pattern of ribosomal protein mRNAs in different human tissues. We show that the more recently evolved RPs, the majority of which are paralogs, show a much greater level of tissue-specific expression. Specifically, we investigated Rpl39l. Rpl39l was previously described to be highly expressed in the testis and upregulated in a number of cancer cell lines.21,24 We extended this study by showing its expression in ESCs and its strong correlation with vascular invasion and high tumor grading in HCC tumors. Its association with more aggressive HCCs makes it a potential biomarker. Upregulation of Rpl39l in ESCs and cancer cells is of significant interest, yet its function remains unknown. These data highlight the potentially significant role the ribosomal protein paralogs may play in tissue-specific gene expression control and disease progression.
RPL39, the paralog of RPL39L, is present in the ribosome at the polypeptide exit tunnel.25 The exit tunnel is a protein-sensitive channel that is believed to play a role in the regulation of translation through recognition of specific sequences.26 The exit tunnel recognizes nascent proteins and interactions between RPs, including RPL39, and the new polypeptide are believed to influence the function of the translocon.27 The translocon is the site on the ER membrane where membrane proteins are synthesized by ribosomes.28 The conserved Arginine to Glutamine Amino Acid change in RPL39L is significant as it results in the loss of a positive charge in a region that could interact with the exiting polypeptide. RPL39L was shown by proteomic analysis to be incorporated into the large ribosomal subunit in the mouse testis, and antibodies specifically targeting RPL39L demonstrated that it is incorporated into the translating ribosome.19,24 Given the similarity in sequence between the two proteins it is highly likely that RPL39L would be present at the same location as RPL39 within the ribosome. RPs are frequently modified so it is possible that this amino acid change could result in a change in protein modification that then alters its function. As a single ribosome can likely only carry one copy of RPL39/RPL39L, it is likely that within one cell different pools of ribosomes exist carrying RPL39L or RPL39. It is possible that RPL39L is incorporated into a subset of ribosomes and functions to regulate protein production or localization of specific proteins dependent on their N-terminal sequence.
We have knocked down Rpl39l in mouse ESCs and in two human HCC cell lines (HuH-7 and PLCPRF5) and have seen no phenotype over several days. This may be due to the incomplete knockdown of the mRNA and the relative stability of the ribosomal proteins.29 In yeast, Rpl39 is non-essential although strains without Rpl39 have a decrease in the accuracy of translation.30 It is possible that prolonged passaging in the absence of Rpl39l would reveal a phenotype stemming from decreased translational efficiency. Rpl39l expression is correlated with more aggressive, invasive HCC tumors, so it is possible that the role of Rpl39l is linked to the specific tumor environment that cannot be recapitulated in a culture dish.
RP mRNAs are coordinately regulated at the level of translation resulting in altered ribosome biogenesis in response to different environmental and cellular situations.31 Altered RP synthesis could also contribute to cell type ribosome heterogeneity. In addition, the presence of a particular RP does not guarantee incorporation into the ribosome as many RP have non-ribosomal functions. Despite the clear role of translational control in moderating RP protein levels, our data suggests a significant role for transcriptional regulation of RP mRNAs in specific tissues. Similar to previous reports, we see that Rpl22l is highly expressed in the liver, and Rpl10l and Rpl39l are highly expressed in the testis.19 We have shown that there is a selection of ribosomal proteins which show a highly tissue-specific expression pattern compared with the majority or RPs (Fig. 3). These RPs evolved more recently and many of them are paralogs of more conserved RP counterparts. For example RPL39L, RPL10L, and RPL36AL are paralogs that are believed to have arisen through retrotransposition from the X chromosome and may be under the control of retroviral promoters.21 We show that the promoter sequences of the RP paralogs show a similar level of conservation as the original RPs suggesting they are under similar selective pressure. We propose that the RP paralogs evolved to provide a greater level of gene expression control to higher eukaryotes. Based on the similarity of the ribosomal protein paralogs (Fig. S3), it is likely that many of them will bind the same location in the ribosome. If this is the case, it is possible that the more conserved paralog functions as the predominant form in the ribosome and the more recently evolved form can be utilized in specific cell types or cellular conditions.
The function of a number of the ribosomal protein paralogs has been described. The rpl22 paralog rpl22l1 has a distinct and essential role during hematopoiesis in zebrafish.16 Rps27l, not rps27, is a p53 target gene and its overexpression promotes etoposide-induced apoptosis while siRNA knock-down inhibits it.32 A selection of the RP paralogs have been shown to be present on the translating ribosome: RPL22L1, RPL10L, and RPL39L were all identified following 2D gel and proteomics analysis of different human tissues,19 suggesting they play a role in translation. Together, these data suggest that the RP paralogs are functional and have evolved specific roles required by the cells that express them.
The concept of the ribosomal code was originally proposed following the observation of paralog-specific phenotypes in budding yeast. Komili et al. proposed that in yeast there are many different forms of functionally distinct ribosomes and these possess specific functions.1 In plants and yeast a role for paralogous RPs has been demonstrated and interestingly phenotypes arising from loss of the paralogs are often only detectable when cells are exposed to stress. It has become clear that RP paralogs have distinct functions in plants, yeast, fruitfly, and zebrafish.4 Examples of paralog-specific functions can be seen with RPL22 and RPL22L1 in zebrafish. These paralogs are 73% identical and contain identical RNA binding domains. Despite this, they are required for differing and opposing roles during hematopoiesis.16 Despite the clear importance of RP paralogs very little is known about their expression patterns or function in mammals. To our knowledge ours is the first study to quantitatively assess the RNA distribution patterns of all the RPs including the paralogs across different tissues. Given their highly specific expression pattern and the importance of the paralogs in lower eukaryotes, it is highly likely these mammalian RP paralogs will have important roles in specific cellular contexts.
Our data highlight the potentially significant role played by RPL39L in HCC tumor progression and its association with more aggressive HCC tumors makes it a potential biomarker. In addition, we have shown that the more recently evolved RP paralogs show a greater level of tissue-specific expression at the RNA level, suggesting they may have significant roles in tissue-specific gene expression control.
Experimental Procedures
Cell culture and RNA extraction
The 10 cancer and two normal epithelial cell lines used in the study were kind gifts given by Dr Boon Tin Chua from Institute of Molecular and Cell Biology (IMCB). Cell lines were cultured according to protocols as suggested from the American Type Culture Collection (ATCC), the Japanese Collection of Research Bioresources, and Lonza. For mouse embryonic stem cells (mESCs), culturing procedures were followed as described earlier.33 All culture reagents were purchased from either Gibco (Life Technologies) or ATCC. Total RNA samples were extracted using Trizol (Life Technologies) extraction followed by purification using an RNeasy Mini Kit (Qiagen). Total RNA from H7.514 was generously provided by Dr Stuart Avery.
RNA samples of H9 human ESC and their differentiated derivatives were kind gifts given by Dr Jinqiu Zhang and Dr Alan Colman.
HCC patient samples
Tumor and adjacent non-tumor liver tissues were collected from 52 HCC patients from the National University Hospital (NUH) in Singapore as previously described,34 in accordance with ethical guidelines.
Semi-quantitative real-time polymerase chain reaction
First strand cDNA was synthesized using a reverse transcription kit (SuperScript III, Invitrogen) as described previously.33 The cDNA samples of normal human tissue panels (First Choice Human total RNA survey panel from Ambion) were a gift from Dr Bruno Reversade. The HCC tissue cDNA panel was provided with the help of Dr Boon Tin Chua and Dr Ho Han Kiat from IMCB and NUS Singapore, respectively. Reverse transcriptions were performed as detailed.34 QRT-PCR was performed using Power SYBR Green master mix (Applied Biosystems). For the human RP gene panel, pre-designed mRNA-specific primers targeting 80 human RP genes and the housekeeping genes (Gapdh, Tbp, Ppia) were purchased from Integrated DNA Technologies. Primers were designed for nine human RP genes were validated using standard curves: Rpl6 v1 (forward:5′-CCTGTCTGCA AGACTTGAGT TC-3′; reverse: 5′-GCCTCTCAGT TAGCCTTGGA-3′), Rpl2b (forward:5′-GGTGTCCACA GCCACATAAA -3′; reverse: 5′-GTGGTCAGCG GAAACTTGAT-3′), Rrpl13l (forward: 5′- CATGATCCTG AAGCCCAACT -3′; reverse: 5′- GGCACCTTAC TATGGGTGGA -3′), Rpl22l1 (forward: 5′- CTTCGAGTGG TTGCATCTGA -3′; reverse: 5′- TGCCTAGTCC TCCGACTCTG -3′), Rplp0 (forward: 5′- TCGACAATGG CAGCATCTAC -3′; reverse: 5′- ATCCGTCTCC ACAGACAAGG -3′), Rps6 (forward: 5′- TGCTCTGAAG AAGCAGCGTA -3′; reverse: 5′- AGTCTGCGTC TCTTCGCAAT -3′), Rps26 (forward: 5′- CCGTCTCCTA AGGATTCTCC -3′; reverse: 5′- GCACAGTTAG TGCAGCGAAT -3′), Rpl10l (forward: 5′- AAGCTTCAGA ACGAGGAGCA -3′; reverse: 5′- CTTCTTGGCC ACCATGTCTT -3′), Rpl30l (Loc100652953) (forward: 5′-CGGCTCCAAC TCATGAAAAG -3′; reverse: 5′- AAAGCTGGGC AGTTGTAAGC -3′). For expression analysis of mouse mRNAs, specific primers were designed with sequences shown below: Rpl39l (forward: 5′-gcaaaatcgtcccattccaca -3′; reverse: 5′- caatttggttcgtctccaatgtc-3′), Rpl39 (forward: 5′-CCTATTCCTC AGTGGATCCG -3′; reverse: 5′-CTCAGTCTTG CCATTGTGTG -3′), and Gapdh (forward: 5′- CAATGTGTCC GTCGTGGAT-3′; reverse: 5′-CCTGCTTCAC CACCTTCTT -3′). qPCR was performed on an ABI PRISM 7900 sequence detection system. Gapdh was used as the normalization control. Any normalized gene expressions with a ≥ 2-fold change relative to the average mean of all were considered differentially expressed.
Data processing, hierarchical clustering, and heatmap presentation
Raw Ct values were converted into expression levels with Ct = 31 as the cutoff. Normalization of expression levels across all samples was performed as follows. Three housekeeping mRNAs (Gapdh, Tbp, Ppia) were used for normalization, and the pairwise normalization factor was determined by the linear regression of the expression levels of the three genes. Gene expression variation of a ribosomal protein was defined as the standard deviation of expression level divided by the mean expression across different tissues. Two-way hierarchical clustering with average linkage was performed. Prior to clustering, the normalized expression data was log2 transformed and then subtracted by the mean of the log2 transformed expression data across all samples for each RP gene. Subsequently, the heatmap was generated from the clustering results. The heatmap presents the log2 expression levels adjusted to the mean value of zero for each RP gene.
Protein sequence alignment
All ribosomal protein sequences were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/protein), Ensembl (http://www.ensembl.org/index.html), and UCSC genome browser (http://genome.ucsc.edu/). Sequence alignment was conducted using the online tool of ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Orthologs and evolutionary stage
Orthologs of a ribosomal protein across different species were identified based on their annotations. The evolutionary stage (E-stages of 1, 2, 3, 4, 5, 6) of a ribosomal protein was defined based on the first appearance of its orthologs in species including (1) achaea, (2) bacteria, (3) protoza or plant or fungi, (4) invertebrates, (5) fish or birds, and (6) mammals, respectively. Evolution distance for a human ribosomal protein was defined as the average distance obtained from the multiple sequence alignment for all orthologs of the ribosomal protein across different species.
Phylogenetic tree of ribosomal proteins and promoter conservation analysis
Clustering of orthologs of different ribosomal proteins across various species was performed based on multiple alignment of their protein sequences. Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) was used to perform the multiple sequence alignment and subsequently to build the corresponding phylogenetic tree. Similarity analysis of the RP promoters was done using the multiple alignment of 1KB of promoter sequence across species using Clustal Omega. The average distance was used as a measure of similarity. The greater the distance the lower the similarity.
Statistical analysis
Expression of Rpl39l in HCC tumors and paired adjacent non-tumor liver samples were compared using the paired Student’s t test. The Mann-Whitney test was used to examine the Rpl39l transcript expression with clinic-pathological features, including tumor grading, cirrhosis, and vascular invasion. The unpaired Student’s t test was used to evaluate the association between Rpl39l level and the presence of α-fetoprotein. The Pearson correlation was used to assess the relationship between the E-stage and expression variation of ribosomal proteins. Student’s t-test was employed to detect the association between expression variation and E-stage of the ribosomal proteins. P < 0.05 was considered statistically significant.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Drs Boon Tin Chua and Ho Han Kiat for making the tumor samples available to us. Drs Lim Chin Yan and Dr Mathijs Voorhoeve for helpful discussion on the work and for experimental assistance. We thank Drs Stuart Avery and Zhang Jinqiu for providing human ESC RNA samples. This work was supported by the Agency for Science Technology and Research (A*STAR), Singapore.
Glossary
Abbreviations:
- ESC
embryonic stem cell
- NPC
neural precursor cell
- RP
ribosomal protein
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/27427
References
- 1.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–71. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert WV. Functional specialization of ribosomes? Trends Biochem Sci. 2011;36:127–32. doi: 10.1016/j.tibs.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–97. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13:355–69. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–9. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 6.Klinge S, Voigts-Hoffmann F, Leibundgut M, Ban N. Atomic structures of the eukaryotic ribosome. Trends Biochem Sci. 2012;37:189–98. doi: 10.1016/j.tibs.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marygold SJ, Roote J, Reuter G, Lambertsson A, Ashburner M, Millburn GH, Harrison PM, Yu Z, Kenmochi N, Kaufman TC, et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007;8:R216. doi: 10.1186/gb-2007-8-10-r216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teng T, Thomas G, Mercer CA. Growth control and ribosomopathies. Curr Opin Genet Dev. 2013;23:63–71. doi: 10.1016/j.gde.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Boria I, Garelli E, Gazda HT, Aspesi A, Quarello P, Pavesi E, Ferrante D, Meerpohl JJ, Kartal M, Da Costa L, et al. The ribosomal basis of Diamond-Blackfan Anemia: mutation and database update. Hum Mutat. 2010;31:1269–79. doi: 10.1002/humu.21383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horos R, Ijspeert H, Pospisilova D, Sendtner R, Andrieu-Soler C, Taskesen E, Nieradka A, Cmejla R, Sendtner M, Touw IP, et al. Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood. 2012;119:262–72. doi: 10.1182/blood-2011-06-358200. [DOI] [PubMed] [Google Scholar]
- 12.Shenoy N, Kessel R, Bhagat TD, Bhattacharyya S, Yu Y, McMahon C, Verma A. Alterations in the ribosomal machinery in cancer and hematologic disorders. J Hematol Oncol. 2012;5:32. doi: 10.1186/1756-8722-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bee A, Brewer D, Beesley C, Dodson A, Forootan S, Dickinson T, Gerard P, Lane B, Yao S, Cooper CS, et al. siRNA knockdown of ribosomal protein gene RPL19 abrogates the aggressive phenotype of human prostate cancer. PLoS One. 2011;6:e22672. doi: 10.1371/journal.pone.0022672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacInnes AW, Amsterdam A, Whittaker CA, Hopkins N, Lees JA. Loss of p53 synthesis in zebrafish tumors with ribosomal protein gene mutations. Proc Natl Acad Sci U S A. 2008;105:10408–13. doi: 10.1073/pnas.0805036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Duc AC, Rao S, Sun XL, Bilbee AN, Rhodes M, Li Q, Kappes DJ, Rhodes J, Wiest DL. Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. Dev Cell. 2013;24:411–25. doi: 10.1016/j.devcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopes AM, Miguel RN, Sargent CA, Ellis PJ, Amorim A, Affara NA. The human RPS4 paralogue on Yq11.223 encodes a structurally conserved ribosomal protein and is preferentially expressed during spermatogenesis. BMC Mol Biol. 2010;11:33. doi: 10.1186/1471-2199-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugihara Y, Sadohara E, Yonezawa K, Kugo M, Oshima K, Matsuda T, Nadano D. Identification and expression of an autosomal paralogue of ribosomal protein S4, X-linked, in mice: potential involvement of testis-specific ribosomal proteins in translation and spermatogenesis. Gene. 2013;521:91–9. doi: 10.1016/j.gene.2013.02.040. [DOI] [PubMed] [Google Scholar]
- 19.Sugihara Y, Honda H, Iida T, Morinaga T, Hino S, Okajima T, Matsuda T, Nadano D. Proteomic analysis of rodent ribosomes revealed heterogeneity including ribosomal proteins L10-like, L22-like 1, and L39-like. J Proteome Res. 2010;9:1351–66. doi: 10.1021/pr9008964. [DOI] [PubMed] [Google Scholar]
- 20.Yoshihama M, Uechi T, Asakawa S, Kawasaki K, Kato S, Higa S, Maeda N, Minoshima S, Tanaka T, Shimizu N, et al. The human ribosomal protein genes: sequencing and comparative analysis of 73 genes. Genome Res. 2002;12:379–90. doi: 10.1101/gr.214202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uechi T, Maeda N, Tanaka T, Kenmochi N. Functional second genes generated by retrotransposition of the X-linked ribosomal protein genes. Nucleic Acids Res. 2002;30:5369–75. doi: 10.1093/nar/gkf696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bortoluzzi S, d’Alessi F, Romualdi C, Danieli GA. Differential expression of genes coding for ribosomal proteins in different human tissues. Bioinformatics. 2001;17:1152–7. doi: 10.1093/bioinformatics/17.12.1152. [DOI] [PubMed] [Google Scholar]
- 23.Ishii K, Washio T, Uechi T, Yoshihama M, Kenmochi N, Tomita M. Characteristics and clustering of human ribosomal protein genes. BMC Genomics. 2006;7:37. doi: 10.1186/1471-2164-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadano D, Notsu T, Matsuda T, Sato T. A human gene encoding a protein homologous to ribosomal protein L39 is normally expressed in the testis and derepressed in multiple cancer cells. Biochim Biophys Acta. 2002;1577:430–6. doi: 10.1016/S0167-4781(02)00445-1. [DOI] [PubMed] [Google Scholar]
- 25.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–9. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 26.Petrone PM, Snow CD, Lucent D, Pande VS. Side-chain recognition and gating in the ribosome exit tunnel. Proc Natl Acad Sci U S A. 2008;105:16549–54. doi: 10.1073/pnas.0801795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–36. doi: 10.1016/S0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- 28.Schnell DJ, Hebert DN. Protein translocons: multifunctional mediators of protein translocation across membranes. Cell. 2003;112:491–505. doi: 10.1016/S0092-8674(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 29.Hirsch CA, Hiatt HH. Turnover of liver ribosomes in fed and in fasted rats. J Biol Chem. 1966;241:5936–40. [PubMed] [Google Scholar]
- 30.Dresios J, Derkatch IL, Liebman SW, Synetos D. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry. 2000;39:7236–44. doi: 10.1021/bi9925266. [DOI] [PubMed] [Google Scholar]
- 31.Caldarola S, De Stefano MC, Amaldi F, Loreni F. Synthesis and function of ribosomal proteins--fading models and new perspectives. FEBS J. 2009;276:3199–210. doi: 10.1111/j.1742-4658.2009.07036.x. [DOI] [PubMed] [Google Scholar]
- 32.He H, Sun Y. Ribosomal protein S27L is a direct p53 target that regulates apoptosis. Oncogene. 2007;26:2707–16. doi: 10.1038/sj.onc.1210073. [DOI] [PubMed] [Google Scholar]
- 33.Zhang D, Zhao T, Ang HS, Chong P, Saiki R, Igarashi K, Yang H, Vardy LA. AMD1 is essential for ESC self-renewal and is translationally down-regulated on differentiation to neural precursor cells. Genes Dev. 2012;26:461–73. doi: 10.1101/gad.182998.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho HK, Pok S, Streit S, Ruhe JE, Hart S, Lim KS, Loo HL, Aung MO, Lim SG, Ullrich A. Fibroblast growth factor receptor 4 regulates proliferation, anti-apoptosis and alpha-fetoprotein secretion during hepatocellular carcinoma progression and represents a potential target for therapeutic intervention. J Hepatol. 2009;50:118–27. doi: 10.1016/j.jhep.2008.08.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.