Abstract
Interleukin-6 (IL-6) is a multifunctional cytokine that is involved in the progression of various inflammatory diseases, such as rheumatoid arthritis and certain cancers; for example, multiple myeloma or hepatocellular carcinoma. To interfere with IL-6-dependent diseases, targeting IL-6 receptor (IL-6R)-presenting tumor cells using aptamers might be a valuable strategy to broaden established IL-6- or IL-6R-directed treatment regimens.
Recently, we reported on the in vitro selection of RNA aptamers binding to the human IL-6 receptor (IL-6R) with nanomolar affinity. One aptamer, namely AIR-3A, was 19 nt in size and able to deliver bulky cargos into IL-6R-presenting cells. As AIR-3A is a natural RNA molecule, its use for in vivo applications might be limited due to its susceptibility to ubiquitous ribonucleases.
Aiming at more robust RNA aptamers targeting IL-6R, we now report on the generation of stabilized RNA aptamers for potential in vivo applications. The new 2'-F-modified RNA aptamers bind to IL-6R via its extracellular portion with low nanomolar affinity comparable to the previously identified unmodified counterpart. Aptamers do not interfere with the IL-6 receptor complex formation. The work described here represents one further step to potentially apply stabilized IL-6R-binding RNA aptamers in IL-6R-connected diseases, like multiple myeloma and hepatocellular carcinoma.
Keywords: SELEX, RNA, Aptamers, Interleukin-6 receptor, Hyper-IL-6, RNA-protein interaction, post-selective modification
Introduction
Aptamers are short (15–100 nt) nucleic acid molecules (DNA or RNA) that are able to interact with target molecules with high affinity and specificity due to defined three-dimensional structures. Aptamers can be enriched by an in vitro selection process, named SELEX (Systematic Evolution of Ligands by EXponential enrichment).1,2 Up to now, aptamers have been evolved for a wide variety of molecules, including therapeutically relevant proteins,3 viruses,4 and even whole cells.5 Dependent on their function, aptamers seem to be suited for therapeutic applications.
Next to their stimulating6 or inhibiting7 properties, aptamers might serve as cell-specific delivery vehicles for drug molecules into target cells or tissues.8 One promising example is an RNA aptamer binding to PSMA (prostate-specific membrane antigen) presented on the surface of prostate cancer cells.9 Upon binding, PSMA aptamers undergo receptor-mediated internalization. Coupling those nucleic acids to cargo molecules, such as toxins,10 siRNAs,11 and nanoparticles,12 improves the cargo delivery into the cells of interest.8 This allows lowering the dose required for effective treatment. Therefore, aptamers are considered to be promising therapeutic approaches, which had already been validated in animal studies.13 Despite aptamers being very promising tools for cell-specific targeting and drug delivery, there are only few appropriate representatives to date.8,14 Therefore, new aptamers specifically binding to cell surface proteins or whole cells are urgently needed. To increase the toolbox of cell-specific drug delivery molecules, we are interested in the development of RNA aptamers binding the human Interleukin-6 receptor (IL-6R).
Interleukin-6 (IL-6) is a multifunctional cytokine that is involved in many immune and inflammatory responses. It belongs to the family of the four-helix bundle cytokines.15,16 On target cells, IL-6 interacts with its receptor, namely IL-6R, and two molecules of glycoprotein 130 (gp130), the transducer of the IL-6 signal. This results in Janus kinase (JAK) activation and phosphorylation of STAT (signal transducer and activator of transcription) proteins, particularly STAT3. The dimerization and translocation of STAT3 into the nucleus result in the activation of various genes, including genes encoding cell survival and cell regulating proteins, but also mediators of angiogenesis and metastasis as well as oncogenes.15
IL-6 and its receptor are involved in the progression of various inflammatory diseases, such as Crohn's disease and rheumatoid arthritis (RA), and certain cancers, such as multiple myeloma or hepatocellular carcinoma (HCC).17 HCC is one of the most common cancers worldwide. The liver is a site of metastasis for several malignancies, e.g., colorectal cancer. Metastatic cells entering the liver can trigger an inflammatory response that entails the release of several cytokines, including TNF-α, a pro-apoptotic factor. However, it has been shown that highly metastatic lung and colon cancer cells can be rescued from apoptosis in the presence of high TNF-α concentrations in vitro.18 The reason is a marked increase in IL-6 production leading to an autocrine gp130- and IL-6R-dependent activation of STAT3, thereby inhibiting caspase-3-mediated apoptosis and promoting tumor cell survival.19 Additionally, IL-6 serves as a paracrine survival factor in the liver, acting on both primary hepatocellular carcinoma and metastases.18 To interfere with IL-6-dependent diseases, such as liver metastases, specific targeting of the IL-6R axis seems to be one promising therapeutic strategy.
Recently, we reported the in vitro selection and characterization of RNA aptamers with high affinity (Kd = 20 nM) for the human IL-6 receptor (IL-6R).20 Due to optimization, we obtained an only 19-nt short RNA aptamer, named AIR-3A, which adopted a parallel G-quadruplex structure and retained all necessary characteristics for high affinity and selective recognition of recombinantly produced IL-6R and as well as the native IL-6R presented on the cell surfaces. Upon binding to IL-6R-presenting cells, this aptamer was rapidly internalized and could be used to deliver bulky cargos into IL-6R-presenting cells.21 Seeing that IL-6 takes an active part in inflammation associated cancers,22,23 selective targeting of IL-6R-presenting tumor cells using drug loaded aptamers might be a valuable strategy to broaden established IL-6 or IL-6R directed treatment regimens.
AIR-3A is composed of naturally occurring nucleotides and adopts a G-quadruplex structure. Advantageously, G-quadruplex structures can exhibit enhanced stabilities against cellular or serum nucleases if compared with unstructured nucleic acid molecules.24 However, for future in vivo applications, additional modifications of the aptamer might be a prerequisite to further improve its affinity, activity, stability, and shelf life in biological environments.
Here we report about the development of stabilized IL-6R binding RNA aptamers. Trials to post-selectively stabilize AIR-3A by incorporation of 2’-F-substituted pyrimidine residues failed. We decided to perform a new in vitro selection experiment aiming at new IL-6R-binding 2’-Fluoro-modified (2'-F-modified) RNA aptamers using a 2’-Fluoro-modified RNA library and Hyper-IL-6—a fusion protein of sIL-6R and IL-6—as target molecule.25 This strategy brought us to stabilized IL-6R-binding RNA aptamers with binding affinities and sequence composition comparable to AIR-3A. We identified FAIR-6, a 2’-F-modified RNA aptamer also comprising a G-quadruplex structure, and thus, containing the same functional motif as AIR-3A. These stabilized RNA aptamers might serve as potential drug delivery vehicles applied in IL-6R-related diseases.
Results
Recently, we identified IL-6R-binding RNA aptamers containing the conserved G-rich motif 5′-GGGGHGGCWG UGGWGWGGG-3′, whereas H encodes A, C, or U and W encodes A or U, respectively.20 It had already been shown that the 19-nt short aptamer AIR-3A (5′-GGGGAGGCUG UGGUGAGGG-3′) was able to bind Hyper-IL-6 with high affinity (Kd ~20 nM) and to bind the human IL-6R presented on target cells. CD spectroscopic investigations revealed that AIR-3A adopts a G-quadruplex structure.
Mutational analyses
Trying to increase the affinity of AIR-3A for its target and to obtain an idea about which nucleotides of AIR-3A are essential for protein binding or structural stability, we decided to generate a series of AIR-3A descendent single nucleotide variants (Table 1). Thereby, all purine residues (A and G) were replaced by U and all pyrimidine residues (U and C) were replaced by A, respectively.
Table 1. Interaction of RNA aptamer AIR-3A and single nucleotide variants with Hyper-IL-6.
| RNA | Sequence | Kd value [nM] | RNA | Sequence | Kd value [nM] |
|---|---|---|---|---|---|
| AIR-3A | GGGGAGGCUG UGGUGAGGG | 23.1 ± 4.0 | U11A | GGGGAGGCUG AGGUGAGGG | 130.9 ± 47.4 |
| G1U | UGGGAGGCUG UGGUGAGGG | 29.6 ± 4.6 | G12U | GGGGAGGCUG UUGUGAGGG | n. d. |
| G2U | GUGGAGGCUG UGGUGAGGG | n. d. | G13U | GGGGAGGCUG UGUUGAGGG | n. d. |
| G3U | GGUGAGGCUG UGGUGAGGG | 109.0 ± 43.0 | U14A | GGGGAGGCUG UGGAGAGGG | 54.4 ± 11.3 |
| G4U | GGGUAGGCUG UGGUGAGGG | n. d. | G15U | GGGGAGGCUG UGGUUAGGG | n. d. |
| A5U | GGGGUGGCUG UGGUGAGGG | 101.1 ± 76.6 | A16U | GGGGAGGCUG UGGUGUGGG | 86.9 ± 19.5 |
| G6U | GGGGAUGCUG UGGUGAGGG | n. d. | G17U | GGGGAGGCUG UGGUGAUGG | n. d. |
| G7U | GGGGAGUCUG UGGUGAGGG | n. d. | G18U | GGGGAGGCUG UGGUGAGUG | n. d. |
| C8A | GGGGAGGAUG UGGUGAGGG | 24.1 ± 12.8 | G19U | GGGGAGGCUG UGGUGAGGU | n. d. |
| U9A | GGGGAGGCAG UGGUGAGGG | 42.7 ± 13.7 | –G19 | GGGGAGGCUG UGGUGAGG | n. d. |
| G10U | GGGGAGGCUU UGGUGAGGG | n. d. |
n.d. none determinable.
The dependence of the aptamer affinity on each single nucleotide exchange was analyzed by filter retention assays (FRA). The results are summarized in Figure 1 and Table 1. AIR-3A revealed the highest affinity for Hyper-IL-6 (Kd = 23.1 ± 4.0 nM).
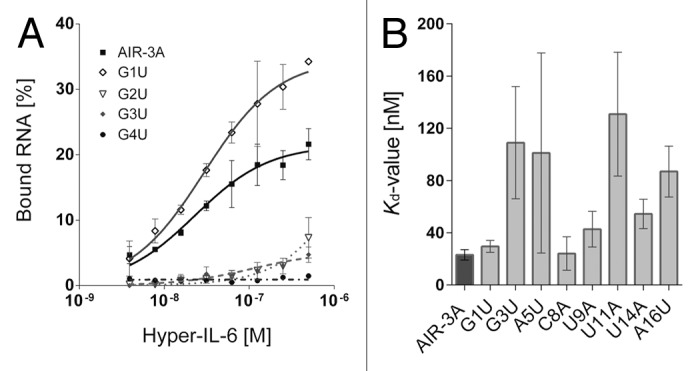
Figure 1. Filter retention analyses of the Hyper-IL-6 binding RNA aptamer AIR-3A and its single nucleotide variants G1U, G2U, G3U, and G4U. (A) Constant amounts (< 1 nM) of individual radioactively labeled RNAs were incubated with increasing amounts of Hyper-IL-6 protein (0–500 nM). Filter-bound RNAs were detected by autoradiography and fractions of Hyper-IL-6-bound RNAs plotted against the concentration of Hyper-IL-6 (logarithmic scale). Data points represent mean values of at least two independent measurements. (B) Kd values of AIR-3A and single nucleotide variants that bind Hyper-IL-6. Remaining variants lost their activity.
The binding affinity of almost all variants comprising a single nucleotide replacement of G to U was impaired. However, the variant G1U depicted an exception of this observation since it still revealed a high affinity for Hyper-IL-6 (Kd = 29.6 ± 4.6 nM). The variant G3U was characterized by a strongly diminished functionality (Kd = 109.0 ± 43.0 nM). Each variant in which an A, C, or U nucleotide was replaced (A5U, C8A, U9A, U11A, U14A, and A16U) maintained its functionality even though a decreased affinity for Hyper-IL-6 was detected (Fig. 1B; Table 1).
Structural analyses of AIR-3A variants
Recently performed CD spectroscopic investigations of AIR-3A confirmed that the aptamer adopts a G-quadruplex structure.20 To answer the question if structural distortion is the reason why most variants lost their affinity for Hyper-IL-6, we performed UV melting transitions at 295 nm. Therefore, RNAs (5 µM) were dissolved in 10 mM Tris buffer (pH 7.5), including increasing amounts of monovalent potassium ions (0 mM, 5 mM, and 100 mM KCl). Figure 2 summarizes the Tm-values of AIR-3A and selected nucleic acid variants at a KCl concentration of 5 mM.
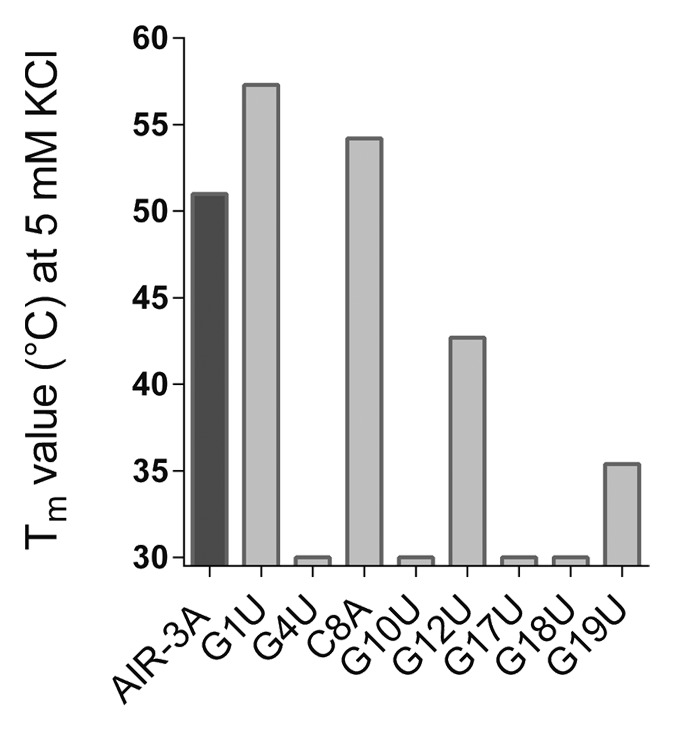
Figure 2. Tm values of AIR-3A and selected variants in presence of 5 mM KCl. It is shown that the structural stability of most G nucleotide variants was clearly reduced if compared with AIR-3A. Except for G1U, for which no structural distortion could be detected. The same could be shown for the C single nucleotide variant C8A.
We confirmed that the UV melting profile of AIR-3A was characterized by a hypochromic shift as typically observed for nucleic acids containing a G-quadruplex structure and we obtained a Tm-value of about 51 °C in the presence of 5 mM KCl. Furthermore, we evaluated the stability of the AIR-3A G-quadruplex by determining its melting temperature at different potassium concentrations (0 mM, and 100 mM, respectively). The melting temperature increased with increasing amounts of KCl (Table S1).
The structural stability of most G nucleotide variants was clearly reduced when compared with AIR-3A. The variant G1U again depicted an exception as it showed a similar UV-melting behavior to AIR-3A and was therefore assumed to adopt a stable G-quadruplex structure at 5 mM KCl. For the selected C and U single nucleotide variants (C8A, U11A, and U14A) no structural distortion in presence of 5 mM or 100 mM KCl could be detected (Fig. 2; Table S1).
Taken into account the results of the FRAs and UV melting transitions, it might only be possible to post-selectively modify AIR-3A at position G1 and/or at non-G nucleotides, respectively, if the base moiety of the nucleotide is considered and not the sugar-phosphate backbone.
Post-selective 2’-Fluoro-modified AIR-3A
To post-selectively increase the stability of AIR-3A, we directly incorporated 2’-F- pyrimidines during in vitro transcription using the T7 RNA polymerase variant Y637F.26 Subsequently, we modified the RNAs by incorporation of only 2’-F-CTP or 2’-F-UTP, followed by a combination of both 2'-F pyrimidine residues. FRAs revealed that all trials to post-selectively stabilize AIR-3A caused a loss of function (Fig. S1).
In vitro selection of 2’-F-modified RNA aptamers specific for Hyper-IL-6
Due to the failure of the successful post-selective stabilization of AIR-3A, we decided to select new 2'-F-modified RNA aptamers that target IL-6R. We performed a new SELEX experiment using Hyper-IL-6 as target molecule. The used RNA library B50 contained 50 nucleotides in random and 2'-F-modified pyrimidine residues for an increased stability against nucleases. 2'-F-modified pyrimidine residues could be incorporated using the T7 RNA polymerase variant Y637F during in vitro transcription.26 After 15 rounds, we cloned the dsDNA library into the TOPO TA vector and sequenced 12 clones (Table 2). We obtained seven different sequence variants that did not show any conserved binding motif. All seven variants were subjected to filter retention analyses (FRAs).
Table 2. 2’-F-modified RNA aptamers binding to Hyper-IL-6.
| Aptamer | Sequence | Frequency | Kd value [nM] |
|---|---|---|---|
| FAIR-1 | AGGAUGGCAA UCAGUGAACG GAAGGUGUAG GGUUAGAGGU GUGGUGGGUA | 1 | 179.1 ± 37.4 |
| FAIR-4 | CUCGAGAGGC GUUGAUCGGA AGGCGGUAGG UGUUAUGGGU GGGAGGGAGC | 2 | 120.0 ± 36.4 |
| FAIR-6 | GUAAGUAGUG UAGGCUGUGG GAGUUAUAGGGGUGGAUGUGGAGUGGGGUG | 4 | 40.9 ± 12.7 |
| FAIR-7 | UGUCAGUAGC ACCAGUGGCG GACAGUAGGG CAGGGUGGAG UGGGUGUCCC | 1 | 79.7 ± 33.2 |
| FAIR-8 | CGAGGCGCCG UGGAACAAGU AGGCUGUUAG GGAAGGGUGG AGCGGGUAGC | 2 | 196.7 ± 27.9 |
| FAIR-14 | GAAGGCAGUG UGUAGUGCGG AGGUAGUUGA GUGGUGGGAG GGUGGAGGUA | 1 | 118.9 ± 63.6 |
| FAIR-15 | GGGAUGACGG UAGGGUAGGG GGGUGGGGGU CAUCAAUUGG GAGGUAG | 1 | 162.9 ± 69.0 |
Sequences are printed in 5′–3′ direction omitting flanking primer-binding sites. Bold letters highlight the conserved G-rich motif 5′-GGGGHGGCWG UGGWGWGGG-3′, which had already been identified in unmodified sIL-6R-binding RNA aptamers.20
The identified 2'-F-modified RNA candidates were screened for Hyper-IL-6 binding in FRAs. We found that all sequenced aptamer candidates were able to bind Hyper-IL-6. Aptamer FAIR-6 revealed the highest affinity for Hyper-IL-6 demonstrated by a Kd-value of 40.9 ± 12.7 nM (Fig. 3; Table 2). The initial RNA library B50 served as a control and did not reveal any unspecific protein binding. Further FRAs revealed that FAIR-6 did neither bind to gp130 nor to IL-6 (data not shown). This indicates that FAIR-6 probably binds within the sIL-6R moiety of Hyper-IL-6 as the latter is a designer cytokine comprising a recombinant fusion of the two proteins sIL-6R and IL-6.25 We performed CD spectroscopy to get information about the structure of FAIR-6. We could observe a negative minimum at 240 nm and a positive maximum at 264 nm, which hints to the formation of a parallel G-quadruplex similar to AIR-3A (Fig. S2).20,27 Additionally, we determined the Tm-value of FAIR-6 by preforming UV-melting analyses at 295 nm as described above. The UV melting profile of FAIR-6 was characterized by a hypochromic shift typically observed for nucleic acids containing a G-quadruplex structure. In the presence of 5 mM KCl we obtained a Tm-value of 54 °C (Table S1).
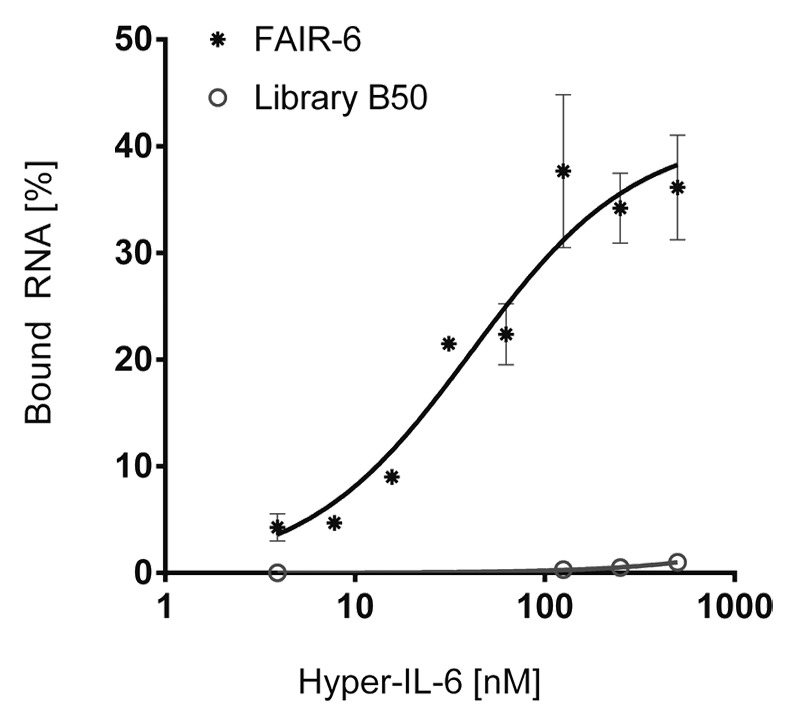
Figure 3. Aptamer FAIR-6 binds Hyper-IL-6 with high affinity. Filter retention assays; constant amounts (< 1 nM) of 32P-radioactively labeled aptamer FAIR-6 (dots) and RNA starting library B50 (open circles) were incubated with increasing amounts of Hyper-IL-6 (0–500 nM). Protein-bound RNA was visualized by autoradiography. Fractions of bound RNA molecules were plotted against the concentration of Hyper-IL-6. Data points represent mean values of two independent measurements.
Stability of FAIR-6 in cell culture medium
Since FAIR-6 turned out to be the most competent 2'-F-modified IL-6R-binding aptamer, it was further analyzed with respect to its stability in DMEM (cell culture medium) and DMEM containing 10% (FCS) at 37 °C. To compare the stability of FAIR-6 with an unmodified counterpart, we used the natural RNA aptamer AIR-3, which is the originally selected longer form of AIR-3A containing 106 nt.20 Incubation of both, the 2’-F-modified RNA aptamer FAIR-6 and the natural RNA aptamer AIR-3, for up to 4 h in cell culture medium did not disclose any significant differences in stability. When incubated in FCS-containing DMEM, however, we observed differences in the aptamers degradation (Fig. S3). In medium containing FCS, AIR-3 was undetectable after seconds, whereas FAIR-6 remained unaffected, indicating a protective effect of the 2'-F-modification.
Influence of FAIR-6 on the interaction between Hyper-IL-6 and gp130
On the basis of sequence comparisons, the isolated FAIR-6 displayed a high degree of similarity to the unmodified AIR-3A, including the same conserved motif (Table 2). Due to the discovery of this sequence element within FAIR-6 (96 nucleotides in length), we hypothesized that FAIR-6 and AIR-3A share the same binding site at Hyper-IL-6. We analyzed the interaction between FAIR-6 and Hyper-IL-6 in the absence or presence of surplus unlabeled AIR-3A as competitor (Fig. S4). We could demonstrate that AIR-3A indeed displaced FAIR-6 binding to Hyper-IL-6 confirming one common or at least similar Hyper-IL-6 binding site.
The ability of FAIR-6 and two other aptamers, FAIR-14 and FAIR-15, to prevent the interaction between Hyper-IL-6 and gp130 was analyzed by electrophoretic mobility shift assays (EMSA). As expected, FAIR-6 did bind to Hyper-IL-6 as demonstrated by a slowed migration rate in the native PAGE when compared with FAIR-6 only (Fig. 4, lanes 1 and 2). Additionally, a stronger shift confirmed the interaction between FAIR-6 and Hyper-IL-6 in complex with gp130 (Fig. 4, lane 3). This indicates that complex formation between Hyper-IL-6 and gp130 was not disrupted by FAIR-6. No unspecific binding of FAIR-6 to gp130 was observed (Fig. 4, lane 4). Additionally, none of the other two aptamers tested (FAIR-14 and FAIR-15) was found to inhibit the interaction between Hyper-IL-6 and gp130 (Fig. S4).
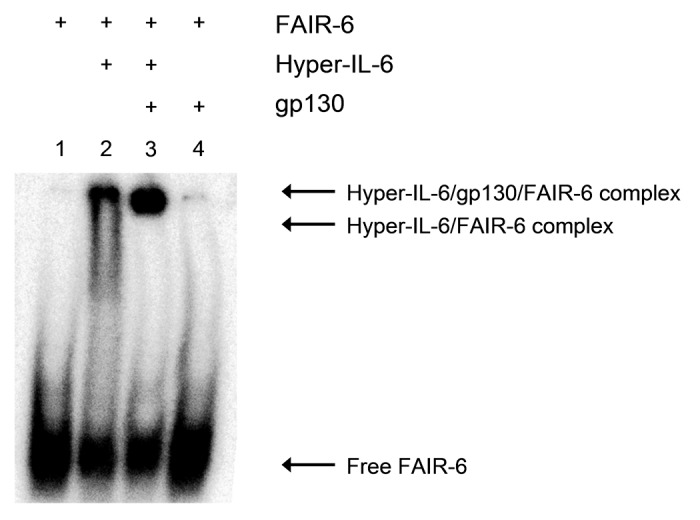
Figure 4. Aptamer FAIR-6 does not compete with gp130 for binding to Hyper-IL-6. Interaction of aptamer FAIR-6 (< 1 nM) with Hyper-IL-6 (500 nM) or with Hyper-IL-6/gp130 complex (1:2 stoichiometry) analyzed by gel-electrophoretic mobility shift assay. Samples: free FAIR-6 (lane 1); aptamer + 500 nM Hyper-IL-6 (2); aptamer + 500 nM Hyper-IL-6 + 1 µM gp130 (3); aptamer + 1 µM gp130 (4).
Minimization of the 2'-F-modified RNA aptamer FAIR-6
Assuming that the affinity of FAIR-6 is constricted to the conserved G-rich binding motif, we subsequently generated a 19-nt short version, named FAIR-6A, and a single nucleotide variant as control (Table S2). Contrary to our expectations based on AIR-3A, FAIR-6A did not show any binding to Hyper-IL-6 at all (Fig. S5). Therefore, the 19 nucleotides do not suffice for functional and/or structural stability.
Discussion
Aptamers for extracellular targets have high potential to be used in medicine and therapy. Aptamers that are able to interact with specific disease-related cell-surface proteins can be used as effectors directly. These aptamers are potential therapeutic compounds because they might cause inhibitory7,28 as well as stimulatory effects.6,29 Furthermore, most cell-surface proteins undergo recycling processes, such as ligand-induced internalization. Aptamers binding to receptors that are subject to internalization can be shuffled into target cells, and thus, serve as vehicles for desired cargo molecules.21,30,31 Recently, we identified RNA aptamers targeting the soluble part of the interleukin-6 receptor with high affinity (Kd ~20 nM).20 It was shown that AIR-3A did bind the human IL-6R presented on target cells and served as cargo delivery agent.
For therapeutic applications, however, RNA aptamers hold one major drawback, which is their susceptibility to nucleases and their chemical instability. Today, various chemical modifications exist that increase the stability of RNAs against nucleases. Modifications can be introduced at the phosphate backbone, such as the replacement by phosphorothioates, or at the nucleobases. However, the most prominent modification of aptamers is the derivatization of the 2'-OH group of the pyrimidine ribo-nucleotides by 2'-fluoro- and 2'-amino-2'-deoxy groups.14 Next to considerably increased nuclease resistance, examples exist whereat the incorporation of 2’-F-modified pyrimidine residues (C and U) into RNA aptamers additionally revealed equal or higher binding affinities for the target molecule if compared with the corresponding natural aptamers.18,19
Stabilizing modifications can be incorporated into already obtained aptamers in a post-selective manner. However, any chemical modification may alter the aptamers' structure, selectivity, and binding affinity without a significant increase in stability. Therefore, the effects of each executed post-selective modification has to be carefully evaluated.
One the other hand, stabilizing modifications can already be incorporated into the starting oligonucleotide library deployed in the SELEX procedure. Recent developments of the SELEX process have led to in vitro selections using a combinatorial library already containing modified bases, yielding nuclease-stabilized aptamers that directly bind tightly to a given ligand.9,32,33 It is a prerequisite that the modifications are compatible with the enzymatic steps required to perform SELEX. Successfully applied modifications include the incorporation of 2'-F- or 2'-OMe-modified pyrimidine nucleotides using variants of the T7 RNA polymerase that are able to incorporate these modified nucleotides during in vitro transcription.34-36
To convert the already known RNA aptamer AIR-3A into a serum-stable counterpart, we first performed mutational analyses to identify nucleotide positions that are alterable to some extend without loss of aptamer function. Exchanges of each single non-G residue fulfilled these requirements. Since it is known that G-rich regions in nucleic acids can adopt G-quadruplex conformations, we wanted to find out whether replacements of guanine nucleotides might cause structural instabilities and are therefore the reason for the loss of aptamer function. UV-melting analyses at 295 nm showed that AIR-3A and selected non-G variants, such as C8A, revealed a G-quadruplex structure in the presence of KCl. The opposite held true for G variants, such as G4U or G17U, which did not fulfill the criteria for the formation of G-quadruplexes as a minimum of four interspersed GG dinucleotides was missing.
The variant G1U constitutes an exception as it fulfilled the above mentioned criteria and adopted a G-quadruplex structure. Structural investigations supported our observation that mutations of nucleotides not involved in quadruplex formation are less harmful on aptamer function.
The post-selective 2'-F-modification of AIR-3A by incorporation of 2'-F-modified C and U residues during T7 transcription led to a complete loss of function. These data demonstrate quite convincingly that even a single modification of an aptamer can have a very strong negative impact on its performance.
Therefore, we decided to evolve novel nuclease-stabilized RNA aptamers with affinity for the extracellular portion of IL-6R. For smooth SELEX performance a variant of the T7 RNA polymerase was developed that can use 2'-Fluoro-pyrimidine nucleotide triphosphates as substrates for in vitro transcription.26,37 After completing 15 rounds of in vitro selection using this variant, we obtained seven different aptamers. All bound Hyper-IL-6 with the best dissociation constants as low as 40 nM. These affinities match with those obtained for the unconstrained aptamers.20 Even though the 2'-F-modified aptamers did not show any conserved binding motif, one of them, aptamer FAIR-6, included the same conserved G-rich binding motif that had previously been identified in unmodified IL-6R-binding RNA aptamers.20 Following our expectations, FAIR-6 formed a G-quadruplex structure like AIR-3A and both shared one common binding site at the IL-6R. Contrary to our expectations, we could not minimize FAIR-6 to the 19-nt short variant FAIR-6A as it has been successfully done for AIR-3 and AIR-3A. In the present case, the truncation resulted in the non-functional variant FAIR-6A. To gain shortened versions of FAIR-6 as well as for the other FAIRs, a more gentle strategy might be necessary, such as the implementation of damage selection procedures to identify all base positions required for binding or structural stability.38
Aptamers binding to cell surface receptors that are subject to internalization can be taken up by target cells, and thus, serve as vehicles for desired cargo molecules. As AIR-3A retained all necessary characteristics for high affinity and selective recognition of IL-6R on cell surfaces, it is conceivable that FAIR-6 might have the same properties, too. Further investigations using cell-based assays might demonstrate whether the 2'-F-modified aptamers are internalized by IL-6R-presenting cells. To go one step further, these aptamers might also be able to deliver cargo molecules to or into specific cells. But these open questions are the subject of future investigations.
Materials and Methods
Chemicals
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich. Buffers were prepared using de-ionized water obtained from a water purification system (Millipore). Selection buffer for SELEX (1x PBS/MgCl2) consisted of 137 mM NaCl; 2.7 mM KCl; 6.5 mM Na2HPO4; 1.5 mM KH2PO4; and 3 mM MgCl2 at pH 7.5.
Oligonucleotides
See Table 3.
Table 3. Oligonucleotides.
| Oligo | Sequence (printed in 5‘-3‘ direction) | Note |
|---|---|---|
| AIR-3A | GGGGAGGCUG UGGUGAGGG | (1) |
| G1U | UGGGAGGCUG UGGUGAGGG | (1) |
| C8A | GGGGAGGAUG UGGUGAGGG | (1) |
| G17U | GGGGAGGCUG UGGUGAUGG | (1) |
| G18U | GGGGAGGCUG UGGUGAGUG | (1) |
| DNA library B50 | AATGCTAATACGACTCACTATAGGAAGAAA GAGGTCTGAG ACATTCT–N50–CTTCTGGAGT TGACGTTGCT T | (2) |
| T7 primer B50 | AATGCTAATACGACTCACTATAGGAAGAAA GAGGTCTGAG ACATT | (2) |
| RT primer B50 | AAGCAACGTCAACTCCAGAAG | (2) |
| G2U-rev | CCCTCACCAC AGCCTCCACT ATAGTGAGTC GTATTAATAC GACTC | (3) |
| G3U-rev | CCCTCACCAC AGCCTCACCT ATAGTGAGTC GTATTAATAC GACTC | (3) |
| G4U-rev | CCCTCACCAC AGCCTACCCT ATAGTGAGTC GTATTAATAC GACTC | (3) |
| A5U-rev | CCCTCACCAC AGCCACCCCT ATAGTGAGTC GTATTAATAC GACTC | (3) |
| G6U-rev | CCCTCACCAC AGCATCCCCT ATAGTGAGTC GTATTAATAC GACTC | (3) |
| G7U-rev | CCCTCACCAC AGACTCCCCT ATAGTGAGTC GTATTAATAC GACTC | (3) |
| U9A-rev | CCCTCACCAC TGCCTCCCCT ATAGTGAGTC GTATTAATAC GACTC | (3) |
| G10U-rev | CCCTCACCAA AGCCTCCCCT ATAGTGAGTC GTATTAATAC GACTC | (3) |
| U11A-rev | CCCTCACCTC AGCCTCCCCT ATAGTGAGTC GTATTAATAC GACTC | (3) |
| G12U-rev | CCCTCACAAC AGCCTCCCCT ATAGTGAGTC GTATTAATAC GACTC | (3) |
| G13U-rev | CCCTCAACAC AGCCTCCCCT ATAGTGAGTC GTATTAATAC GACTC | (3) |
| U14A-rev | CCCTCTCCAC AGCCTCCCCT ATAGTGAGTC GTATTAATAC GACTC | (3) |
| G15U-rev | CCCTAACCAC AGCCTCCCCT ATAGTGAGTC GTATTAATAC GACTC | (3) |
| A16U-rev | CCCACACCAC AGCCTCCCCT ATAGTGAGTC GTATTAATAC GACTC | (3) |
| G19U-rev | ACCTCACCAC AGCCTCCCCT ATAGTGAGTC GTATTAATAC GACTC | (3) |
| (-G19)-rev | CCTCACCACA GCCTCCCCTA TAGTGAGTCG TATTAATACG ACTC | (3) |
| FAIR-6 | CCCACTCCAC ATCCACCCCT ATAGTGAGTC GTATTAATAC GACTC | (3) |
| FAIR-6_G18U | CACACTCCAC ATCCACCCCT ATAGTGAGTC GTATTAATAC GACTC | (3) |
(1) All RNAs were synthesized and purified by IBA. (2) DNA library B50, containing 50 randomized nucleotides (N50), corresponding primers were purchased from Metabion. The T7 promoter region is underlined. (3) All additionally listed ssDNAs were purchased from Life Technologies GmbH.
Production of IL-6, sgp130Fc, sIL-6R, and Hyper-IL-6
The proteins IL-6,39 sgp130Fc,40 and Hyper-IL-625 were produced as previously described.20
Hybridization and in vitro transcription to produce the unmodified RNAs AIR-3A and its variants
First, a primer containing the T7-promotor sequence was hybridized with the reverse complementary strand encoding the RNA of interest. For this hybridization reaction, equal amounts (5 µM) of both oligonucleotides were mixed in 1 × PCR buffer B (Solis BioDyne), heated to 80 °C for 5 min and slowly cooled down to RT. Hybridization products (0.1 µM) were directly used for in vitro transcription by wild type T7 RNA polymerase for 3 h at 37 °C in transcription buffer (40 mM TRIS-HCl, pH 7.9) containing T7 RNA polymerase (0.25 U/µL), NTPs (2.5 mM each) and MgCl2 (15 mM). Resulting RNAs were purified on 10% denaturing polyacrylamide gels.
Fill in reaction and in vitro transcription to produce the 2’-Fluoro-modified RNA library B50
The initial single stranded DNA library B50 contained 50 randomized nucleotides (N50) flanked by two constant primer-binding sites. The forward primer (T7 primer B50) contained the T7 RNA polymerase promoter region. The DNA library B50 was converted into dsDNA by a two-step fill in reaction. First the ssDNA library was hybridized with the reverse primer in a hybridization reaction: equal amounts (1 µM) of DNA library B50 and reverse primer (RT primer B50) were mixed in 1 × PCR buffer B (Solis BioDyne), heated to 80 °C for 5 min, and slowly cooled down to RT. The second strand synthesis was completed using the Klenow fragment (Thermo Fisher Scientific, EP0051) under the following conditions: 1.25 U Klenow fragment per 100 µL; 0.5 µM hybridization product; 500 µM of each dNTP in Klenow buffer (Thermo Fisher Scientific); temperature profile: 1 h 37 °C, 10 min 75 °C. Double stranded nucleic acids (0.1 µM) were directly used for in vitro transcription using the T7 RNA polymerase variant Y639F for 3 h at 37 °C in transcription buffer (40 mM TRIS-HCl, pH 7.9) containing T7 RNA polymerase Y639F (0.25 U/µL),26 ATP and GTP (1 mM each), 2’-Fluoro-modified CTP and 2’-Fluoro-modified UTP (1 mM each), and MgCl2 (15 mM). The derived RNA library B50 was purified on 8% denaturing polyacrylamide gels.
Biotinylation of Hyper-IL-6 and its immobilization on streptavidin-coated dynabeads
For immobilization of a biotinylated target protein on Streptavidin-coated magnetic beads (Dynabeads® M-280 Streptavidin, Invitrogen, 112.06D), 100 µg of Hyper-IL-6 were mixed with a 3-fold molar excess of Sulfo-NHS-LC-Biotin (Thermo Fisher Scientific, 10538723) in a final volume of 100 µL selection buffer followed by incubation on ice for 15 min and further 15 min at RT.41 The excess of non-reacted and hydrolyzed biotin reagent was removed by dialysis against selection buffer using a Slide-A-Lyzer® dialysis cassette (MWCO 10K; Thermo Fisher Scientific). The biotinylated protein was immobilized on 5 mg Dynabeads and suspended in selection buffer (including 1.25 µg BSA/µL).
In vitro selection procedure
In the first round of the in vitro selection process 500 pmol of the 2’-Fluoro-modified RNA library B50 (~1013 molecules) were incubated with 100 pmol Hyper-IL-6 immobilized on magnetic beads in selection buffer (containing 1 µg BSA/µL) for 30 min at 37 °C. Unbound RNA molecules were removed by magnetic separation. After washing with 200 µL selection buffer, bound RNA molecules were eluted in 50 µL water by heating the sample to 80 °C for 3 min and amplified by reverse transcription and polymerase chain reaction (RT-PCR). Therefore the following RT-PCR reaction was prepared: 1 × PCR buffer B; 0.2 × First-Strand Buffer (Invitrogen); forward and reverse primer (1 µM each); 1.5 mM MgCl2; 0.3 mM dNTPs; 2 mM DTT. For reverse primer hybridization the mixture was heated to 65 °C for 5 min and cooled down on ice. The RT-PCR was started after addition of 15 U SuperScriptTM III Reverse Transcriptase (Invitrogen, 18080-044) and 5 U FIREPol® (Solis BioDyne, 01-01-02000) per 100 µL reaction. Following settings were used: 10 min at 54 °C for the reverse transcription; for PCR amplification: 30 s at 95 °C; 30 s at 60 °C, and 30 s at 72 °C for an appropriate number of PCR cycles. For the subsequent rounds of selection the derived dsDNAs were transcribed into an RNA library as described above. To increase the stringency during the following rounds of selection, the number of washing steps was raised by one each round. After 15 rounds of this in vitro selection process the dsDNA library was cloned via TOPO TA Cloning (pCR2.1, Invitrogen, K456001) and individual clones were sequenced.
Filter retention assay (FRA)
To investigate the binding of RNA molecules to the target protein, filter retention assays were performed. Nucleic acids were radioactively labeled at their 5′-termini using γ-[32P]-ATP (3000 Ci/mmol, Hartmann Analytic, FP-301TD) and polynucleotide kinase. After gel purification, constant amounts of labeled RNA (< 1 nM) were incubated with increasing amounts of the target protein (0–500 nM) in 1 x selection buffer. After incubation the samples were filtered through a pre-equilibrated nitrocellulose membrane (0.45 µm, Carl Roth) on a vacuum manifold (Minifold® I Dot-Blot-System; Schleicher&Schuell) and washed four times with selection buffer. The nitrocellulose membrane was dried and exposed to a phosphor-imaging screen (Bio-Rad). The amount of radioactively labeled RNA on the filter was quantified using the Quantity One® software (Version 4.6.6, Bio-Rad) and used for the calculation of the bound RNA fraction. For the determination of dissociation constants (Kd) and maximal binding (Bmax) data were fitted using a one site-binding model with the aid of the program GraphPad Prism applying the following equation: RNAbound = (Bmax × cProtein)/(Kd + cProtein).
Electrophoretic mobility shift assay (EMSA)
RNA–protein interactions were investigated using a native electrophoretic mobility shift assay. Radioactively labeled RNAs (< 1 nM) were incubated with proteins of interest in selection buffer for 30 min at room temperature. Six × DNA Loading Dye (Thermo Fisher Scientific, R0611) was added and samples were loaded on 3.5% non-denaturing polyacrylamide gels (acrylamide/bisacrylamide 37.5:1). The samples were then electrophoretically separated at 60 V for 2–3 h in 1 × TBE buffer. The gel was dried on a vacuum dryer at 70 °C for 2 h and exposed to a phosphor imager screen overnight. Bands were detected as described above.
UV spectroscopy
For UV-melting experiments, RNAs (2.5 µM) were dissolved in 10 mM TRIS-HCl (pH 7.5) optionally containing 0 mM, 5 mM, or 100 mM KCl. UV-melting studies of prepared RNAs were conducted on a Varian Cary Bio 300 UV-Visible Spectrophotometer with a temperature controller. Each sample (1200 µL) was filled into a quartz cuvette (1-cm path length), covered with a thin layer of mineral oil, transferred to the spectrophotometer, heated to 80 °C and cooled down to 20 °C for two times with a heating or cooling rate of 0.5 °C min−1. Absorbance was recorded at 295 nm every 30 s. Each melting curve was analyzed using the method of van’t Hoff to determine the Tm value.42,43
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft [SFB 877, projects A1, A6 to Grötzinger J and Rose-John S] and the cluster of excellence “inflammation at interfaces” [to Grötzinger J and Rose-John S].
Glossary
Abbreviations:
- IL-6
Interleukin-6
- IL-6R
Interleukin-6 receptor
- sIL-6R
soluble Interleukin-6 receptor
- gp130
glycoprotein 130
- sgp130Fc
fusion protein of soluble gp130 and Fc fragment of immunoglobulin G1
- Hyper-IL-6
a fusion protein of sIL-6R and IL-6, SELEX, Systematic Evolution of Ligands by EXponential enrichment
- FRA
filter retention assay
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/27447
References
- 1.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–10. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–22. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Zhou J, Swiderski P, Li H, Zhang J, Neff CP, Akkina R, Rossi JJ. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009;37:3094–109. doi: 10.1093/nar/gkp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gopinath SC, Sakamaki Y, Kawasaki K, Kumar PK. An efficient RNA aptamer against human influenza B virus hemagglutinin. J Biochem. 2006;139:837–46. doi: 10.1093/jb/mvj095. [DOI] [PubMed] [Google Scholar]
- 5.Raddatz MS, Dolf A, Endl E, Knolle P, Famulok M, Mayer G. Enrichment of cell-targeting and population-specific aptamers by fluorescence-activated cell sorting. Angew Chem Int Ed Engl. 2008;47:5190–3. doi: 10.1002/anie.200800216. [DOI] [PubMed] [Google Scholar]
- 6.Dollins CM, Nair S, Boczkowski D, Lee J, Layzer JM, Gilboa E, Sullenger BA. Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem Biol. 2008;15:675–82. doi: 10.1016/j.chembiol.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mann AP, Somasunderam A, Nieves-Alicea R, Li X, Hu A, Sood AK, Ferrari M, Gorenstein DG, Tanaka T. Identification of thioaptamer ligand against E-selectin: potential application for inflamed vasculature targeting. PLoS One. 2010;5:5. doi: 10.1371/journal.pone.0013050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer C, Hahn U, Rentmeister A. Cell-specific aptamers as emerging therapeutics. J Nucleic Acids. 2011;2011:904750. doi: 10.4061/2011/904750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62:4029–33. [PubMed] [Google Scholar]
- 10.Chu TC, Marks JW, 3rd, Lavery LA, Faulkner S, Rosenblum MG, Ellington AD, Levy M. Aptamer:toxin conjugates that specifically target prostate tumor cells. Cancer Res. 2006;66:5989–92. doi: 10.1158/0008-5472.CAN-05-4583. [DOI] [PubMed] [Google Scholar]
- 11.McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–15. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 12.Dhar S, Kolishetti N, Lippard SJ, Farokhzad OC. Targeted delivery of a cisplatin prodrug for safer and more effective prostate cancer therapy in vivo. Proc Natl Acad Sci U S A. 2011;108:1850–5. doi: 10.1073/pnas.1011379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO, 2nd, Giangrande PH. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol. 2009;27:839–49. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer G. The chemical biology of aptamers. Angew Chem Int Ed Engl. 2009;48:2672–89. doi: 10.1002/anie.200804643. [DOI] [PubMed] [Google Scholar]
- 15.Rose-John S, Scheller J, Elson G, Jones SA. Interleukin-6 biology is coordinated by membrane-bound and soluble receptors: role in inflammation and cancer. J Leukoc Biol. 2006;80:227–36. doi: 10.1189/jlb.1105674. [DOI] [PubMed] [Google Scholar]
- 16.Jones SA, Scheller J, Rose-John S. Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J Clin Invest. 2011;121:3375–83. doi: 10.1172/JCI57158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Febbraio MA, Rose-John S, Pedersen BK. Is interleukin-6 receptor blockade the Holy Grail for inflammatory diseases? Clin Pharmacol Ther. 2010;87:396–8. doi: 10.1038/clpt.2010.1. [DOI] [PubMed] [Google Scholar]
- 18.Li S, Wang N, Brodt P. Metastatic cells can escape the proapoptotic effects of TNF-α through increased autocrine IL-6/STAT3 signaling. Cancer Res. 2012;72:865–75. doi: 10.1158/0008-5472.CAN-11-1357. [DOI] [PubMed] [Google Scholar]
- 19.Grivennikov S, Karin M. Autocrine IL-6 signaling: a key event in tumorigenesis? Cancer Cell. 2008;13:7–9. doi: 10.1016/j.ccr.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 20.Meyer C, Eydeler K, Magbanua E, Zivkovic T, Piganeau N, Lorenzen I, Grötzinger J, Mayer G, Rose-John S, Hahn U. Interleukin-6 receptor specific RNA aptamers for cargo delivery into target cells. RNA Biol. 2012;9:67–80. doi: 10.4161/rna.9.1.18062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li N, Larson T, Nguyen HH, Sokolov KV, Ellington AD. Directed evolution of gold nanoparticle delivery to cells. Chem Commun (Camb) 2010;46:392–4. doi: 10.1039/b920865h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schiechl G, Bauer B, Fuss I, Lang SA, Moser C, Ruemmele P, Rose-John S, Neurath MF, Geissler EK, Schlitt HJ, et al. Tumor development in murine ulcerative colitis depends on MyD88 signaling of colonic F4/80+CD11b(high)Gr1(low) macrophages. J Clin Invest. 2011;121:1692–708. doi: 10.1172/JCI42540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Klöppel G, Yoshimura A, Reindl W, Sipos B, Akira S, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19:456–69. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 24.Reyes-Reyes EM, Teng Y, Bates PJ. A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010;70:8617–29. doi: 10.1158/0008-5472.CAN-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer M, Goldschmitt J, Peschel C, Brakenhoff JP, Kallen KJ, Wollmer A, Grötzinger J, Rose-John S. I. A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat Biotechnol. 1997;15:142–5. doi: 10.1038/nbt0297-142. [DOI] [PubMed] [Google Scholar]
- 26.Sousa R, Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995;14:4609–21. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J Biol Chem. 1994;269:21858–69. [PubMed] [Google Scholar]
- 28.Gutsaeva DR, Parkerson JB, Yerigenahally SD, Kurz JC, Schaub RG, Ikuta T, Head CA. Inhibition of cell adhesion by anti-P-selectin aptamer: a new potential therapeutic agent for sickle cell disease. Blood. 2011;117:727–35. doi: 10.1182/blood-2010-05-285718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNamara JO, Kolonias D, Pastor F, Mittler RS, Chen L, Giangrande PH, Sullenger B, Gilboa E. Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J Clin Invest. 2008;118:376–86. doi: 10.1172/JCI33365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther. 2008;16:1481–9. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J, Swiderski P, Li H, Zhang J, Neff CP, Akkina R, Rossi JJ. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009;37:3094–109. doi: 10.1093/nar/gkp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jellinek D, Green LS, Bell C, Lynott CK, Gill N, Vargeese C, Kirschenheuter G, McGee DP, Abesinghe P, Pieken WA, et al. Potent 2′-amino-2′-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry. 1995;34:11363–72. doi: 10.1021/bi00036a009. [DOI] [PubMed] [Google Scholar]
- 33.Klussmann S, Nolte A, Bald R, Erdmann VA, Fürste JP. Mirror-image RNA that binds D-adenosine. Nat Biotechnol. 1996;14:1112–5. doi: 10.1038/nbt0996-1112. [DOI] [PubMed] [Google Scholar]
- 34.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 35.Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–90. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 36.Ibach J, Dietrich L, Koopmans KR, Nöbel N, Skoupi M, Brakmann S. Identification of a T7 RNA polymerase variant that permits the enzymatic synthesis of fully 2′-O-methyl-modified RNA. J Biotechnol. 2013;167:287–95. doi: 10.1016/j.jbiotec.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 37.Sousa R. Use of T7 RNA polymerase and its mutants for incorporation of nucleoside analogs into RNA. Methods Enzymol. 2000;317:65–74. doi: 10.1016/S0076-6879(00)17006-5. [DOI] [PubMed] [Google Scholar]
- 38.Burgstaller P, Kochoyan M, Famulok M. Structural probing and damage selection of citrulline- and arginine-specific RNA aptamers identify base positions required for binding. Nucleic Acids Res. 1995;23:4769–76. doi: 10.1093/nar/23.23.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Dam M, Müllberg J, Schooltink H, Stoyan T, Brakenhoff JP, Graeve L, Heinrich PC, Rose-John S. Structure-function analysis of interleukin-6 utilizing human/murine chimeric molecules. Involvement of two separate domains in receptor binding. J Biol Chem. 1993;268:15285–90. [PubMed] [Google Scholar]
- 40.Jostock T, Müllberg J, Ozbek S, Atreya R, Blinn G, Voltz N, Fischer M, Neurath MF, Rose-John S. Soluble gp130 is the natural inhibitor of soluble interleukin-6 receptor transsignaling responses. Eur J Biochem. 2001;268:160–7. doi: 10.1046/j.1432-1327.2001.01867.x. [DOI] [PubMed] [Google Scholar]
- 41.Mayer G, Wulffen B, Huber C, Brockmann J, Flicke B, Neumann L, Hafenbradl D, Klebl BM, Lohse MJ, Krasel C, et al. An RNA molecule that specifically inhibits G-protein-coupled receptor kinase 2 in vitro. RNA. 2008;14:524–34. doi: 10.1261/rna.821908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bugaut A, Balasubramanian S. A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry. 2008;47:689–97. doi: 10.1021/bi701873c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marky LA, Breslauer KJ. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–20. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


