Abstract
Lectins are valuable tools for detecting specific glycans in biological samples, but the interpretation of the measurements can be ambiguous due to the complexities of lectin specificities. Here we present an approach to improve the accuracy of interpretation by converting lectin measurements into quantitative predictions of the presence of various glycan motifs. The conversion relies on a database of analyzed glycan array data that provides information on the specificities of the lectins for each of the motifs. We tested the method using measurements of lectin binding to glycans on glycan arrays and found that the combined measurements from several lectins are more accurate than individual measurements for predicting the presence or absence of motifs. We then applied the method to predicting motifs on the protein MUC1 expressed in eight different pancreatic cancer cell lines. Each cell line expressed a unique pattern of MUC1 glycoforms, and the glycoforms significantly differed between MUC1 collected from conditioned media and MUC1 collected from cell lysates. This new method could provide more accurate analyses of glycans in biological sample and make the use of lectins more practical and effective for a broad range of researchers.
Introduction
Molecular biomarkers are becoming more important in cancer care. Because cancers with outwardly similar appearances have major differences at the molecular level [1], physicians need strategies to detect, diagnose, and treat cancers of defined molecular subtypes. Molecular biomarkers are the tools needed to apply the optimized strategies. Given the diversity between cancers in molecular characteristics and clinical needs, the ongoing requirements for new biomarkers will be extensive [2]. For example, for certain cancers, physicians may struggle with the decision to perform surgery or the choice between treatment options. For other cancers, the physicians may have great difficulty differentiating cancers from benign conditions. Researchers are devoting significant resources to identifying molecular biomarkers that provide more precise information. These efforts have produced a variety of new tests, but in general the generation of effective biomarkers has been slow and difficult.
An approach to developing accurate biomarkers is to detect the glycan modifications on specific glycoproteins [3]. The carbohydrate modifications on a protein can influence the protein's structure and function in healthy and disease conditions [4-7]. Certain cell types can modify the glycosylation on a protein in response to changing conditions without altering the level of protein production. For that reason, the detection of certain glycoforms of a protein can provide more accurate information about a disease than the detection of the total protein abundance. Several research groups have demonstrated the potential for improved biomarkers based on this concept [8-19].
An important step in developing biomarkers based on glycan alterations is to characterize the glycosylation on individual proteins in clinical specimens. Such information would help researchers to optimize the detection of the molecular features most associated with a particular condition. But obtaining that information for individual proteins derived from clinical samples is difficult using conventional methods, such as those involving enzymatic digestions, chromatography, and mass spectrometry [7]. More protein is required than typically available from clinical samples, and because of the many processing steps involved, precise comparisons between samples in the levels of protein glycoforms are not possible. An alternate approach for studying protein glycosylation is to use affinity reagents, such as lectins and glycan-binding antibodies [20-22]. Lectins are proteins that bind specific glycans, so they are useful as probes to measure the level of a glycan structure on a protein or in a sample. Assays based on affinity reagents are well suited to biomarker research because they can provide precise measurements over many samples using a small amount of each sample.
A limitation in the use of lectins to detect glycans is the ambiguity in the interpretation of the measurements. Each lectin has a unique set of glycans that it binds. A lectin's specificity usually is represented as the primary, simplified glycan motif that it binds. For example, the specificity of the lectin from aleuria aurantia typically is defined as alpha-linked fucose. When a researcher uses a lectin to detect a glycan, the researcher typically infers the presence or absence of the primary target of the lectin based on the amount of lectin binding. However, the specificities of most lectins are more complex than indicated by the simplified primary target. Certain lectins strongly bind a specific glycan motif but also bind other, related motifs more weakly. For example, the lectin from the snail species helix pomatia binds terminal, alpha-linked N-acetylgalactosamine but also binds terminal, alpha-linked N-acetylglucosamine [23]. Other lectins do not always bind their primary target, depending on the nature of the complete glycan structure. In such cases, significant uncertainty might remain about which glycans are present in a sample.
A strategy to provide more precision in the interpretation of lectin measurements is to use quantified specificities of each lectin. For example, when interpreting the binding of a lectin to a sample, instead of making a judgment based on experience and personal knowledge, the researcher could use an algorithm to give the probabilities that various glycan motifs are present in the sample. Such quantitative interpretation could more accurately account for the complexities in lectin specificities and would remove the burden from researchers for acquiring a detailed knowledge of the subtleties of each lectin. In addition, quantitative interpretation could enable the use of combined measurements from multiple lectins to get more information about a sample. An individual lectin can give good information about the presence of a motif but with some ambiguity, because most lectins bind a few related motifs. The use of several lectins together, chosen to probe a particular motif, could account for some of the ambiguities of individual lectins.
Here we tested whether an algorithm to interpret lectin binding based on quantified lectin specificities gives better accuracy than the qualitative approach currently used. We obtained the quantified lectin specificities from our previous analyses of lectin binding to glycan arrays [24-26]. We optimized and tested the method using glycan array data and then applied it to the study of glycans on individual proteins in cell culture.
Materials and Methods
Glycan array data and analysis
We obtained and analyzed glycan array data from the Consortium for Functional Glycomics website (www.functionalglycomics.org) as previously described [26]. We developed the algorithms for motif prediction in IPython (http://ipython.org) and the scripts for processing antibody array data in Octave (http://wiki.octave.org) and Microsoft Office Excel. We produced the graphs using Matplotlib (http://matplotlib.org/citing.html).
The method for selecting panels of lectins that specifically detected each motif was based on negative selection. Starting with all lectins with motif scores > = 3 for a particular motif, a script calculated the accuracy for all possible panels of size N-1, where N is the number of starting lectins. (The accuracy was the percentage of glycans on the array that were correctly identified as containing or not containing the motif.) We chose the panel with the best accuracy and repeated until reaching a panel of 5 lectins. Next we further reduced the panel size if one of the smaller panels gave better accuracy than the larger panel.
Antibodies and lectins
The antibodies and lectins were obtained from various sources (Table 1). All antibodies were dialyzed (Slide-A-Lyzer Mini Dialysis Units, Pierce Biotechnology) against pH 7.2 PBS at 4 degrees for 2 hours and ultracentrifuged at 47,000 × g at 4 °C for 1 hour. The antibodies were prepared at 250 μg/ml in 1X PBS with 0.1% Tween-20 prior to printing.
Table 1.
Lectins used for motif prediction in the antibody array experiments.
| Name | Vendor | Catalog # |
|---|---|---|
| Erythrina cristagalli Lectin (ECL) | Vector Labs | L-1140 |
| Solanum tuberosumLectin (STL) | Vector Labs | BK-3000 |
| Jacalin | Vector Labs | BK-3000 |
| Griffonia simplicifolia Lectin II (GSL II) | Vector Labs | BK-3000 |
| Datura stramonium Lectin (DSL) | Vector Labs | BK-3000 |
| Phaseolus vulgaris Leucoagglutinin (PHA-L) | Vector Labs | B-1115 |
| Phaseolus vulgaris Erythroagglutinin (PHA-E) | Vector Labs | B-1125 |
| Psophocarpus Tetragonolobus Lectin (PTL-1) | Vector Labs | B-1365 |
| Wheat Germ Agglutinin (WGA) | Vector Labs | B-1025 |
| Soybean Agglutinin (SBA) | Vector Labs | BK-1000 |
| Peanut Agglutinin (PNA) | Vector Labs | BK-1000 |
| Lycopersicon esculentum Lectin (LEL) | EY Labs | BA-7001-1 |
| Helix aspersa Agglutinin (HAA) | Sigma Aldrich | L8764 |
| Helix pomatia Agglutinin (HPA) | Sigma Aldrich | L6512 |
| Hemagglutinin A (H5N1)(A/Vietnam/1203/2004) | Immune Technology | IT-003-0051p |
| Hemagglutinin A (H2N2)(A/Japan/305/1957) (aa 17-529) | e.Enzyme | IA-0032W-005P |
| MUC1 Antibody (CM1) | GeneTex | GTX10114 |
Cell culture and sample preparation
The cell lines BxPC-3, Su86.86, Capan2, AsPC-1, MIAPaCa2, PANC1 and Hs766T were purchased from American Type Culture Collection (Manassas, VA), and PSN1 was purchased from Sigma-Aldrich (St. Louis, MO). BxPC-3, Su86.86, Capan2, and AsPC-1 were grown in RPMI1640 (GIBCO, Grand Island, NY) supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin (GIBCO). MIAPaCa2, PANC1, Hs766T and PSN1 were grown in DMEM (GIBCO) supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. We grew the each cell line to ~80% confluency before harvesting the conditioned media and the cell lysates. All the cells were lysed in RIPA buffer (Cell Signaling Technology, Boston, MA) according to the manufacturer's protocol.
Antibody array assays
Forty-eight identical antibody arrays were printed onto glass microscope slides coated with ultra-thin nitrocellulose (PATH Slides, Grace BioLabs, Inc) using a contact printer (Aushon 2470, Aushon BioSystems). Each antibody was printed with 6 replicates and randomized within each array. The individual arrays were spaced by 4.5 mm in a 4 × 12 arrangement [27, 28]. After printing, hydrophobic borders were imprinted onto the slides (SlideImprinter, The Gel Company, San Francisco, CA) to segregate the arrays and allow for individual incubations on each array. The arrays were blocked using 1% bovine serum albumin (BSA) in phosphate buffered saline (PBS) plus 0.5% Tween-20 for one hour at room temperature.
The cell lysates were adjusted to 400 μg/ml and the cell media were diluted two-fold into PBS containing 0.1% Tween-20, 0.1% Brij-35, an IgG blocking cocktail (200 μg/ml mouse and rabbit IgG and 100 μg/ml goat and sheep IgG (Jackson ImmunoResearch Inc.)) and protease inhibitor (Complete Mini EDTA-free Tablet, Roche Applied Science). After briefly rinsing the arrays in PBS/0.1% Tween-20, the samples were incubated on the arrays overnight at 4 °C. The arrays were washed in three changes of PBS/0.1% Tween-20 for three minutes each and dried by centrifugation (Eppendorf 5810R, rotor A-4-62, 1500 × g for three minutes), and a biotinylated lectin detection (1 μg/ml in PBS with 0.1% BSA and 0.1% Tween-20) was incubated for one hour at room temperature. After washing and drying the arrays as above, Cy5-conjugated streptavidin (Invitrogen) prepared at 2 μg/ml in PBS with 0.1% BSA and 0.1% Tween-20 was incubated for one hour at room temperature, followed by a final wash and dry.
We generated standard curves for each antibody-lectin pair using pooled cell lysates ranging from 400 μg/ml to 3.125 μg/ml. We converted the relative fluorescence unit (RFU) measurements from each antibody-lectin pair to concentration units by calibrating to the standard curve, using MasterPlex 2010 (Hitachi Solutions America, San Francisco, CA). The data analysis and preparation were performed using Microsoft Excel, MultiExperiment Viewer (www.tm4.org), and Deneba Canvas XII.
Prior to running antibody-lectin sandwich experiments, it was important to assess whether lectin binding directly to the glycans on the spotted antibodies could skew the measurements. An approach to reducing lectin binding to the antibodies is to oxidize and chemically modify the glycans on the antibodies [8]. We examined the standard curves both with modification of the antibodies and without. Most of the lectins used in this study showed very low direct binding to the MUC1 antibody. Those that did show weak binding to the antibody did not bind the glycans on MUC1, and we did not observe an improvement in the standard curve after antibody modification (not shown). Therefore we chose not to chemically modify the antibodies for the experiments described here.
Results
Quantitative prediction of motifs in individual glycans
To develop an algorithm to quantitatively predict glycan motifs based on lectin binding, we needed detailed experimental data on the specificity of each lectin. Glycan array data provided that information. In a glycan array experiment, a lectin is incubated on a microarray containing hundreds of different glycans, resulting in parallel measurements of the binding of the lectin to each glycan [29]. In order to quantitatively and objectively determine lectin specificities from a glycan array experiment, we previously developed a method called Motif Segregation [24, 25]. Motif Segregation determines the statistical significance of a motif as an explanation of the pattern of lectin binding to the glycans on the array. We calculate the significance, termed the “Motif Score,” for many different motifs. The motifs with the highest scores represent the binding determinants of the lectin, according to the information available from the glycan array.
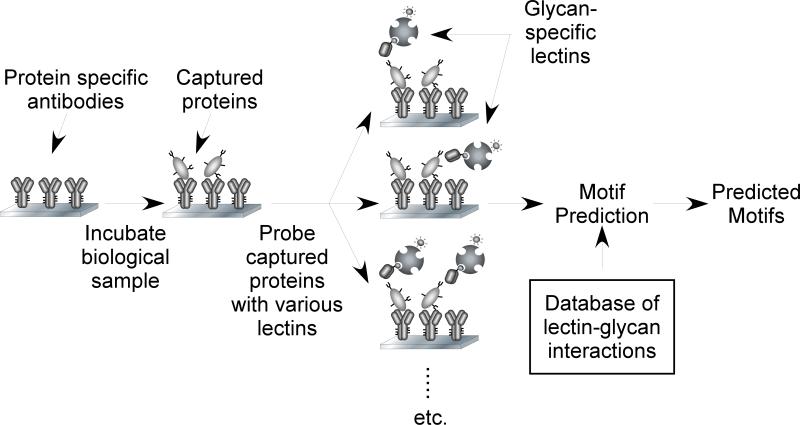
A wealth of glycan array data is available through the Consortium for Functional Glycomics (CFG) [30]. The latest version of the CFG glycan array contains over 500 different glycans that are especially relevant to human and mammalian biology. Participating investigators have sent lectins or other types of samples to the CFG, and the CFG performed the experiments and primary analyses. We downloaded the entire CFG set of almost 3,000 experiments and assembled the analyzed data into a database [26]. This database and analysis tool enables searches for lectins with defined specificities, global studies of lectin-glycan interactions, and detailed investigations of the specificities of individual lectins. A potential experiment, in which a researcher would like to know what glycans are on a protein in clinical samples, could proceed as follows. The researcher uses immobilized antibodies to isolate the protein and incubates a series of lectins on the protein to probe for the presence of selected glycans. A computer algorithm interprets the data. The input is the level of binding of each lectin, and the output is a list of glycan motifs likely present on the protein, with a probability score for each motif (Fig. 1).
Figure 1. Predicting glycan motifs on proteins using multiple lectin measurements.
We begin with an array of antibodies immobilized on a solid support. We then incubate a biological solution on the antibody array, and the antibodies capture their target glycoproteins. In parallel experiments, we use a series of lectins to probe the glycans on the captured proteins. The motif prediction algorithm uses these measurements and information from a database of lectin-glycan interactions to calculate the likelihood of the presence of various glycan motifs.
We began by developing and testing motif prediction algorithms on a simple model system for which we knew the glycan compositions: glycan arrays. Glycan array data provided measurements of the binding of many different lectins to each glycan on the array. Because we knew the structure of each glycan, and because we had measurements of the binding of multiple lectins to each of the glycans, we could determine if we could accurately predict the presence or absence of a motif in a glycan. We began with the basic logic that the binding of a lectin indicates the presence of at least one of the lectin's target motifs, and a lack of binding indicates that none of its target motifs is present. Here we made use of the database of motif scores derived from the glycan array data [26]. The motif scores indicate the likelihood that a lectin binds each of the defined motifs. Based on that information, if we observe the binding of a lectin to a glycan, we know the corresponding likelihood that each of the target motifs is present. The accuracy of the prediction directly relates to the accuracy with which we have defined the lectin specificity.
We first tested the prediction accuracy based on the binding of individual lectins. We sought to determine the presence or absence in each glycan of four motifs that can be difficult to distinguish using conventional methods: terminal alpha-linked GalNAc, terminal beta-linked GalNAc, terminal alpha-linked GlcNAc, and terminal beta-linked GlcNAc. We identified in the database the lectins with the highest motif scores for each of these motifs and then evaluated how accurately each of the lectins detected the motifs in the glycans on the array. Some of the lectins had good, but not perfect, accuracy. For example, the lectins from helix aspersa (HAA) and griffonia simplicifolia (GSL-II) each detected the terminal alpha-GlcNAc motif with ~95% accuracy at optimized thresholds (Fig. 2). The lectins targeting the other motifs each had inaccuracies (Table 2), either by detecting glycans not containing the motif or by not detecting glycans that do contain it (termed outlier glycans [25]).
Figure 2. Prediction accuracy using individual and combined lectins.
A) Motif prediction scores for terminal, alpha-GlcNAc. Using only GSL-2 (top left) and only HAA (top right), we calculated the motif prediction score for every glycan on the array for the terminal, alpha-GlcNAc motif and plotted the scores for the glycans without the motif (n = 602) and the glycans with the motif (n = 9). Both lectins had high scores for some glycans that did not have terminal, alpha-GlcNAc. When we calculated the motif prediction scores using both lectins (bottom, calculated by adding the scores from the two lectins for each glycan), all the glycans with the motif had higher scores that all the glycans without the motif. The size of each circle indicates the number of glycans with values in the region. We chose the thresholds to give maximum in discriminating between the groups. B) Motif prediction scores for distinct sets of glycans. We separately analyzed glycans that contained terminal beta-GlcNAc (left), terminal alpha-GalNAc (middle), or terminal alpha-GlcNAc. For each glycan, we calculated the motif prediction score for the terminal alpha-GlcNAc motif using either HAA alone, GSL-2 alone, or both HAA and GSL-2. Ideally, only glycans containing terminal alpha-GlcNAc (the glycans in the right box) should have high scores. GSL-2 but not HAA reacts with glycans containing terminal beta-GlcNAc; HAA but not GSL-2 reacts with glycans containing terminal alpha-GalNAc; and both lectins react with glycans containing the targeted motif of terminal alpha-GlcNAc. Therefore, the glycans with the targeted motif can be differentiated from the other glycans using the combined motif prediction score.
Table 2. Comparison of individual and combined lectins for predicting motifs.
For each of the four motifs, we calculated motif prediction scores for every glycan on the array using either a single lectin or the combination of all listed lectins. The area-under-the-curve (AUC) statistic from receiver-operator characteristic analysis indicates the degree of separation in motif prediction scores between the glycans that contain the motif and those that do not contain the motif.
| Lectin | Abbreviation | AUC | |
|---|---|---|---|
| Terminal GlcNAcα | Helix aspersa Agglutinin | HAA | 0.98 |
| Griffonia simplicifolia Lectin II | GSL-2 | 0.97 | |
| Panel | 1.00 | ||
| Terminal GlcNAcβ | Wheat Germ Agglutinin | WGA | 0.77 |
| Phaseolus vulgaris Leucoagglutinin | PHA-L | 0.60 | |
| Phaseolus vulgaris Erythroagglutinin | PHA-E | 0.63 | |
| Jacalin | Jacalin | 0.77 | |
| Griffonia simplicifolia Lectin II | GSL-2 | 0.81 | |
| Panel | 0.87 | ||
| Terminal GalNAcα | Psophocarpus Tetragonolobus Lectin | PTL-1 | 0.86 |
| Helix aspersa Agglutinin | HAA | 0.98 | |
| Clitocybe nebularis Lectin | CNL | 0.95 | |
| Panel | 0.98 | ||
| Terminal GalNAcβ | Vicia Villosa Lectin | VVL | 0.77 |
| Soybean Agglutinin | SBA | 0.75 | |
| Clitocybe nebularis Lectin | CNL | 0.81 | |
| Panel | 0.82 | ||
Prediction by multi-lectin binding
The next step was to test the hypothesis that the use of additional information from more lectins adds to the final accuracy. Two pieces of information are available for each lectin measurement: the previously-determined likelihood that the lectin binds each motif (the motif scores from glycan array data), and the amount of binding to the unknown sample. If a lectin has a strong motif score for a particular motif, a high amount of binding strongly predicts the presence of the motif, whereas weak binding points to the absence of the motif. In contrast, if a lectin has a weak motif score for the motif in question, the amount of binding is not predictive of the presence or absence of the motif. Thus, for each lectin measurement and for each motif of interest, we multiplied the measurement of lectin binding by the motif score for that lectin.
To arrive at a final prediction score for each motif, we added the contributions from each lectin. (We reasoned that the contributions from the individual lectins would be additive, given that each lectin is independent.) Therefore, the final motif prediction (MP) score for each motif, indicating the likelihood that a motif is present in a glycan, was MPM1 = (IL1 × MM1,L1) + (IL2 × MM1,L2) + (IL3 × MM1,L3) + ... etc. for additional lectins, where MPM1 is the motif prediction score for motif 1, IL1 is the intensity (amount of binding) of lectin 1, MM1,L1 is the motif score for motif 1 and lectin 1, and so on for additional lectins.
We asked whether the MP scores from multiple lectins used in combination were more accurate than MP scores from individual lectins for predicting the presence/absence of motifs in glycans on the array. For each of the four targeted motifs, we identified the combination of lectins that worked best together for discriminating glycans with the motif from those without. This identification was done by negative selection (see Methods). For each motif, the MP scores calculated from the lectin panels were more accurate than the MP scores from individual lectins (Table 2). For example, the MP scores using the combined measurements of HAA and GSL-2 perfectly identified the terminal, alpha-GlcNAc motif among the glycans on the array (Fig. 2A) We observed similar improvements using combinations of lectins for the other three motifs (not shown).
In order to better understand the requirements for improved motif prediction, we examined the glycans that were incorrectly called by individual lectins but correctly called by the panels. Of the glycans that did not contain terminal, alpha-GlcNAc but that had high scores by GSL-2, nearly all had terminal beta-GlcNAc (a known target of GSL-2), and nearly all of the glycans not containing terminal, alpha-GlcNAc but with high scores by HAA had terminal alpha-GalNAc (a main target of HAA) (Fig. 2B). None of these glycans was bound by both GSL-2 and HAA. However, all of the 9 glycans that had the terminal, alpha-GlcNAc motif were bound by both GSL-2 and HAA, yielding a higher score than the glycans bound by only one of the lectins (Fig. 2B). Therefore, the key to the improved accuracy of the panel was the common binding of multiple lectins to the target motif but not to off-target motifs. We observed a similar source of improvement for the other panels (not shown).
Prediction of glycan motifs on MUC1 produced by pancreatic cancer cell lines
Next we applied this method to the analysis of glycans on mucins in pancreatic cancer cell lines. Mucins are large, heavily glycosylated proteins providing protection and control of epithelial cell surfaces [31, 32]. The glycosylation of mucins can change in neoplastic transformation and cytokine signaling [33], so specific glycoforms of mucins could be accurate biomarkers [32]. Pancreatic cancer cell lines provide an in vitro model of the diversity of histologies and cellular phenotypes observed in vivo. As observed in many primary pancreatic tumors, cultured pancreatic cancer cell lines can acquire either epithelial-like or mesenchymal-like characteristics [33-35]. Markers that differentiate these phenotypes could be useful for prognosis or for diagnosing subtypes of cancer with divergent behaviors [35, 36].
We selected eight different cell lines: three with epithelial characteristics, three with mesenchymal, and two with mixed phenotypes. We applied the cell lysates and the conditioned media from each cell line to antibody arrays targeting the mucins MUC1, MUC5AC, and MUC16, and we probed the glycans on the captured proteins with various lectins. The lectins comprised most of those identified above plus others known to bind related structures (Table 1). We focused on the glycans associated with MUC1, because all the cell lines express MUC1 [33].
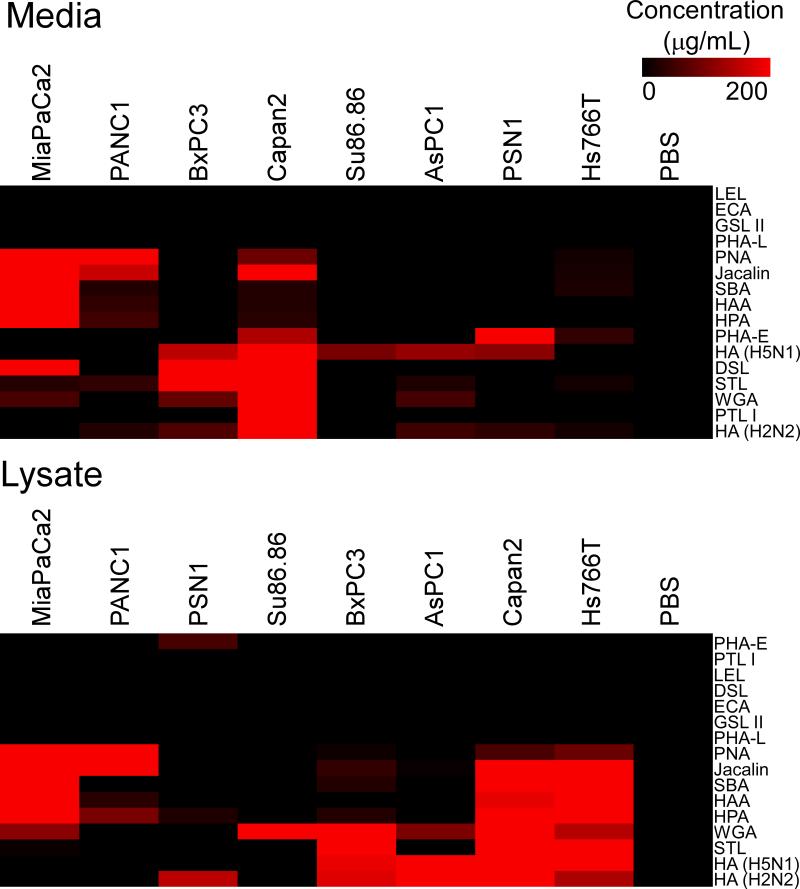
We observed major differences between the cell lines in lectin binding (Fig. 3). We did not see consistent differences based on epithelial or mesenchymal characteristics; each cell line appeared to have a unique signature. Some lectin reactivities were more abundant in the lysates than in the media, as with the Hs766T cells, and some were higher in the media, as with the DSL-reactive form of MUC1 in the MiaPaca-2 cells.
Figure 3. Lectin probing of MUC1 in cell media and lysates.
Using the scheme of Fig. 1, we incubated cell media (top) and lysate (bottom) from various cell lines (indicated by the column labels) on antibody arrays and probed the glycans on the captured proteins with various lectins (indicated by the row labels). We quantified the signal at the MUC1 capture antibody and calibrated the raw fluorescence values to the concentration of the media or lysates (scale given by the color bar) based on dilution curves (see Methods). The rows and columns are ordered according to similarity by hierarchical clustering.
We then calculated motif prediction scores from the lectin measurements. We calculated the scores for the four motifs mentioned above (Table 2) and for other motifs that could be detected by the lectins included in the panel. To determine if a motif could be detected by the panel of lectins, we calculated the maximum potential score for each motif, which was the score that would be achieved if every lectin bound at the maximum amount. Motifs with potential scores within 80% of the potential score of terminal, beta-GalNAc were included in the analysis, after removal of highly similar motifs.
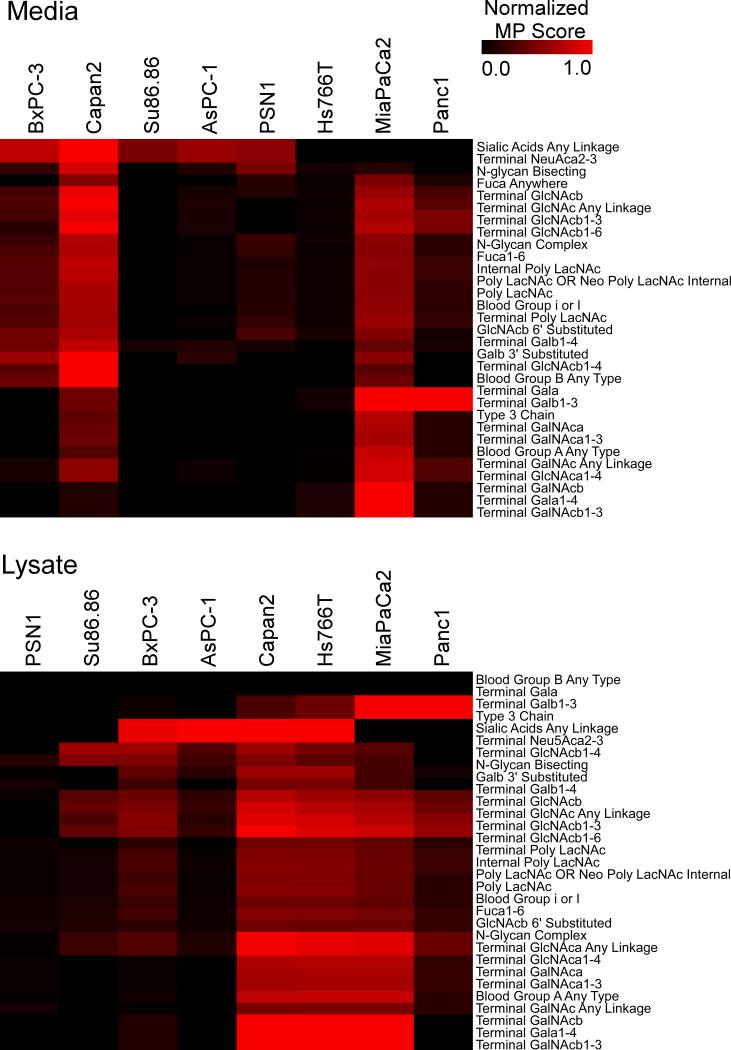
Each cell line had a unique pattern of MUC1 glycoforms and some overlap with other cell lines (Fig. 4). In the media, Capan2 was notable for its high terminal, beta-GlcNAc and sialic acids; MiaPaCa2 was unique in its high scores for terminal, beta-GalNAc and shared terminal galactose with Panc1; Su86.86, PSN1, and AsPC-1 primarily scored high for sialic acids, and Hs766T showed no MUC1 glycoforms. Many patterns were the same in the lysates, except Hs766T showed many MUC1 glycoforms; Su86.86 displayed terminal, beta-GlcNAc glycoforms; and Capan2 and Hs766T showed terminal GalNAc glycoforms.
Figure 4. MUC1 glycoforms in cell media and lysates.
Using the data from Fig. 3, we converted the lectin measurements to motif predictions. The motif names appear in the row labels, and the cell line names are in the column labels. Each cell indicates the normalized motif prediction (MP) score on the scale in the color bar. To normalize the motif prediction scores, we divided each motif prediction score by the maximum potential score for the motif. The rows and columns are ordered according to similarity by hierarchical clustering.
These analyses revealed major differences between the cell lines in the glycoforms of MUC1 they produce. We also uncovered significant differences between media and lysates within certain cell lines in the MUC1 glycoforms. The conversion from lectin measurements into motif predictions aided the analysis and interpretation of the experiment. Future research could further characterize the glycoforms and address the origins and functional consequences of the differences in glycoforms.
Discussion
We demonstrated a new strategy for analyzing measurements obtained using lectins. This method could give more precise information than manual interpretation because it better accounts for complexities in lectin specificities. The integration of detailed information from multiple lectins, which would be extremely difficult if done manually, could further improve accuracy. Another benefit of this approach is that it could remove the burden for researchers to gain a detailed knowledge of the binding specificities of each lectin; researchers could instead rely on the algorithm to interpret lectin experiments. This assistance would broaden the range of researchers who could effectively use lectins and would increase the number of different lectins that each researcher could reliably use.
The quantitative interpretation of lectin binding could be useful several types of studies, especially those involving relationships between glycans and phenotypes in clinical samples. For example, researchers could examine whether unusual protein glycoforms have increased levels in pre-neoplastic tissue; whether the glycoforms are more abundant in individuals with specific genotypes; whether the expression of particular glycosyltransferases is linked to protein glycosylation; if differences in protein glycosylation exist between humans and mice or other model systems; or whether drug or cytokine treatments affect protein glycosylation. Furthermore, manufacturers of protein and antibody drugs are interested in characterizing the effects of protein glycosylation on drug activity and retention [37]. With further development of the capabilities suggested here, drug researchers could retrieve a protein from the in vivo setting and determine the relative amounts of each glycoform. Such analyses could be effectively performed on lectin arrays [38, 39].
The method currently has limitations and areas requiring improvement, since here we simply sought to establish feasibility and potential value. One potential problem is that the glycan array from which we determined lectin specificity does not contain all the important glycans encountered in a biological setting, which could limit the accuracy of the interpretation. One way to address this limitation is to continue to acquire more glycan array data for each lectin. The new versions of the CFG array and other glycan array platforms should provide complementary information that we could incorporate into our database. Another potential problem is difficulty in handing more complex glycans, a problem we would recognize if we achieve good accuracy for purified glycans but not for more complex glycoproteins. In that case it may be necessary to modify the experimental system to analyze simpler glycans, perhaps by enzymatically digesting proteins prior to antibody capture and lectin probing. The capture and detection of glycopeptides rather than whole glycoproteins would provide a simpler set of glycans.
It will be important to improve the various components of this method to get the accuracy as high as possible. In some cases, the failure of the algorithm may be due to inaccuracies in the motif definitions for particular lectins. For many lectins, our best definition of the binding determinant (indicated by the motif with the top motif score) is not perfectly accurate. In some cases the lectin does not bind glycans that contain the motif, and in others the lectin binds glycans without the motif. Because of this persistent lack of accuracy, the predictions will not be accurate for such “outlier” glycans [25]. We hope to mitigate this effect through the use of multiple lectins, but we can also increase prediction accuracy by further improving the accuracy of the motif definitions for each lectin. To this end, we could make use of new glycan array data being generated on arrays with ever increasing complexity. The new information could lead to a better understanding of the specificity of each lectin. In addition, we could further train our motif definitions on the outlier glycans found in each dataset to better account for the nuances in specificity of each lectin [25]. We are currently pursuing automated approaches for this training process [26, 40]. Potentially we could incorporate additional algorithms that have been developed for defining the specificities of lectins [41-43].
The prediction accuracy may be further improved by optimizing the algorithm for combining information from multiple lectins. Part of this optimization includes using the right selection of lectins to predict each particular motif. It may be valuable to search the database for lectins that bind specified motifs, using completely flexible terms for the motifs that include wildcards, exclusions, or combinations of motifs. For example, if we are targeting motif A, and if lectin 1 binds motif A and motif B, we could search for a second lectin that binds motif B but not motif A. Taking that strategy further, we could find another lectin that overlaps with lectin 2 but not with lectin 1. This capability will be important when we do not have a lectin that exclusively binds a glycan motif of interest. The proper combination of lectins, along with our analysis of combinations of lectins, should allow the indirect detection of that motif.
In summary, we demonstrated a new tool for quantitatively interpreting lectin measurements. The method promises to provide more detail and accuracy than the current manual approach and should be particularly valuable for researchers who do not have expert knowledge of lectins. We anticipate further enhancements to improve the accuracy of the method and software to make it broadly accessible.
Acknowledgements
We gratefully acknowledge support of this work by the NCI (Alliance of Glycobiologists for Cancer Detection, U01CA168896; Early Detection Research Network, U01CA152653) and the Van Andel Research Institute.
References
- 1.Perou CM, Sorlie T, Eisen MB, van de Rijn M, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Ransohoff DF. The process to discover and develop biomarkers for cancer: a work in progress. Journal of the National Cancer Institute. 2008;100:1419–1420. doi: 10.1093/jnci/djn339. [DOI] [PubMed] [Google Scholar]
- 3.Dube DH, Bertozzi CR. Glycans in cancer and inflammation--potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 4.Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nature reviews. Molecular cell biology. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 6.Lau KS, Dennis JW. N-Glycans in cancer progression. Glycobiology. 2008;18:750–760. doi: 10.1093/glycob/cwn071. [DOI] [PubMed] [Google Scholar]
- 7.McGowan ED, Bowman K. Services, D. o. H. a. H. (Ed.) 2011 [Google Scholar]
- 8.Chen S, LaRoche T, Hamelinck D, Bergsma D, et al. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nature methods. 2007;4:437–444. doi: 10.1038/nmeth1035. [DOI] [PubMed] [Google Scholar]
- 9.Yue T, Goldstein IJ, Hollingsworth MA, Kaul K, et al. The prevalence and nature of glycan alterations on specific proteins in pancreatic cancer patients revealed using antibody-lectin sandwich arrays. Mol Cell Proteomics. 2009;8:1697–1707. doi: 10.1074/mcp.M900135-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haab BB, Porter A, Yue T, Li L, et al. Glycosylation Variants of Mucins and CEACAMs as Candidate Biomarkers for the Diagnosis of Pancreatic Cystic Neoplasms. Annals of surgery. 2010;251:937–945. doi: 10.1097/SLA.0b013e3181d7738d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue T, Maupin KA, Fallon B, Li L, et al. Enhanced discrimination of malignant from benign pancreatic disease by measuring the CA 19-9 antigen on specific protein carriers. PLoS ONE. 2011;6:e29180. doi: 10.1371/journal.pone.0029180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kreunin P, Zhao J, Rosser C, Urquidi V, et al. Bladder cancer associated glycoprotein signatures revealed by urinary proteomic profiling. Journal of proteomeresearch. 2007;6:2631–2639. doi: 10.1021/pr0700807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Simeone D, Brenner D, Anderson MA, et al. Pancreatic Cancer Serum Detection Using a Lectin/Glyco-Antibody Array Method. Journal of proteome research. 2009;8:483–492. doi: 10.1021/pr8007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Tao SC, Bova GS, Liu AY, et al. Detection and verification of glycosylation patterns of glycoproteins from clinical specimens using lectin microarrays and lectin-based immunosorbent assays. Analytical chemistry. 2011;83:8509–8516. doi: 10.1021/ac201452f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabares G, Radcliffe CM, Barrabes S, Ramirez M, et al. Different glycan structures in prostate-specific antigen from prostate cancer sera in relation to seminal plasma PSA. Glycobiology. 2006;16:132–145. doi: 10.1093/glycob/cwj042. [DOI] [PubMed] [Google Scholar]
- 16.Block TM, Comunale MA, Mehta A. 2010.
- 17.Aoyagi Y, Mita Y, Suda T, Kawai K, et al. The fucosylation index of serum alpha-fetoprotein as useful prognostic factor in patients with hepatocellular carcinoma in special reference to chronological changes. Hepatol Res. 2002;23:287. doi: 10.1016/s1386-6346(01)00184-x. [DOI] [PubMed] [Google Scholar]
- 18.Thompson S, Cantwell BM, Cornell C, Turner GA. Abnormally-fucosylated haptoglobin: a cancer marker for tumour burden but not gross liver metastasis. British journal of cancer. 1991;64:386–390. doi: 10.1038/bjc.1991.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuyama N, Ide Y, Nakano M, Nakagawa T, et al. Fucosylated haptoglobin is a novel marker for pancreatic cancer: A detailed analysis of the oligosaccharide structure and a possible mechanism for fucosylation. International journal of cancer. 2006;118:2803–2808. doi: 10.1002/ijc.21728. [DOI] [PubMed] [Google Scholar]
- 20.Hirabayashi J. Lectin-based structural glycomics: glycoproteomics and glycan profiling. Glycoconjugate journal. 2004;21:35–40. doi: 10.1023/B:GLYC.0000043745.18988.a1. [DOI] [PubMed] [Google Scholar]
- 21.Sharon N. Lectins: carbohydrate-specific reagents and biological recognition molecules. The Journal of biological chemistry. 2007;282:2753–2764. doi: 10.1074/jbc.X600004200. [DOI] [PubMed] [Google Scholar]
- 22.Van Damme EJM, Peumans WJ, Pusztai A, Bardocz S. Handbook of plant lectins: properties and biomedical applications. John Wiley & Sons; Chichester: 1998. [Google Scholar]
- 23.Markiv A, Peiris D, Curley GP, Odell M, Dwek MV. Identification, cloning, and characterization of two N-acetylgalactosamine-binding lectins from the albumen gland of Helix pomatia. The Journal of biological chemistry. 2011;286:20260–20266. doi: 10.1074/jbc.M110.184515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porter A, Yue T, Heeringa L, Day S, et al. A motif-based analysis of glycan array data to determine the specificities of glycan-binding proteins. Glycobiology. 2010;20:369–380. doi: 10.1093/glycob/cwp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maupin KA, Liden D, Haab BB. The fine specificity of mannose-binding and galactose-binding lectins revealed using outlier-motif analysis of glycan array data. Glycobiology. 2011;22:160–169. doi: 10.1093/glycob/cwr128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kletter D, Singh S, Bern M, Haab BB. Global comparisons of lectin-glycan interactions using a database of analyzed glycan array data. Molecular & cellular proteomics : MCP. 2013;12:1026–1035. doi: 10.1074/mcp.M112.026641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forrester S, Kuick R, Hung KE, Kucherlapati R, Haab BB. Low-volume, high-throughput sandwich immunoassays for profiling plasma proteins in mice: identification of early-stage systemic inflammation in a mouse model of intestinal cancer. Molecular Oncology. 2007;1:216–225. doi: 10.1016/j.molonc.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haab BB, Yue T. High-throughput studies of protein glycoforms using antibody-lectin sandwich arrays. Methods in molecular biology (Clifton, N.J. 2011;785:223–236. doi: 10.1007/978-1-61779-286-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang PH, Wu CY, Greenberg WA, Wong CH. Glycan arrays: biological and medical applications. Current opinion in chemical biology. 2008;12:86–92. doi: 10.1016/j.cbpa.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blixt O, Head S, Mondala T, Scanlan C, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 32.Moniaux N, Andrianifahanana M, Brand RE, Batra SK. Multiple roles of mucins in pancreatic cancer, a lethal and challenging malignancy. British journal of cancer. 2004;91:1633–1638. doi: 10.1038/sj.bjc.6602163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu YM, Nowack DD, Omenn GS, Haab BB. Mucin glycosylation is altered by pro-inflammatory signaling in pancreatic-cancer cells. Journal of proteome research. 2009;8:1876–1886. doi: 10.1021/pr8008379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arumugam T, Ramachandran V, Fournier KF, Wang H, et al. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer research. 2009;69:5820–5828. doi: 10.1158/0008-5472.CAN-08-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maupin K, Sinha A, Eugster E, Miller J, et al. Glycogene Expression Alterations Associated with Pancreatic Cancer Epithelial-Mesenchymal Transition in Complementary Model Systems. PLoS ONE. 2010;5:e13002. doi: 10.1371/journal.pone.0013002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collisson EA, Sadanandam A, Olson P, Gibb WJ, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nature medicine. 2011 doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Astronomo RD, Burton DR. Carbohydrate vaccines: developing sweet solutions to sticky situations? Nat Rev Drug Discov. 2010;9:308–324. doi: 10.1038/nrd3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pilobello KT, Krishnamoorthy L, Slawek D, Mahal LK. Development of a lectin microarray for the rapid analysis of protein glycopatterns. Chembiochem. 2005;6:985–989. doi: 10.1002/cbic.200400403. [DOI] [PubMed] [Google Scholar]
- 39.Kuno A, Uchiyama N, Koseki-Kuno S, Ebe Y, et al. Evanescent-field fluorescence-assisted lectin microarray: a new strategy for glycan profiling. Nature methods. 2005;2:851–856. doi: 10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- 40.Kletter D, Cao Z, Bern M, Haab B. Determining Lectin Specificity from Glycan Array Data using Motif Segregation and GlycoSearch Software. Current Protocols in Chemical Biology. 2013;5:1–13. doi: 10.1002/9780470559277.ch130028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hizukuri Y, Yamanishi Y, Nakamura O, Yagi F, et al. Extraction of leukemia specific glycan motifs in humans by computational glycomics. Carbohydr Res. 2005;340:2270–2278. doi: 10.1016/j.carres.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 42.Xuan P, Zhang Y, Tzeng TR, Wan XF, Luo F. A quantitative structure-activity relationship (QSAR) study on glycan array data to determine the specificities of glycan-binding proteins. Glycobiology. 2012;22:552–560. doi: 10.1093/glycob/cwr163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cholleti SR, Agravat S, Morris T, Saltz JH, et al. Automated motif discovery from glycan array data. Omics : a journal of integrative biology. 2012;16:497–512. doi: 10.1089/omi.2012.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]