Abstract
The development of efficient strategies for generating fully human monoclonal antibodies with unique functional properties that are exploitable for tailored therapeutic interventions remains a major challenge in the antibody technology field. Here, we present a methodology for recovering such antibodies from antigen-encountered human B cell repertoires. As the source for variable antibody genes, we cloned immunoglobulin G (IgG)-derived B cell repertoires from lymph nodes of 20 individuals undergoing surgery for head and neck cancer. Sequence analysis of unselected “LYmph Node Derived Antibody Libraries” (LYNDAL) revealed a naturally occurring distribution pattern of rearranged antibody sequences, representing all known variable gene families and most functional germline sequences. To demonstrate the feasibility for selecting antibodies with therapeutic potential from these repertoires, seven LYNDAL from donors with high serum titers against herpes simplex virus (HSV) were panned on recombinant glycoprotein B of HSV-1. Screening for specific binders delivered 34 single-chain variable fragments (scFvs) with unique sequences. Sequence analysis revealed extensive somatic hypermutation of enriched clones as a result of affinity maturation. Binding of scFvs to common glycoprotein B variants from HSV-1 and HSV-2 strains was highly specific, and the majority of analyzed antibody fragments bound to the target antigen with nanomolar affinity. From eight scFvs with HSV-neutralizing capacity in vitro, the most potent antibody neutralized 50% HSV-2 at 4.5 nM as a dimeric (scFv)2. We anticipate our approach to be useful for recovering fully human antibodies with therapeutic potential.
Keywords: fully human antibodies, combinatorial libraries, phage display, lymph nodes, herpes simplex virus, neutralizing antibodies, scFv, immune libraries
Introduction
Monoclonal antibodies (mAbs) have become an integral part of modern treatment concepts for cancer, inflammation, immunological disorders, and infectious diseases. Fully human mAbs, carrying no xenogenic components and displaying most preferable pharmacokinetic profiles, have emerged as the fastest-growing group of therapeutic antibodies entering clinical trials.1,2 Since the first report of the production of a mouse mAb through employment of B cell hybridoma fusion techniques,3 several powerful technologies for the generation of entirely human antibodies have been developed.4,5 These include non-combinatorial methodologies for the efficient immortalization of human B cells,6,7 the generation of stable human hybridomas,8 and expression-cloning of physiological variable (V) gene pairings from blood-derived human B cells of infected individuals.9 The vast majority of clinically-investigated fully human mAbs, however, are produced by either immunization of transgenic mice that are equipped with the human antibody gene repertoire,10,11 or by preparing human antibody phage display libraries. In the latter approach, random pairs of the variable genes of the heavy and light antibody chains (VH and VL, respectively) are cloned as combinatorial libraries involving bacteriophages for presentation of selectable antibody fragments.12 Since the initial description of antibody phage display,13 this technology has evolved into the most successful in vitro selection platform for human antibodies because it is robust, inexpensive, and offers great potential for automation.14,15 The first fully human phage display-derived mAbs have recently been approved by the FDA for the treatment of rheumatic and chronic inflammatory bowel diseases (adalimumab), systemic lupus erythematosus (belimumab), and inhalational anthrax infection (raxibacumab). Other phage display library-selected mAbs are currently at advanced stages of clinical development and may reach market approval within the next few years.
Over the past 20 years, a plethora of phage-derived antibody libraries have been constructed that differ in design, origin, diversity, and method of generation. Nevertheless, depending on the gene sources employed, all antibody libraries can be grouped into naïve, immune, and (semi-) synthetic libraries. Naïve libraries were amplified from naturally rearranged V genes of primary B-cell repertoires of healthy donors,16-19 whereas immune libraries were created from V genes from infected or immunized donors against a wide range of disease-related target antigens.20-32 The design of synthetic libraries involved computational approaches and gene synthesis33-37 that were partially completed by segments from natural sources.38-42 Nowadays, almost all existing commercial libraries are based on non-immune repertoires of high complexity because selection of high-affinity binders is expected to directly correlate with the size of the functional repertoire. To theoretically allow the selection of mAbs against a virtually unlimited number of target antigens, latest state-of-art libraries reach complexities of up to 1011–1012 clones and thus may even exceed the size of individual human B cell repertoires.17,34,37 Although these libraries are highly successful in recovering target-specific and high affinity antibodies against a wide range of different antigens, they do not usually contain specificities that are developed during the course of a humoral immune response in vivo. It is nonetheless conceivable that antibodies derived from secondary and hyperimmune responses may have advantageous functional properties and improved fine-specificities over immunologically unchallenged repertoires.43,44
To exploit this particular antibody pool for therapeutic purposes, we aimed at generating a library collection of independently combinable IgG repertoires from B cells that have previously encountered defined target antigen(s) in the course of disease. In the present study, we employed lymph-node derived B cell repertoires for cloning 20 individualized phage display libraries in the scFv antibody format. Seven libraries deriving from donors with high serum titers against herpes simplex virus (HSV) were employed for panning against the viral glycoprotein B (gB) of HSV-1 that serves as an entry receptor for viral transmission.45 Our data show that high affinity scFvs with HSV-neutralizing properties could successfully be selected by this approach.
Results
Library construction
The LYNDAL concept is based on the cloning of individual lymph node IgG donor repertoires. In total, 20 donor lymph nodes were included for generation of 40 individual sublibraries (20 VH/VL-kappa and 20 VH/VL-lambda repertoires, respectively). Sizes of all individual LYNDAL are listed in Table 1. On average, each sublibrary consisted of 1.1 × 108 independent members and thus 2.2 × 108 for each donor repertoire. The total size of the final LYNDAL library collection comprising 40 sublibraries was approximately 4.4 × 109 clones.
Table 1. Number of independent antibody clones within LNYDAL.
| Library Size [x107] | |||
|---|---|---|---|
| Donor | HC/VL-kappa | HC/VL-lambda | Total |
| 1 | 0.7 | 12.1 | 12.8 |
| 2 | 4.4 | 2.0 | 6.4 |
| 3 | 1.0 | 1.1 | 2.1 |
| 4 | 4.7 | 1.1 | 5.8 |
| 5 | 1.1 | 2.9 | 4.0 |
| 6 | 4.0 | 28.1 | 32.1 |
| 7 | 2.7 | 2.6 | 5.3 |
| 8 | 4.8 | 4.3 | 9.1 |
| 9 | 8.6 | 16.6 | 25.2 |
| 10 | 16.3 | 5.2 | 21.5 |
| 11 | 9.6 | 15.7 | 25.3 |
| 12 | 14.2 | 7.7 | 21.9 |
| 13 | 3.9 | 13.1 | 17.0 |
| 14 | 2.9 | 15.4 | 18.3 |
| 15 | 7.3 | 4.2 | 11.5 |
| 16 | 29.3 | 2.6 | 31.9 |
| 17 | 89.5 | 54.3 | 143.8 |
| 18 | 8.5 | 10.7 | 19.2 |
| 19 | 3.9 | 3.9 | 7.8 |
| 20 | 4.5 | 11.3 | 15.8 |
Single sublibrary sizes were determined by preparing serial dilutions from two randomly chosen transformations and extrapolation to the number of grown colonies.
Characterization of LYNDAL repertoires
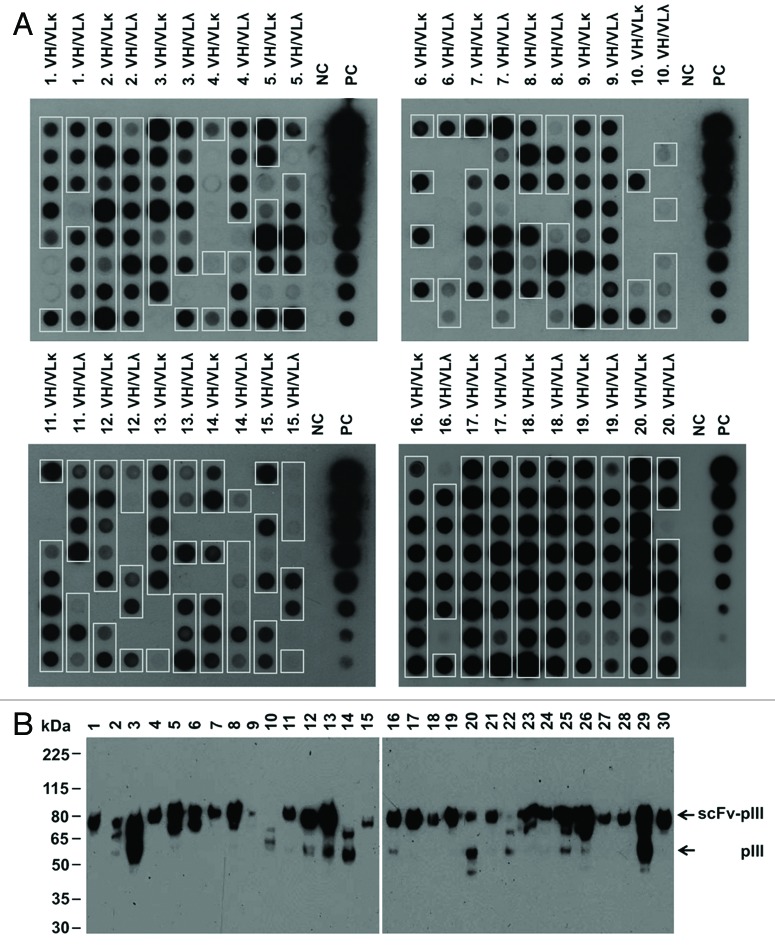
Selection of specific binders from phage display libraries requires expression of correctly folded scFv fragments on the phage surface. The quality of the LYNDAL repertoire was therefore characterized on both the gene and protein level. Of 1460 sequenced clones from 20 donors, 67% contained full-length scFv inserts with a similar proportion of kappa and lambda sublibraries (71% and 65%, respectively; Table S1). To further evaluate the number of solubly expressed scFvs by E. coli, we analyzed the induced periplasmic preparations of randomly picked clones from the LYNDAL collection by dot blot and western blot analysis. Eighty-two percent of LYNDAL clones investigated by dot blot screening were expressed as soluble antibody-pIII fusion proteins (Fig. 1A) with comparable expression rates of kappa and lambda light chain sublibraries (80% and 83%, respectively; Table S2). The proportion of detectable antibody-pIII fusions correlated well with that of complete inserts as analyzed by colony PCR (r = 0.75). Similar results were obtained in anti-pIII western blotting. Expression of intact scFv-pIII fusion proteins was shown for 27 of 30 clones chosen at random from positive dot blot signals (Fig. 1B). Thus, in total 73% of the LYNDAL clones could be expressed as complete scFv-pIII fusion proteins. On the basis of these analyses the current LYNDAL collection can be estimated to consist of 3.1 × 109 independent clones.
Figure 1.
LYNDAL antibody-pIII fusion protein expression. Induced periplasmic preparations of individual clones from 40 sublibraries were analyzed by (A) dot blot and, (B) western blot. (A) Dot blot analysis using the anti-c-myc-tag antibody 9E10 showed that 261/320 clones (82%) expressed detectable amounts of antibody-pIII fusion proteins (framed boxes). Negative control (NC) growth medium; positive control (PC) purified scFv (5 µg in 1:2 dilutions). (B) Immunoblotting with monoclonal anti-pIII antibody of 30 random clones with detectable protein expression in dot blot revealed for all clones with exception of clones 10, 14 and 22 (90%) protein bands migrating at an apparent molecular weight of approximately 80 kDa corresponding to scFv-pIII fusion proteins. Size of molecular weight markers are indicated (kDa).
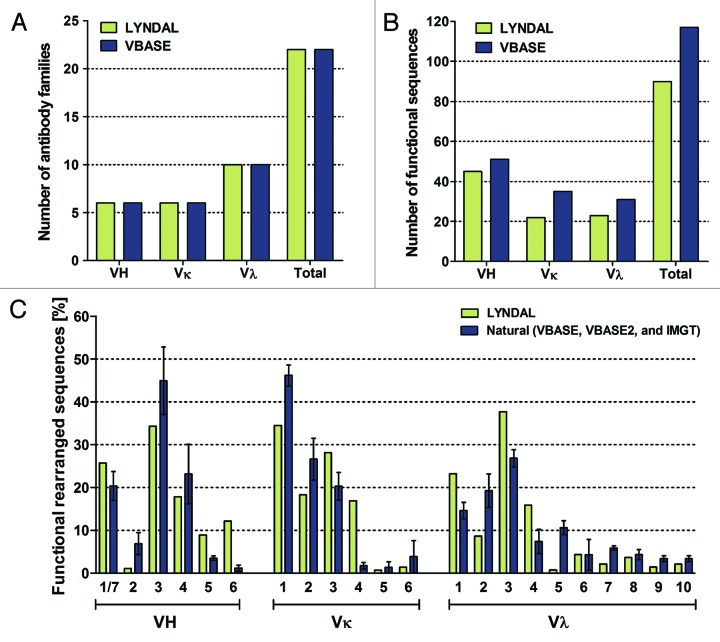
The primer set used for LYNDAL cloning was designed for PCR amplification of all known functional human antibody alleles and families as represented in the antibody gene database VBASE. Sequence analysis of 280 randomly picked clones (142 VH/VL-kappa and 138 VH/VL-lambda) with verified scFv insert was performed to analyze distribution and usage of germline genes and gene families actually being amplified by this primer set. We identified 90 out of the 117 VBASE annotated functional antibody genes (Fig. 2A–C). All known 22 human V gene families (Fig. 3A) and most of the VH (45 out of 51), VL-kappa (22 out of 35), and VL-lambda (23 out of 31) functional sequences were identified (Fig. 3B). The distribution pattern of the functional sequences in LYNDAL was comparable to that of three large antibody gene databases (VBASE, VBASE2, IMGT) considered to closely represent the naturally occurring distribution (Fig. 3C). Apparent differences were found for 4 families that occurred more frequently in either LYNDAL (i.e., VH6, VK4) or in the database entries (i.e., VH2, VL5).
Figure 2.
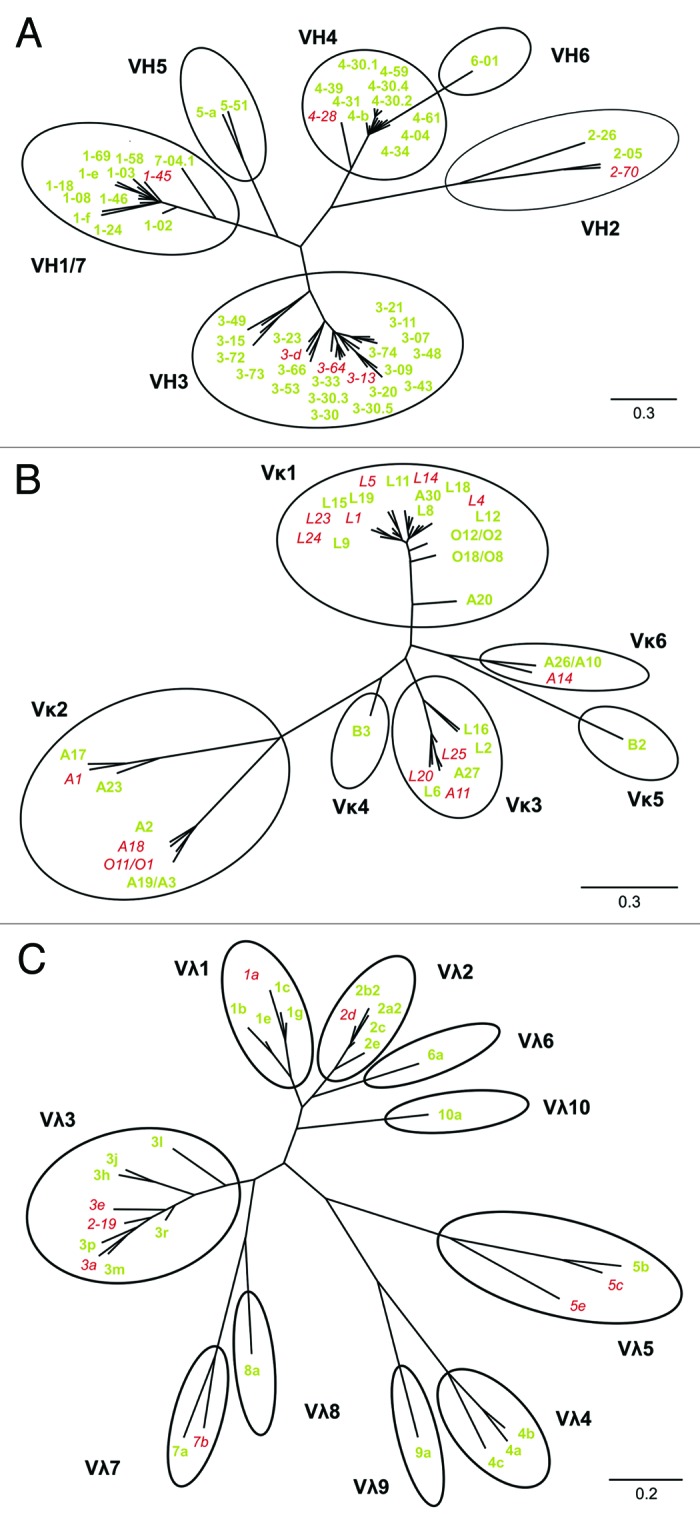
Phylogenetic analysis of LYNDAL sequences. Germline sequences of 280 randomly sequenced clones containing verified scFv genes were determined. Functional sequences from the VBASE database were employed for drawing three unrooted phylogenetic trees for the VH (A), VL-kappa (B) and VL-lambda (C) subset by means of the Phylogeny.fr web tool, followed by grouping the sequences to corresponding antibody families (black cycles). All functional sequences that were identified within the analyzed sample are marked green (77%), and non-represented germline sequences are labeled red. The nucleotide distance scales are indicated with a value of 30% distance for VH and VL-kappa, and 20% distance for VL-lambda.
Figure 3.
Diversity of LYNDAL antibody sequences. Sequence analysis of 280 randomly chosen clones from all sublibraries against the VBASE database identified all antibody V gene families (A) and most of the functional VH (88%), VL-kappa (63%) and VL-lambda (74%) germline sequences (B). Comparison of LYNDAL functional germline gene usage against the three independent human antibody databases VBASE, VBASE2, and IMGT (C). Error bars are shown as standard deviations (SD) of mean values.
Antibody selection from LYNDAL
In a next step, we assessed whether the pool of affinity-matured antibodies within the LYNDAL collection was directly accessible for antibody selection. We chose envelope glycoprotein B of HSV type 1 (gB-1) as the target for antibody selection because of the high prevalence of HSV-1 in the adult world population (50–85%).46,47 To exclusively use HSV-experienced B cell repertoires for screening of LYNDAL, only donor libraries from patients with confirmed serum antibody response in ELISA were included. Of 17 tested serum samples, 14 LYNDAL donors (82%) were gB-reactive with OD signals at least five times greater than those of non-immunized controls (OD 0.25; SD ± 0.04). Seven libraries (No. 1, 3–8) from donors with high IgG antibody titers against gB-1, containing a total of 7.1 × 108 clones, were combined for selection. Specific binders to gB-1 were successfully enriched (Fig. 4A). ELISA screening with monoclonal phages from round 2 and 3 revealed increases of target-specific scFv phage antibodies of 21% (20 out of 96) (Fig. 4B) and 34% (33 out of 96) (Fig. 4C), respectively. Subsequent fingerprint analyses of clones with verified scFv genes (Fig. S1) resulted in the identification of 34 individual binders from 192 screened colonies, i.e., every 6th screened colony (17.7%) encoded for a gB-specific antibody with unique gene sequence.
Figure 4.
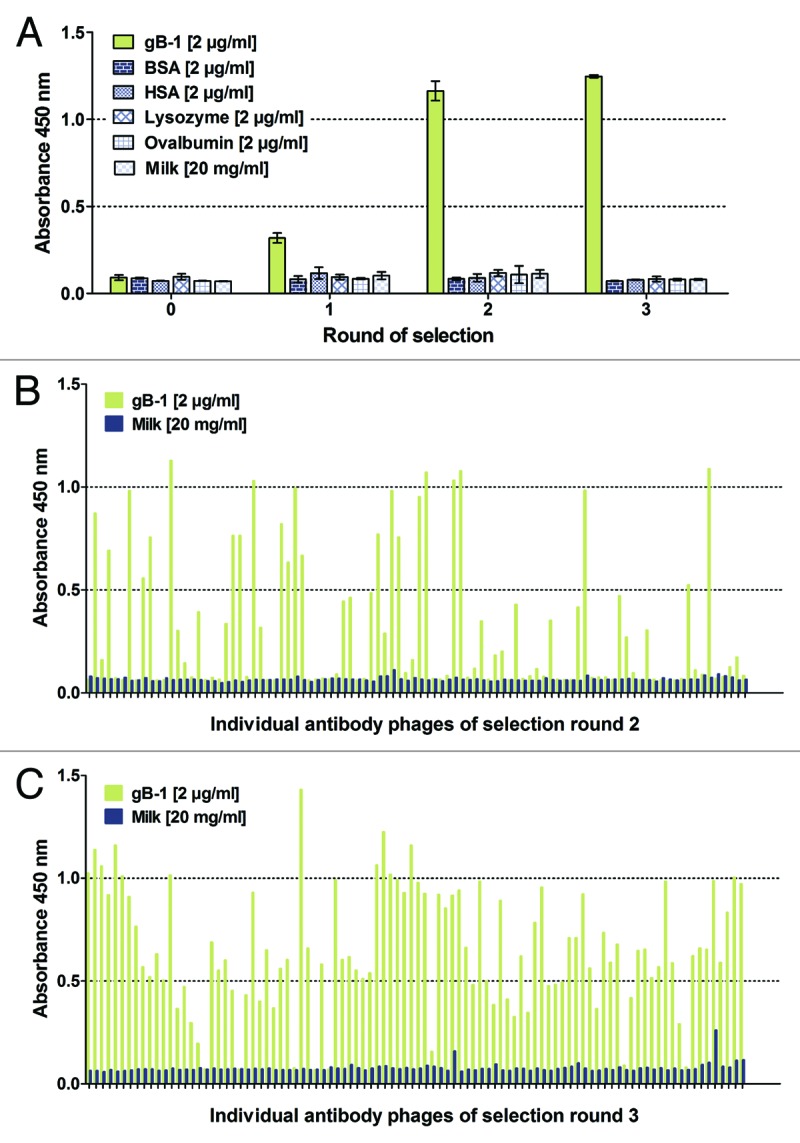
Enrichment of gB-specific LYNDAL antibodies. LYNDAL of seven donors with high anti-HSV titers (No. 1, 3 - 8) were combined for panning against target gB-1. Polyclonal phage ELISA (A) confirmed enrichment of target-specific scFv phage antibodies during selection rounds 1 to 3. The error bars represent standard deviations of duplicates. Screening of individual clones by monoclonal phage ELISA revealed an increase of specific binders between round 2 (B) and 3 (C). Phage antibodies were detected using an anti-phage peroxidase conjugate in combination with colorimetric substrate TMB.
Sequence analysis of HSV-specific LYNDAL antibodies
Sequence analysis revealed that the variable genes of selected scFvs derived from various germline sequences (18 out of 117) and antibody families (9 out of 22) with a predominant use of VH1 and VH3 gene families and lambda light chains (Fig. 5). By analyzing respective VH/VL pairings, 22 out of 34 scFvs were partially clonally related. To further analyze antigen-driven affinity maturation of enriched scFvs, the number of somatic hypermutations was determined on both the nucleotide (Fig. 6A) and the amino acid level (Fig. 6B). On average, the VH domain possessed more amino acid exchange mutations than the VL domain (17.4 vs. 8.6). Considering the number of nucleotide mutations (VH: 32.5, VL: 14.8), the frequency for non-silent mutations was comparable for both variable genes (VH: 54%, VL: 58%). When excluding the CDR3 and FR4 regions for analysis, the exchange mutations predominantly accumulated within the CDRs with the highest mutation frequency for CDRH2 and CDRH1, followed by CDRL2 and CDRL1 (Fig. 6C).
Figure 5.
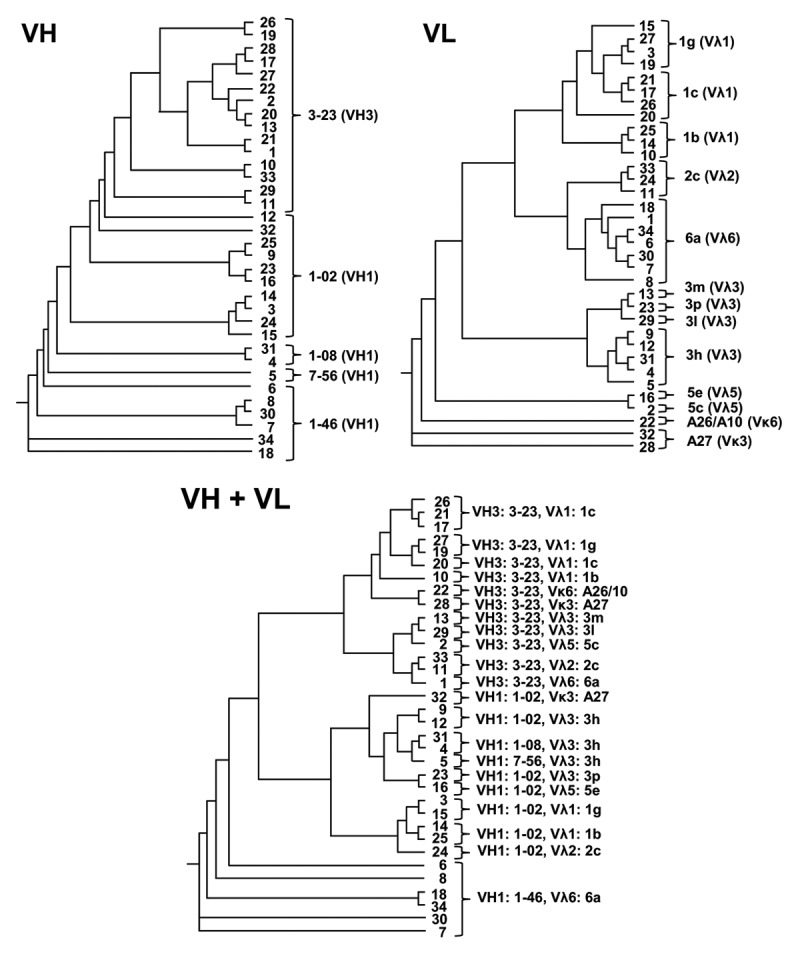
Sequence analysis of gB-specific LYNDAL antibodies. Germline sequences of the 34 selected scFvs were determined and phylogenic relationships analyzed by drawing phenograms employing the Phylogenic.fr web tool. The germline sequences as well as corresponding antibody families are shown for each clone.
Figure 6.
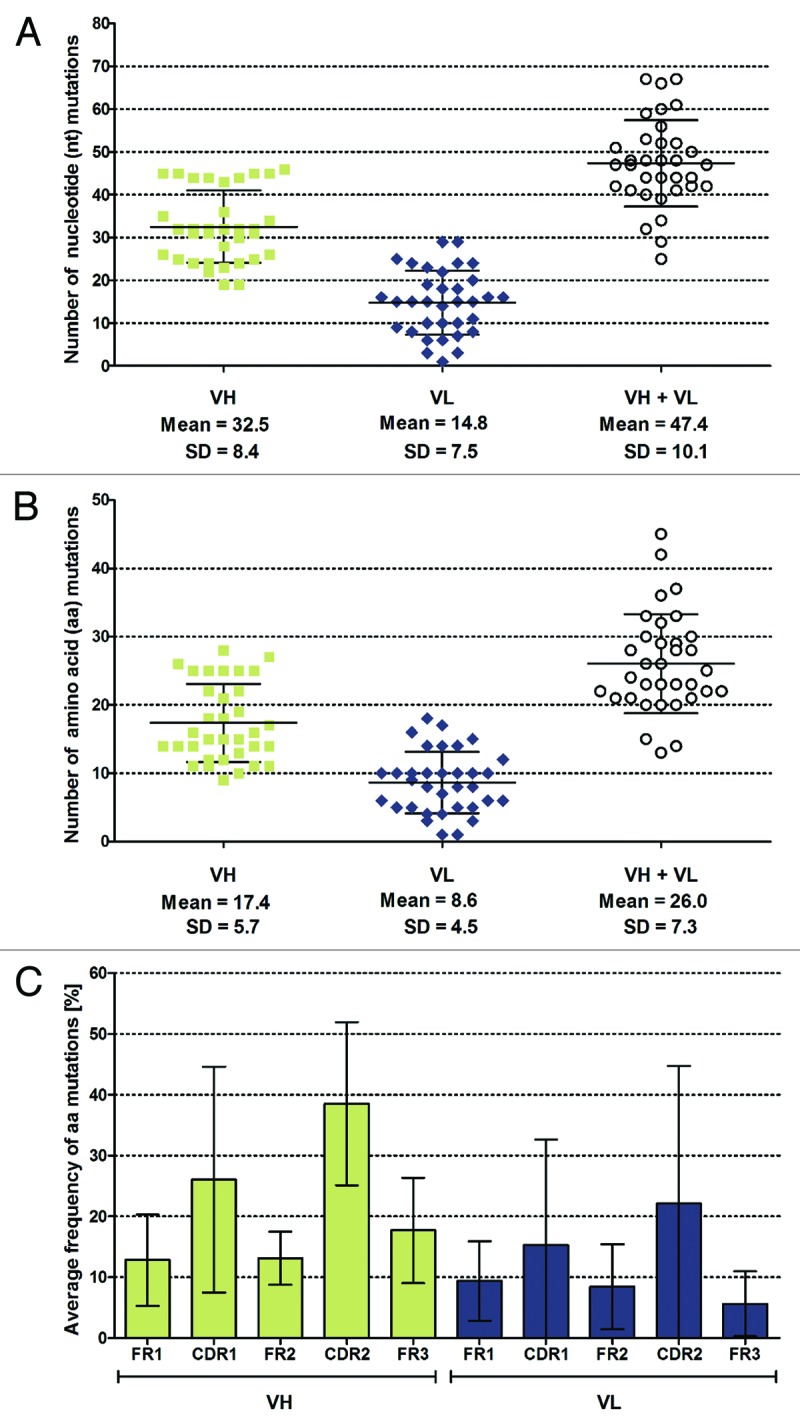
Analysis of somatic mutations within enriched LYNDAL antibodies. Variable genes of the 34 gB-specific scFvs were aligned to the closest respective germline sequences and the number of nucleotide (A) and amino acid (B) mutations was determined. Mean values and corresponding standard deviation are shown for VH, VL, and combined VH/VL genes. The distribution of amino acid mutations was analyzed separately for the VL and VH domains (C). The number of mutations in the framework regions 1 - 3 and complementarity determining regions 1 and 2 were determined and normalized to the length of each corresponding region. Results are presented as mean mutation frequency, i.e., the average probability of observing an amino acid mutation within the investigated segment when compared with its corresponding germline sequence. Error bars represent standard deviations of the mean values.
Functional characterization of HSV-specific antibodies
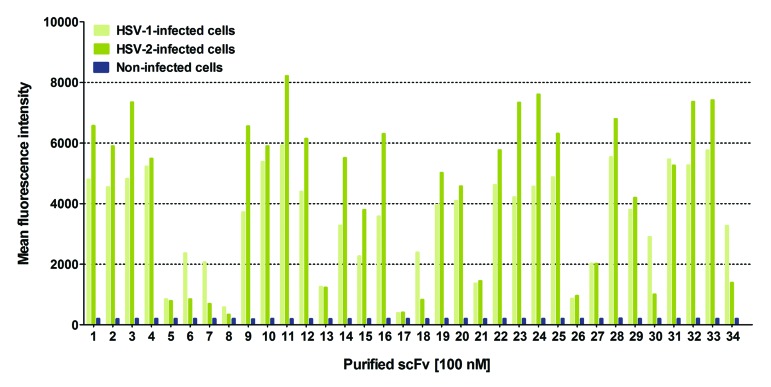
For further analyses, scFvs were solubly expressed in the periplasm of TG1 E. coli cells, purified by immobilized metal ion affinity chromatography (IMAC) (Fig. S2), and subjected to size exclusion chromatography (SEC). The vast majority of the investigated antibody fragments eluted predominantly as scFv monomers (Fig. S3). In flow cytometry, using Vero cells either infected with HSV-1 or HSV-2, all scFvs bound specifically to membrane-associated gB of both members of the herpes virus family, while no binding was found on uninfected Vero cells (Fig. 7). Flow cytometric competition assays of four randomly selected scFvs in the presence of a 10-fold molar excess of recombinant gB-1 further confirmed antigen specificity of LYNDAL scFvs (Fig. S4). Obtained immunofluorescence signals tended to be stronger on HSV-2 infected cells probably due to the known higher genome copy number of HSV-2 compared with that of HSV-1.48 Notably, 5 of 34 scFvs showed an almost three times greater reactivity toward HSV-1 than to HSV-2 infected cells, which could indicate binding to a non-shared epitope of gB (Fig. 7).
Figure 7.
Binding analysis of gB-specific antibodies from LYNDAL. Specificity of scFvs for binding to cell surface glycoprotein B of HSV-1 and HSV-2 infected Vero cells was analyzed by flow cytometry.
To accurately measure equilibrium constants (KD) of scFvs for binding to the target antigen in its natural context, we first performed flow cytometric affinity measurements of 12 randomly chosen scFv monomers on HSV-1 infected cells (Table 2). Of these, 11 scFvs bound to the target antigen with KD values in the nanomolar range. Subsequent surface plasmon resonance (SPR) affinity measurements of these clones confirmed the tight binding (r = 0.90) of selected scFvs (Table 2; Fig. S5). To evaluate the scFvs for mediating therapeutically relevant antiviral activity, we next tested their ability to prevent HSV infection in vitro using a standard plaque neutralization assay. Of eight scFvs with HSV-neutralizing activity, clone 28 exhibited the highest antiviral potency and was therefore further analyzed. We have previously shown that the valency of gB-specific antibodies may have a strong effect on their HSV-neutralizing capacity.48 Clone 28 eluted in gel filtration chromatography on a calibrated Superdex 75 column in two peaks at retention times correlating to the size of a monomer (~34 kDa, 84%) and a non-covalently associated dimeric (scFv)2 fragment (~57 kDa, 16%). Both antibody fractions were therefore separated by preparative size exclusion chromatography (Fig. S6) and further independently characterized.
Table 2. Affinity of monomeric scFvs.
| Flow cytometrya | Surface plasmon resonanceb | |||
|---|---|---|---|---|
| ScFv | KD ± SE [nM] | kon ± SD [105 M−1s−1] | koff ± SD [10−4 s−1] | KD ± SD [nM] |
| 1 | 19.1 ± 1.0 | 0.73 ± 0.07 | 8.67 ± 0.36 | 12.0 ± 1.6 |
| 4 | 6.1 ± 0.6 | NC | NC | NC |
| 5 | 587.0 ± 25.7 | ND | ND | ND |
| 9 | 3.2 ± 0.2 | 7.45 ± 2.05 | 4.83 ± 0.35 | 0.7 ± 0.2 |
| 10 | 11.6 ± 0.8 | 3.58 ± 0.26 | 15.30 ± 0.28 | 4.3 ± 0.4 |
| 11 | 7.0 ± 0.5 | 1.62 ± 0.15 | 6.43 ± 0.01 | 4.0 ± 0.4 |
| 22 | 28.9 ± 3.2 | 0.95 ± 0.01 | 40.45 ± 2.33 | 42.6 ± 2.3 |
| 28 | 15.5 ± 2.3 | 2.22 ± 0.08 | 16.90 ± 0.28 | 7.6 ± 0.2 |
| 30 | 8.5 ± 0.4 | 4.27 ± 0.59 | 3.95 ± 0.43 | 0.9 ± 0.1 |
| 31 | 6.6 ± 0.7 | NC | NC | NC |
| 33 | 11.1 ± 0.8 | 1.03 ± 0.07 | 12.25 ± 0.07 | 12.0 ± 0.6 |
| 34 | 7.2 ± 0.3 | 2.41 ± 0.95 | 8.82 ± 1.24 | 4.1 ± 2.1 |
a Binding affinities (KD) of SEC-purified scFvs to gB on the cell surface of HSV-1 infected cells were calculated from the equilibrium-binding curves as measured by flow cytometry. SE, standard error. bAssociation and dissociation rate constants of monomeric scFvs were determined by SPR using amine coupled gB-1 as ligand. Affinity constants were calculated as KD = koff/kon. Constants and errors were averaged from two independent determinations. SD, standard deviation; NC, not calculated; ND, not determined.
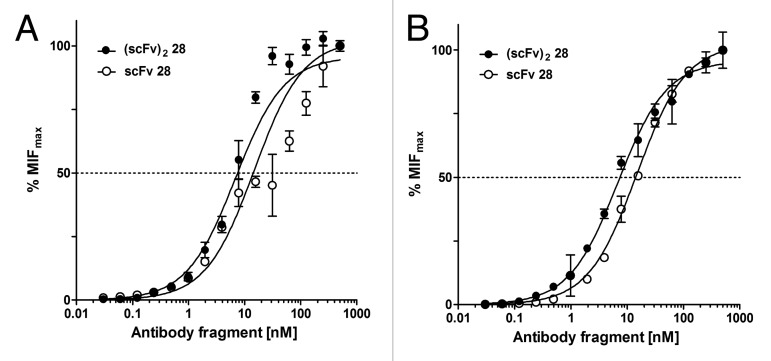
Bivalent binding of the dimeric (scFv)2 28 resulted in increased avidity to glycoprotein B on the surface of HSV infected cells with apparent equilibrium constants of 7.3 nM for HSV-1 and 6.8 nM for HSV-2. The monomeric scFv 28 showed a 2-fold weak binding (KD 15.5 nM for HSV-1 and 14.8 nM for HSV-2, respectively, Fig. 8). Similar affinities of scFv 28 and (scFv)2 28 for binding to both HSV-1 and HSV-2 infected cells indicated that this antibody must recognize an epitope that is shared by both strains. Evaluation of HSV-1 and HSV-2 neutralizing activity showed that the dimeric (scFv)2 28 indeed neutralized both HSV serotypes significantly better than the monomeric scFv (Fig. 9). Surprisingly, both monomeric scFv and dimeric (scFv)2 neutralized HSV-2 with a 14-fold and 7-fold higher efficacy than HSV-1. The efficacy of the dimeric (scFv)2 28 for neutralizing HSV-2 was even more favorable than that of a humanized mAb currently being developed for clinical applications in acyclovir-resistant disease.49
Figure 8.
Equilibrium-binding curves for antibody 28. Binding activities of monovalent scFv and bivalent (scFv)2 was measured on either (A) HSV-1 or (B) HSV-2 infected Vero cells by flow cytometry. Error bars represent standard deviations of the mean values. MFImax, maximum median fluorescence intensity.
Figure 9.
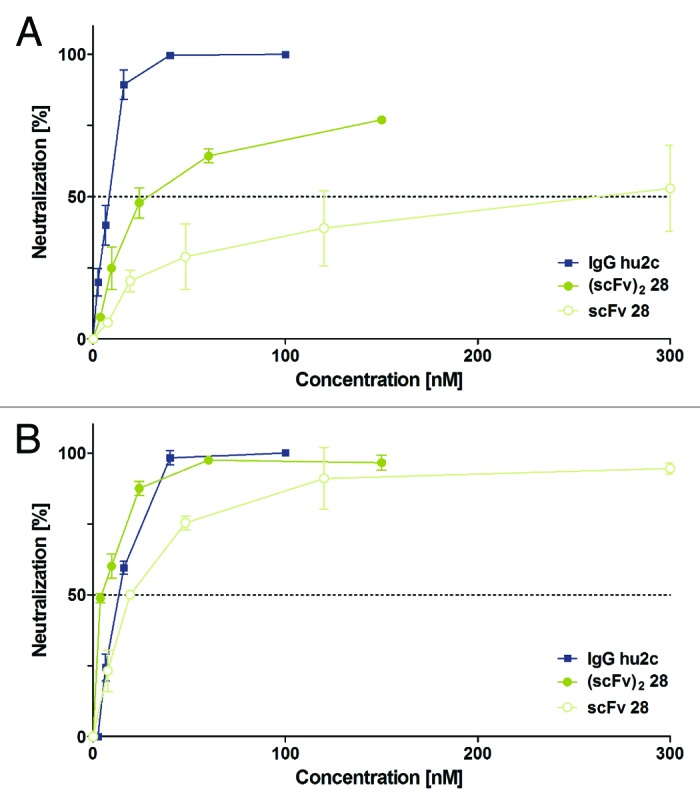
In vitro neutralization of HSV by LYNDAL-selected antibody. Efficacy of clone 28 for neutralizing HSV-1 strain F (A) or HSV-2 strain G (B) was studied by plaque reduction neutralization tests (PRNT) using Vero cells. Neutralization capacity was assessed with serial dilutions of either monovalent scFv or bivalent (scFv)2 and concentrations were determined that neutralized 50% of viruses (PRNT50). The neutralizing humanized monoclonal antibody, IgG hu2c, was used as a control and comparator. Experiments were performed in duplicate. Error bars represent standard deviations of the mean values.
Discussion
Humoral immune responses play a critical role in acquiring immunity against infectious disease. As demonstrated by single B cell expression cloning, single antibodies developed in the natural course of infection in patients may exhibit strong capacity for broadly neutralizing viral pathogens such as HIV-150-52 or influenza virus.53 Despite the still limited understanding of how these antibodies contribute to the actual control of especially chronic infectious diseases in a living organism, it seems clear that they undergo affinity maturation in lymphoid tissue.51 Retrieval of such particular V gene repertoires as a source for library construction is highly attractive because these repertoires may enable discovery and development of antibodies with potentially unique functional properties.
Access to human immune repertoires is generally restricted to peripheral blood and bone marrow. Here, we described an alternative approach for recovering functional mAbs from combinatorial antibody phage display libraries derived from lymph nodes as a source of immune repertoires. The underlying idea of the LYNDAL concept is based on the assumption that human immune libraries cloned from antigen-encountered antibody repertoires may have unique antigen-specific biological properties that can be exploited for therapeutic interventions.
Lymph nodes appear to be a highly valuable source for immune library cloning54 because fundamental processes of the humoral immunity take place within their germinal centers, such as somatic hypermutation, class switch recombination, and differentiation of clonally expanded B-cells into memory B and plasma cells.55,56 Moreover, lymph nodes contain a high number of antigen-encountered and activated B lymphocytes,57,58 including a conceptually interesting subset of re-entered and re-activated IgG positive memory cells harboring extensive somatic hypermutation.59 Since the selection of HIV-neutralizing mAbs from antigen-specific memory B cells has been reported,51 the memory B cell population in general might represent a crucial antibody gene pool for selecting antiviral antibodies with unique biological properties. Based on these considerations, we employed lymph node-derived (memory) B cell populations from 20 donors for cloning 20 independently combinable IgG repertoires. With a combined size of 3.2 × 109 independent antibody clones, LYNDAL represents, to our knowledge, one of the largest lymph node-derived antibody libraries to date.
To demonstrate the proof-of-concept for extracting high affinity antiviral antibodies from the combinatorial repertories of immunized donors, we combined sublibraries from individuals with high target-specific IgG titers for panning. HSV was chosen as the target for antibody selection because of the high infection prevalence in the population and the previously reported successful antibody selection from HSV-seropositive bone marrow libraries.60 Here, we have shown that screening of LYNDAL from donors with humoral response to HSV resulted in a high rate of unique high affinity binders with a broad representation of different germline sequences from various antibody families. Notably, variable regions of isolated scFvs had accumulated a particularly high degree of somatic hypermutations. As a result, the average number of mutated amino acids was higher than that reported for hyperimmunized vaccinated individuals (26.0 vs. 20.5) hypothesized to be representative for a fully matured humoral immune response.61 Thus, the LYNDAL concept allowed the selection of HSV-specific antibodies being matured during the course of a natural occurring immune response. To assess whether the high degree of somatic hypermutations would also translate into favorable functional properties of the selected antibodies, we performed HSV neutralization assays. Despite the quite limited number of analyzed clones, several scFvs exhibited potent antiviral activity. Most notably, neutralization properties of one clone (#28) were so remarkable that further exploration of the antiviral efficacy in HSV infected mice is warranted.
We conclude that antigen-encountered lymph node B cell repertoires from several donors may provide a valuable alternative for selecting high affinity antibodies with immunologically unique functional properties. It remains to be shown in the future if this may also translate into antibodies with superior therapeutic activity when compared to antibodies obtained from very large synthetic or semi-synthetic libraries. By showing the value of lymph node derived-immune libraries for isolating in vivo matured mAbs with promising virus-neutralizing properties, we expect the LYNDAL concept to be also extendable to other disease-specific targets.
Materials and Methods
Ethics statement and lymph node collection
The study was approved by the ethics committee of the Faculty of Medicine, Heidelberg University. All patients provided written informed consent. Lymph nodes were obtained from 20 donors undergoing surgery. Dissected lymph nodes were transferred directly into RNAlater reagent (Qiagen, 76163) and stored at −20 °C. Individual lymph nodes were thawed to room temperature (RT) and homogenized using a TissueRuptor (Qiagen). Total RNA was isolated using the RNeasy Lipid Tissue Midi Kit (Qiagen, 75842), and single stranded cDNA was prepared using a First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, K1612).
Construction of LYNDAL
Libraries were generated by randomly combining amplified human VH and VL genes into phagemid vector pHENIS, a derivative of pHEN,62 containing a hexahistidine and a c-myc-tag encoding sequence. Variable domain genes were cloned as scFv fragments in VH/VL orientation linked by a 15 amino acid encoding peptide (G4S)2GGSAQ. IgG V region repertoires of human donors were amplified in a two-step, semi-nested PCR strategy using primers with a maximum of two degenerated nucleotide positions. Forward primer design was based on all functional variable gene sequences as represented in VBASE (http://www2.mrc-lmb.cam.ac.uk/vbase/). Reverse primers bound to constant genes (kappa, lambda, and CH1) or to J encoding segments and were designed on the basis of sequence information of the Kabat63 and VBASE database, respectively. VH, VL-kappa, and VL-lambda encoding gene repertoires were separately amplified from prepared cDNA in several independent PCRs using 2 units/reaction of Pfu polymerase (Thermo Fisher Scientific, EP0502). Antibody genes were PCR-amplified during the first set of PCRs (1st PCRs, 30 cycles), followed by re-amplification of PCR products (2nd PCRs, 15 cycles) to introduce cloning restriction sites by using the same PCR program: 94 °C for 3 min for initial denaturation, followed by cycles of 94 °C for 30 s, 55 °C for 1 min, 72 °C for 2 min, and 72 °C for 10 min final elongation. Amplified VH gene repertoires were subcloned as SfiI/XhoI-fragments, followed by electroporation of phagemids into electrocompetent TG1 E. coli cells (Agilent Technologies, 200123). Light chain kappa or lambda repertoires were separately cloned as ApaLI/NotI-fragments into the VH-containing phagemid. Library-containing phagemid vectors were transformed by electroporation (n = 88–132 per donor) into E. coli, thus creating two independent libraries (VH/VL-kappa and VH/VL-lambda). Transformed bacteria were spread on 145 mm 2xYT agar plates containing 2% (w/v) glucose, and 100 µg/ml ampicillin (2xYT-GA). After overnight (o/n) growth at 30 °C, individual donor libraries were stored as glycerol (15% final) stocks in aliquots at −80 °C.
Characterization of LYNDAL
To determine LYNDAL sizes, two transformed samples per sublibrary were randomly chosen, incubated for 1 h at 37 °C before 10-fold serial dilutions of cells were plated on 2xYT-GA agar plates. The next day, the respective library size was calculated by extrapolating the average bacterial number from dilutions with countable colonies. Up to 40 randomly picked transformants per sublibrary were further analyzed by colony PCR for successful integration of both heavy and light chain genes (fragment size ~1kb). Sequences from at least five intact, randomly chosen scFvs per sublibrary were determined and analyzed by sequence alignment software DNAPLOT (http://www2.mrc-lmb.cam.ac.uk/vbase/dnaplot2.php) to identify the closest germline V, (D), and J segment genes as deposited in VBASE. Complementarity-determining regions (CDRs) were additionally analyzed by using the Fab Analysis tool (http://www.vbase2.org/vbscAb.php) by aligning the scFv sequences to the VBASE2 database.64 Relative gene family occurrences of sequenced LYNDAL clones were calculated and compared with the mean distribution of all functional V gene entries of human antibody databases VBASE, VBASE2, and IMGT-GENE.65 Clones with non-functional reading frames, orphans, and pseudogenes, were excluded from the analysis.
Each sublibrary was analyzed for soluble expression of antibody-pIII fusions in E. coli by dot blot and western blot analysis. Individual colonies were inoculated in 96-well plates in 2xYT-GA medium and grown o/n at 37 °C on a shaker. Induction plates containing 150 µl 2xYT-A with 0.1% glucose were inoculated with 3 µl o/n cultures and incubated for an additional 3 h until soluble expression of antibody-pIII fusions was induced by adding 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). After o/n expression at 28 °C, pelleted bacteria (3000 xg, 10 min, 4 °C) were resuspended in periplasmic preparation buffer (30 mM TRIS-HCl, pH 8.0, 1 mM EDTA, 20% sucrose) containing 50 µg/ml freshly prepared lysozyme and incubated for 30 min on ice. Then MgSO4 was added to a final concentration of 10 mM. Clarified periplasmic fractions (3000 xg, 20 min, 4 °C) were either applied onto nitrocellulose membranes using a Minifold I Dot Blot system (Whatman) for dot blot analysis (100 µl/dot) or onto 12% SDS-PAGE under reducing conditions for western blot analysis. Antibody-pIII fusions were detected using either peroxidase-conjugated 9E10 anti-c-myc antibody (1 µg/ml; Roche Diagnostics, 11814150001) or mouse anti-pIII antibody (0.5 µg/ml; MoBiTec, PSKAN3) as first and goat anti-mouse IgG peroxidase conjugate (0.02 µg/ml; Jackson ImmunoResearch Laboratories, 115-035-008) as secondary antibody. Peroxidase activity was detected by chemiluminescence using ECL substrate (Thermo Fisher Scientific, 32106) and chemical film Curix HT1.000G Plus (Agfa HealthCare).
Antibody selection
Selection of herpes simplex virus specific scFvs, using antigen-coated immunotubes, was performed as described previously66 with the following modifications. Antibody repertoires of seven LYNDAL donors were separately packaged by inoculating 2× YT-GA medium (200 ml/donor) with corresponding library glycerol stocks, followed by superinfection with helper phage VCSM13 (Agilent Technologies) at a multiplicity of infection of 15. Expression of antibody-pIII fusions was induced by resuspending the pelleted bacteria in induction medium (2× YT, 100 µg/ml ampicillin, 25 µg/ml kanamycin, 0.5 mM IPTG) and incubation of combined libraries on a shaker at 28 °C for 6 h. Phages were purified during two PEG-precipitation steps by incubation o/n at 4 °C and 1 h on ice, respectively. After pre-incubation of 2 × 1013 t.u. (transducing units) in 2% MPBS for 1 h at RT, phages were applied without depletion step to a gB(724tHis)-coated67 (20 µg/ml in PBS) and blocked immunotube. Binding of phages to target protein gB-1 occurred during roll-over shaking for 90 min and 30 min rest at RT. After washing (5× PBST and 5× PBS), bound phage antibodies were eluted with acid solution (0.1 M glycine-HCl, 0.5 M NaCl, pH 2.2) by incubation for 8 min at RT under rotation. Eluted phage antibodies were neutralized (pH 7.4) with 1 M TRIS-HCl pH 9.5 and added to log-phase TG1. To rescue non-eluted phages, the selection tube was additionally filled with log-phase bacteria. After infection at 37 °C, both cultures were combined before continuing as described.66 Two further selection rounds were performed by preparing 160 ml start cultures and increasing the stringency during panning by employing less gB-1 antigen for coating (5 µg/ml) and more washing cycles (10× for round 2 and 15x for round 3 with PBS and PBST, respectively).
Phage ELISA
Polyclonal phage ELISA (ppELISA) was performed as described previously66 by adding 1011 t.u./well of rescued phages to the antigen-coated microtiter plates. For monoclonal phage ELISA (mpELISA), single colonies from positively enriched panning rounds were inoculated into 96-well microtiter plates containing 100 µl of 2× YT-GA. Cultures were shaken o/n at 37 °C and used for inoculation of an infection plate with fresh medium; the master clones were stored by adding glycerol. After incubation of infection plates for 2 h at 37 °C, bacteria were superinfected with 108 t.u./well of helper phages at 37 °C, followed by re-suspension of the pellets in induction medium for antibody-pIII expression o/n at 28 °C under shaking. The next day, each clone was analyzed for binding to coated selection antigen (1 µg/ml gB-1 in PBS) and control antigen (2% MPBS), respectively, by incubation of blocked wells (2% MPBS for 1 h at RT) with 100 µl/well phage supernatant in 2% MPBS for 1 h at RT. Detection using an HRP-conjugated anti-M13 antibody (GE Healthcare, 27-9421-01) was performed as described.66 The absorbance was read at 450 nm using a GENios Plus reader (Tecan). Readings for gB-1 ten times higher than the average signal for milk protein were considered to be antigen-specific binding.
Characterization and expression of enriched scFvs
The integrity of the scFv gene from specific binders was assessed by colony PCR and re-amplified genes were digested with BstNI for fingerprint analysis. After sequencing, germline sequences and respective antibody gene families were determined by DNAPLOT, followed by the construction of phylogenetic trees with the Phylogeny.fr web tool.68 Somatic hypermutation analysis of selected scFvs was performed by employing the IMGT/V-QUEST online tool.69
ScFv encoding genes were PCR-amplified and subcloned as SfiI/NotI fragments into expression vector pAB170 for soluble protein expression in the bacterial periplasm (o/n at 18 °C) as described previously.71 Hexahistidine-tagged scFvs were purified by IMAC with Ni-Sepharose 6 Fast Flow (GE Healthcare, 17-5318-01) according to the manufacturer’s recommendations. The oligomeric state of IMAC-purified scFvs was analyzed after buffer exchange to PBS by SEC on a Superdex 75 10/300 GL column using an ÄKTA FPLC system (both GE Healthcare). The column was calibrated using gel filtration low molecular weight standards (Amersham Biosciences, 17-0442-01). Elution profiles were recorded by monitoring the absorbance at 280 nm and elution volumes determined with the Unicorn software 5.11. If necessary, scFvs were further purified to homogeneity by preparative SEC on a HiLoad 16/60 Superdex 75 column (GE Healthcare). Purified scFvs were concentrated with centrifugal filter units, filter sterilized, and concentrations were measured spectrophotometrically.
Target binding analysis on virus infected cells
Specific binding of purified scFvs to native gB protein was evaluated by flow cytometry using the African green monkey kidney cell line Vero (ECACC, 84113001) either infected or non-infected with HSV-1(F) or HSV-2(G). Infection and fluorescence measurements were performed as described previously48 by detection of bound scFvs (100 nM) with mouse anti-c-myc IgG (5 µg/ml) and goat anti-mouse fluorescein isothiocyanate (FITC) conjugate (7.5 µg/ml, Jackson ImmunoResearch Laboratories, 115–095–008). Fluorescence was measured on a FACSCanto II flow cytometer and mean fluorescence intensity calculated using FACS Diva software (Becton Dickinson). For determination of equilibrium-binding curves, HSV-1 or HSV-2 infected Vero cells were incubated in triplicate with scFvs dilutions (0.03–1000 nM) followed by detection as described above. Background fluorescence was subtracted from measured median fluorescence intensities, and relative affinities were calculated by nonlinear regression using GraphPad Prism version 5.0 (GraphPad Software).
SPR biosensor binding analysis
Binding kinetics were determined by SPR on a BIACORE 2000 system (GE Healthcare). A standard coupling protocol was employed for immobilizing gB(724tHis) via exposed primary amines on a CM5 sensor chip (GE Healthcare, BR-1003–99). Surface activation of flow cells No.1 and No.2 for 7 min was performed with a 1:1 mixture of 0.1 M NHS (N-hydroxysuccinimide) and 0.4 M EDC (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride) at a flow rate of 5 µl/min. After immobilization of the ligand (100 µg/ml in 10 mM sodium acetate, pH 4.7; 7 min, flow rate 5 µl/min) at a density of 230 RU on flow cell, 2 residual reactive sites were blocked by injection of 1 M ethanolamine, pH 8.5 for 7 min at 5 µl/min. Flow cell 1 served as a reference surface. To measure kinetics, increasing concentrations (1 nM, 10 nM, 50 nM, 100 nM, 200 nM, 500 nM, and 1000 nM) of purified monomeric scFvs were injected in running buffer (10 mM HEPES, 150 mM NaCl, 0.05% P20, pH 7.0) at a flow rate of 10 µl/min and 25 °C. Analyte-ligand association and dissociation phases were monitored for 3 min and 30 min, respectively. Surfaces were regenerated in running buffer for further 30 min. The data were globally fitted with the Langmuir 1:1 binding model using the BIAevaluation 4.1.1 software.
Neutralization of HSV-1 and HSV-2
Neutralizing activity of LYNDAL antibodies was determined in a plaque reduction assay as described previously.49 The humanized version of a highly potent HSV-neutralizing antibody was used as positive control.49
Supplementary Material
Acknowledgments
We thank Roselyn J. Eisenberg and Gary H. Cohen for generous donation of the recombinant gB-1 protein. This work has in part been funded by the Bundesministerium für Bildung und Forschung (BMBF; 01EZ0934), and the Klaus Tschira Stiftung (KTS, 00.150.2009).
Glossary
Abbreviations:
- CDR
complementarity-determining region
- FR
framework region
- gB
glycoprotein B of herpes simplex virus
- HSV
herpes simplex virus
- IgG
immunoglobulin G
- LYNDAL
LYmph Node Derived Antibody Libraries
- PRNT
plaque reduction neutralization test
- scFv
single-chain variable fragment
- VH
variable region of the heavy chain
- VL
variable region of the light chain
Disclosure of Potential Conflicts of Interest
The authors declare no conflict of interest.
Supplemental Materials
Supplemental materials may be found here: https://www.landesbioscience.com/journals/mabs/article/27236/
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/27236
References
- 1.Nelson AL, Dhimolea E, Reichert JM. Development trends for human monoclonal antibody therapeutics. Nat Rev Drug Discov. 2010;9:767–74. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- 2.Reichert JM. Antibodies to watch in 2013: Mid-year update. MAbs. 2013;5:513–7. doi: 10.4161/mabs.24990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. 1975. J Immunol. 2005;174:2453–5. [PubMed] [Google Scholar]
- 4.Sullivan M, Kaur K, Pauli N, Wilson PC. Harnessing the immune system’s arsenal: producing human monoclonal antibodies for therapeutics and investigating immune responses. F1000 Biol Rep. 2011;3:17. doi: 10.3410/B3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson PC, Andrews SF. Tools to therapeutically harness the human antibody response. Nat Rev Immunol. 2012;12:709–19. doi: 10.1038/nri3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwakkenbos MJ, Diehl SA, Yasuda E, Bakker AQ, van Geelen CM, Lukens MV, van Bleek GM, Widjojoatmodjo MN, Bogers WM, Mei H, et al. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med. 2010;16:123–8. doi: 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Traggiai E, Becker S, Subbarao K, Kolesnikova L, Uematsu Y, Gismondo MR, Murphy BR, Rappuoli R, Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–5. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu X, Tsibane T, McGraw PA, House FS, Keefer CJ, Hicar MD, Tumpey TM, Pappas C, Perrone LA, Martinez O, et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455:532–6. doi: 10.1038/nature07231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–7. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 10.Green LL. Antibody engineering via genetic engineering of the mouse: XenoMouse strains are a vehicle for the facile generation of therapeutic human monoclonal antibodies. J Immunol Methods. 1999;231:11–23. doi: 10.1016/S0022-1759(99)00137-4. [DOI] [PubMed] [Google Scholar]
- 11.Lonberg N. Human antibodies from transgenic animals. Nat Biotechnol. 2005;23:1117–25. doi: 10.1038/nbt1135. [DOI] [PubMed] [Google Scholar]
- 12.Bradbury AR, Marks JD. Antibodies from phage antibody libraries. J Immunol Methods. 2004;290:29–49. doi: 10.1016/j.jim.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 13.McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–4. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 14.Geyer CR, McCafferty J, Dübel S, Bradbury AR, Sidhu SS. Recombinant antibodies and in vitro selection technologies. Methods Mol Biol. 2012;901:11–32. doi: 10.1007/978-1-61779-931-0_2. [DOI] [PubMed] [Google Scholar]
- 15.Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol. 2005;23:1105–16. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- 16.de Haard HJ, van Neer N, Reurs A, Hufton SE, Roovers RC, Henderikx P, de Bruïne AP, Arends JW, Hoogenboom HR. A large non-immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. J Biol Chem. 1999;274:18218–30. doi: 10.1074/jbc.274.26.18218. [DOI] [PubMed] [Google Scholar]
- 17.Lloyd C, Lowe D, Edwards B, Welsh F, Dilks T, Hardman C, Vaughan T. Modelling the human immune response: performance of a 1011 human antibody repertoire against a broad panel of therapeutically relevant antigens. Protein Eng Des Sel. 2009;22:159–68. doi: 10.1093/protein/gzn058. [DOI] [PubMed] [Google Scholar]
- 18.Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222:581–97. doi: 10.1016/0022-2836(91)90498-U. [DOI] [PubMed] [Google Scholar]
- 19.Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, McCafferty J, Hodits RA, Wilton J, Johnson KS. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol. 1996;14:309–14. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- 20.Burton DR, Barbas CF, 3rd, Persson MA, Koenig S, Chanock RM, Lerner RA. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci U S A. 1991;88:10134–7. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai X, Garen A. Anti-melanoma antibodies from melanoma patients immunized with genetically modified autologous tumor cells: selection of specific antibodies from single-chain Fv fusion phage libraries. Proc Natl Acad Sci U S A. 1995;92:6537–41. doi: 10.1073/pnas.92.14.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chapal N, Chardès T, Bresson D, Pugnière M, Mani JC, Pau B, Bouanani M, Péraldi-Roux S. Thyroid peroxidase autoantibodies obtained from random single chain FV libraries contain the same heavy/light chain combinations as occur in vivo. Endocrinology. 2001;142:4740–50. doi: 10.1210/en.142.11.4740. [DOI] [PubMed] [Google Scholar]
- 23.de Carvalho Nicacio C, Williamson RA, Parren PW, Lundkvist A, Burton DR, Björling E. Neutralizing human Fab fragments against measles virus recovered by phage display. J Virol. 2002;76:251–8. doi: 10.1128/JVI.76.1.251-258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabibov AG, Belogurov AA, Jr., Lomakin YA, Zakharova MY, Avakyan ME, Dubrovskaya VV, Smirnov IV, Ivanov AS, Molnar AA, Gurtsevitch VE, et al. Combinatorial antibody library from multiple sclerosis patients reveals antibodies that cross-react with myelin basic protein and EBV antigen. FASEB J. 2011;25:4211–21. doi: 10.1096/fj.11-190769. [DOI] [PubMed] [Google Scholar]
- 25.Kashyap AK, Steel J, Oner AF, Dillon MA, Swale RE, Wall KM, Perry KJ, Faynboym A, Ilhan M, Horowitz M, et al. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc Natl Acad Sci U S A. 2008;105:5986–91. doi: 10.1073/pnas.0801367105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SJ, Jang MH, Stapleton JT, Yoon SO, Kim KS, Jeon ES, Hong HJ. Neutralizing human monoclonal antibodies to hepatitis A virus recovered by phage display. Virology. 2004;318:598–607. doi: 10.1016/j.virol.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 27.Kramer RA, Marissen WE, Goudsmit J, Visser TJ, Clijsters-Van der Horst M, Bakker AQ, de Jong M, Jongeneelen M, Thijsse S, Backus HH, et al. The human antibody repertoire specific for rabies virus glycoprotein as selected from immune libraries. Eur J Immunol. 2005;35:2131–45. doi: 10.1002/eji.200526134. [DOI] [PubMed] [Google Scholar]
- 28.Mao S, Gao C, Lo CH, Wirsching P, Wong CH, Janda KD. Phage-display library selection of high-affinity human single-chain antibodies to tumor-associated carbohydrate antigens sialyl Lewisx and Lewisx. Proc Natl Acad Sci U S A. 1999;96:6953–8. doi: 10.1073/pnas.96.12.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Persson MA, Caothien RH, Burton DR. Generation of diverse high-affinity human monoclonal antibodies by repertoire cloning. Proc Natl Acad Sci U S A. 1991;88:2432–6. doi: 10.1073/pnas.88.6.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raats JM, Wijnen EM, Pruijn GJ, van den Hoogen FH, van Venrooij WJ. Recombinant human monoclonal autoantibodies specific for citrulline-containing peptides from phage display libraries derived from patients with rheumatoid arthritis. J Rheumatol. 2003;30:1696–711. [PubMed] [Google Scholar]
- 31.Thie H, Toleikis L, Li J, von Wasielewski R, Bastert G, Schirrmann T, Esteves IT, Behrens CK, Fournes B, Fournier N, et al. Rise and fall of an anti-MUC1 specific antibody. PLoS One. 2011;6:e15921. doi: 10.1371/journal.pone.0015921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zebedee SL, Barbas CF, 3rd, Hom YL, Caothien RH, Graff R, DeGraw J, Pyati J, LaPolla R, Burton DR, Lerner RA, et al. Human combinatorial antibody libraries to hepatitis B surface antigen. Proc Natl Acad Sci U S A. 1992;89:3175–9. doi: 10.1073/pnas.89.8.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, Hoess A, Wölle J, Plückthun A, Virnekäs B. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J Mol Biol. 2000;296:57–86. doi: 10.1006/jmbi.1999.3444. [DOI] [PubMed] [Google Scholar]
- 34.Prassler J, Thiel S, Pracht C, Polzer A, Peters S, Bauer M, Nörenberg S, Stark Y, Kölln J, Popp A, et al. HuCAL PLATINUM, a synthetic Fab library optimized for sequence diversity and superior performance in mammalian expression systems. J Mol Biol. 2011;413:261–78. doi: 10.1016/j.jmb.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 35.Rothe C, Urlinger S, Löhning C, Prassler J, Stark Y, Jäger U, Hubner B, Bardroff M, Pradel I, Boss M, et al. The human combinatorial antibody library HuCAL GOLD combines diversification of all six CDRs according to the natural immune system with a novel display method for efficient selection of high-affinity antibodies. J Mol Biol. 2008;376:1182–200. doi: 10.1016/j.jmb.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 36.Silacci M, Brack S, Schirru G, Mårlind J, Ettorre A, Merlo A, Viti F, Neri D. Design, construction, and characterization of a large synthetic human antibody phage display library. Proteomics. 2005;5:2340–50. doi: 10.1002/pmic.200401273. [DOI] [PubMed] [Google Scholar]
- 37.Tiller T, Schuster I, Deppe D, Siegers K, Strohner R, Herrmann T, Berenguer M, Poujol D, Stehle J, Stark Y, et al. A fully synthetic human Fab antibody library based on fixed VH/VL framework pairings with favorable biophysical properties. MAbs. 2013;5:5. doi: 10.4161/mabs.24218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbas CF, 3rd, Bain JD, Hoekstra DM, Lerner RA. Semisynthetic combinatorial antibody libraries: a chemical solution to the diversity problem. Proc Natl Acad Sci U S A. 1992;89:4457–61. doi: 10.1073/pnas.89.10.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, Crosby WL, Kontermann RE, Jones PT, Low NM, Allison TJ, et al. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 1994;13:3245–60. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoet RM, Cohen EH, Kent RB, Rookey K, Schoonbroodt S, Hogan S, Rem L, Frans N, Daukandt M, Pieters H, et al. Generation of high-affinity human antibodies by combining donor-derived and synthetic complementarity-determining-region diversity. Nat Biotechnol. 2005;23:344–8. doi: 10.1038/nbt1067. [DOI] [PubMed] [Google Scholar]
- 41.Pini A, Viti F, Santucci A, Carnemolla B, Zardi L, Neri P, Neri D. Design and use of a phage display library. Human antibodies with subnanomolar affinity against a marker of angiogenesis eluted from a two-dimensional gel. J Biol Chem. 1998;273:21769–76. doi: 10.1074/jbc.273.34.21769. [DOI] [PubMed] [Google Scholar]
- 42.Söderlind E, Strandberg L, Jirholt P, Kobayashi N, Alexeiva V, Aberg AM, Nilsson A, Jansson B, Ohlin M, Wingren C, et al. Recombining germline-derived CDR sequences for creating diverse single-framework antibody libraries. Nat Biotechnol. 2000;18:852–6. doi: 10.1038/78458. [DOI] [PubMed] [Google Scholar]
- 43.Kalinke U, Bucher EM, Ernst B, Oxenius A, Roost HP, Geley S, Kofler R, Zinkernagel RM, Hengartner H. The role of somatic mutation in the generation of the protective humoral immune response against vesicular stomatitis virus. Immunity. 1996;5:639–52. doi: 10.1016/S1074-7613(00)80277-0. [DOI] [PubMed] [Google Scholar]
- 44.Kalinke U, Oxenius A, Lopez-Macias C, Zinkernagel RM, Hengartner H. Virus neutralization by germ-line vs. hypermutated antibodies. Proc Natl Acad Sci U S A. 2000;97:10126–31. doi: 10.1073/pnas.97.18.10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat Rev Microbiol. 2011;9:369–81. doi: 10.1038/nrmicro2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hellenbrand W, Thierfelder W, Müller-Pebody B, Hamouda O, Breuer T. Seroprevalence of herpes simplex virus type 1 (HSV-1) and type 2 (HSV-2) in former East and West Germany, 1997-1998. Eur J Clin Microbiol Infect Dis. 2005;24:131–5. doi: 10.1007/s10096-005-1286-x. [DOI] [PubMed] [Google Scholar]
- 47.Xu F, Markowitz LE, Gottlieb SL, Berman SM. Seroprevalence of herpes simplex virus types 1 and 2 in pregnant women in the United States. Am J Obstet Gynecol. 2007;196:e1–6. doi: 10.1016/j.ajog.2006.07.051. [DOI] [PubMed] [Google Scholar]
- 48.Krawczyk A, Krauss J, Eis-Hübinger AM, Däumer MP, Schwarzenbacher R, Dittmer U, Schneweis KE, Jäger D, Roggendorf M, Arndt MA. Impact of valency of a glycoprotein B-specific monoclonal antibody on neutralization of herpes simplex virus. J Virol. 2011;85:1793–803. doi: 10.1128/JVI.01924-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krawczyk A, Arndt MA, Grosse-Hovest L, Weichert W, Giebel B, Dittmer U, Hengel H, Jäger D, Schneweis KE, Eis-Hübinger AM, et al. Overcoming drug-resistant herpes simplex virus (HSV) infection by a humanized antibody. Proc Natl Acad Sci U S A. 2013;110:6760–5. doi: 10.1073/pnas.1220019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mouquet H, Klein F, Scheid JF, Warncke M, Pietzsch J, Oliveira TY, Velinzon K, Seaman MS, Nussenzweig MC. Memory B cell antibodies to HIV-1 gp140 cloned from individuals infected with clade A and B viruses. PLoS One. 2011;6:e24078. doi: 10.1371/journal.pone.0024078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–40. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 52.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–7. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–71. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yip YL, Hawkins NJ, Clark MA, Ward RL. Evaluation of different lymphoid tissue sources for the construction of human immunoglobulin gene libraries. Immunotechnology. 1997;3:195–203. doi: 10.1016/S1380-2933(97)00013-4. [DOI] [PubMed] [Google Scholar]
- 55.Shlomchik MJ, Weisel F. Germinal center selection and the development of memory B and plasma cells. Immunol Rev. 2012;247:52–63. doi: 10.1111/j.1600-065X.2012.01124.x. [DOI] [PubMed] [Google Scholar]
- 56.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–57. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 57.McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-specific memory B cell development. Annu Rev Immunol. 2005;23:487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
- 58.Tangye SG, Tarlinton DM. Memory B cells: effectors of long-lived immune responses. Eur J Immunol. 2009;39:2065–75. doi: 10.1002/eji.200939531. [DOI] [PubMed] [Google Scholar]
- 59.Bende RJ, van Maldegem F, Triesscheijn M, Wormhoudt TA, Guijt R, van Noesel CJ. Germinal centers in human lymph nodes contain reactivated memory B cells. J Exp Med. 2007;204:2655–65. doi: 10.1084/jem.20071006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williamson RA, Burioni R, Sanna PP, Partridge LJ, Barbas CF, 3rd, Burton DR. Human monoclonal antibodies against a plethora of viral pathogens from single combinatorial libraries. Proc Natl Acad Sci U S A. 1993;90:4141–5. doi: 10.1073/pnas.90.9.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poulsen TR, Jensen A, Haurum JS, Andersen PS. Limits for antibody affinity maturation and repertoire diversification in hypervaccinated humans. J Immunol. 2011;187:4229–35. doi: 10.4049/jimmunol.1000928. [DOI] [PubMed] [Google Scholar]
- 62.Hoogenboom HR, Griffiths AD, Johnson KS, Chiswell DJ, Hudson P, Winter G. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 1991;19:4133–7. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson G, Wu TT. Kabat Database and its applications: future directions. Nucleic Acids Res. 2001;29:205–6. doi: 10.1093/nar/29.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Retter I, Althaus HH, Münch R, Müller W. VBASE2, an integrative V gene database. Nucleic Acids Res. 2005;33:D671–4. doi: 10.1093/nar/gki088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giudicelli V, Chaume D, Lefranc MP. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33:D256–61. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kontermann RE. Immunotube Selections. In: Kontermann RE, Dübel S, eds. Antibody Engineering 2010:127-37. [Google Scholar]
- 67.Bender FC, Samanta M, Heldwein EE, de Leon MP, Bilman E, Lou H, Whitbeck JC, Eisenberg RJ, Cohen GH. Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J Virol. 2007;81:3827–41. doi: 10.1128/JVI.02710-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465-9. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503-8. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kontermann RE, Martineau P, Cummings CE, Karpas A, Allen D, Derbyshire E, Winter G. Enzyme immunoassays using bispecific diabodies. Immunotechnology. 1997;3:137–44. doi: 10.1016/S1380-2933(97)00010-9. [DOI] [PubMed] [Google Scholar]
- 71.Müller D, Karle A, Meissburger B, Höfig I, Stork R, Kontermann RE. Improved pharmacokinetics of recombinant bispecific antibody molecules by fusion to human serum albumin. J Biol Chem. 2007;282:12650–60. doi: 10.1074/jbc.M700820200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.