Abstract
The activin/inhibin system regulates follicle-stimulating hormone (FSH) synthesis and release by pituitary gonadotrope cells in mammals. In vitro cell line data suggest that activins stimulate FSH β-subunit (Fshb) transcription via complexes containing the receptor-regulated SMAD proteins SMAD2 and SMAD3. Here, we used a Cre-loxP approach to determine the necessity for SMAD2 and/or SMAD3 in FSH synthesis in vivo. Surprisingly, mice with conditional mutations in both Smad2 and Smad3 specifically in gonadotrope cells are fertile and produce FSH at quantitatively normal levels. Notably, however, we discovered that the recombined Smad3 allele produces a transcript that encodes the entirety of the SMAD3 C-terminal Mad homology 2 (MH2) domain. This protein behaves similarly to full-length SMAD3 in Fshb transcriptional assays. As the truncated protein lacks the N-terminal Mad homology 1 (MH1) domain, these results show that SMAD3 DNA-binding activity as well as SMAD2 are dispensable for normal FSH synthesis in vivo. Furthermore, the observation that deletion of proximal exons does not remove all SMAD3 function may facilitate interpretation of divergent phenotypes previously described in different Smad3 knockout mouse lines.—Fortin, J., Boehm, U., Weinstein, M. B., Graff, J. M., Bernard, D. J. Follicle-stimulating hormone synthesis and fertility are intact in mice lacking SMAD3 DNA binding activity and SMAD2 in gonadotrope cells.
Keywords: pituitary, reproduction, activin, knockout
Follicle-stimulating hormone (FSH), a dimeric glycoprotein secreted by pituitary gonadotrope cells, is a critical regulator of gonadal function and is required for fertility in mammals (1, 2). Multiple factors derived from the brain, the gonads, and the pituitary regulate the expression of the FSH β-subunit (Fshb), the rate-limiting step in FSH synthesis and release (3). Classical studies identified inhibins and activins as critical negative and positive regulators of FSH synthesis (4–8). Activins, derived from the pituitary gland, bind to their cognate receptors on the cell surface of gonadotrope cells, initiating a signaling cascade that culminates in the activation of Fshb transcription (3, 9). In contrast, inhibins, secreted from the gonads, suppress FSH synthesis by competitively binding to activin type II receptors (10).
Much effort has been directed toward the molecular dissection of the signaling pathways by which activins stimulate Fshb transcription. Canonically, activins signal through heteromeric assemblies of type I and type II receptors, which phosphorylate the effector proteins, mammalian homolog of Drosophila mothers against decapentaplegic; family member 2 (SMAD2) and SMAD3. These proteins then partner with SMAD4 and accumulate in the nucleus, where they act as transcription factors (11, 12). Several lines of evidence implicate SMAD2/3/4 complexes as central components of activin-induced FSH synthesis. Studies in murine immortalized gonadotrope-like (LβT2) and heterologous cell lines indicate that SMAD2/3/4 complexes directly bind to the Fshb promoters of several mammalian species, including mouse, and activate their transcription (13–19). Furthermore, a number of SMAD2/3-interacting transcription factors directly bind to the Fshb promoter and cooperatively activate Fshb transcription (20–23). One such factor, forkhead box L2 (FOXL2), was recently confirmed to be a critical regulator of Fshb expression, FSH synthesis, and fertility in vivo in mice (24, 25).
Despite the large amount of data indicating that activins operate through a canonical SMAD-dependent signaling pathway to stimulate Fshb transcription in vitro, evidence demonstrating necessary roles for SMADs 2 and/or 3 in FSH synthesis in vivo is lacking. Mice harboring a global deletion of the 8th of the 9 exons in Smad3 have modestly decreased pituitary Fshb transcript levels (26). However, Smad3 is broadly expressed and these animals display a range of reproductive defects, including intrinsic gonadal dysfunction, making it difficult to ascertain whether the Fshb deficiency is the result of cell-autonomous loss of SMAD3 function in gonadotropes (27, 28). To investigate the roles of SMADs 2 and 3 in FSH synthesis in vivo, we used a Cre-loxP approach to produce loss of function mutations in Smad2 and/or Smad3 selectively in gonadotrope cells of mice.
MATERIALS AND METHODS
Mouse lines
The Smad2fl, Smad3fl, gonadotropin-releasing hormone receptor (Gnrhr)GRIC and ROSA26eYFP alleles and corresponding genotyping primers (Supplemental Table S1) were described previously (29–32). To generate Smad2/3 conditional knockout (S2/3cKO) mice, Smad2fl/+;Smad3fl/+;GnrhrGRIC/+ females were mated with Smad2fl/fl;Smad3fl/fl males, yielding S2/3cKO (Smad2fl/fl;Smad3fl/fl;GnrhrGRIC/+) and control (Smad2fl/fl;Smad3fl/fl;Gnrhr+/+) mice at a frequency of 1/8 for each genotype. To generate mice with genetically labeled gonadotropes for subsequent fluorescence-activated cell sorting (FACS) purification (see below), Smad2fl/fl;Smad3fl/fl;ROSA26eYFP/eYFP males were mated with Smad2fl/fl;Smad3fl/fl;GnrhrGRIC/+ females to yield Smad2fl/fl;Smad3fl/fl;ROSA26eYFP/+;GnrhrGRIC/+ offspring at a frequency of 1/2. Control gonadotropes were obtained from ROSA26eYFP/+;GnrhrGRIC/+ mice. All animal experiments were performed in accordance with federal guidelines and were approved by McGill University's Institutional Animal Care and Use Committee (Animal Use Protocol No. 5204).
Puberty and estrous cycle assessment
To determine the onset of puberty, female mice were examined daily for vaginal opening starting from the day of weaning (postnatal day 21). Estrous cyclicity was assessed for ≥21 consecutive days starting at 7 wk of age. Vaginal cells, obtained every morning (09:00–10:00) using a cotton swab dampened with sterile saline, were smeared on glass slides, stained with 0.1% methyl blue, and examined under a microscope. Stages were assigned following published guidelines (33). Because of high similarity in cell types, no distinction was made between metestrus and diestrus. A complete estrous cycle was defined as sequential mestrus/diestrus, proestrus, and estrus, regardless of the number of days spent in each stage.
Breeding trials
Mating trials were initiated 1 wk after the completion of estrus cycle assessment. S2/3cKO or control females were paired with one 8-wk-old C57BL/6J male for a period of 6 mo. Starting from 20 d after pairing, cages were examined daily for the presence of newborn mice. As soon as a new litter was observed, pups were counted. Pups were left in the cage for 2 wk before removal.
Organ analyses and sperm count
Reproductive organs were harvested from 10-wk-old males (testes, seminal vesicles) or mestestrus/diestrus females (uterus, ovaries) and weighted on a precision balance. Homogenization-resistant epididymal sperm count was assessed as described previously (25). For ovarian histology, formalin-fixed tissues were paraffin-embedded and serial 5 μM sections collected after sectioning on a microtome. For corpora lutea (CL) counting, every 7th section was hematoxylin and eosin (H&E)-stained and imaged by microscopy, which allowed tracking of individual CLs across several sections. One ovary was sectioned and analyzed per mouse. For testicular histology, testes were fixed in Bouin's overnight and washed in 95 and 70% ethanol before paraffin embedding. Transverse sections (7 μm) were obtained in the middle of the tissue, H&E stained, and imaged by microscopy.
Pituitary and ovarian RNA extraction and quantitative polymerase chain reaction (qPCR)
Pituitaries and ovaries were collected from 10-wk-old males and metestrus/diestrus females and immediately frozen on dry ice. Individual pituitaries and ovaries were homogenized in 500 μl TriZol and RNA extracted following the manufacturer's protocol (Invitrogen, Carlsbad, CA, USA). RNA from pituitaries (1.5 μg) or ovaries (2 μg) was reverse transcribed using Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega, Madison, WI, USA) as described previously (33) in a final volume of 40 μl. cDNA (1 μl) was assayed in triplicate qPCR reactions using Platinum qPCR Supermix-UDG (Invitrogen, Carlsbad, CA, USA) on a Corbett Rotorgene 6000 instrument (Corbett Life Science, Mortlake, NSW, Australia). Gene expression was determined relative to that of the housekeeping gene Rpl19 using the 2−ΔΔCt method (34) and the primers described in Supplemental Table S1.
Hormone assays
Blood was collected from 10-wk-old males and metestrus/diestrus females by cardiac puncture, left to coagulate for 15 min at room temperature, and centrifuged at 3000 g for 10 min. Serum was collected and stored at −20°C. Luteinizing hormone (LH) and FSH levels were measured by multiplex ELISA at the Ligand Assay and Analysis Core of the Center for Research in Reproduction at the University of Virginia (Charlottesville, VA, USA).
Ovariectomy
Adult (≥8-wk-old) S2/3cKO and control females in metestrus/diestrus morning were injected subcutaneously with 5 μg/g body weight of carprofen. Mice were deeply anesthetized using isoflurane and placed on a heating pad. The back skin was shaved and cleaned, and a single midline incision was performed. Small incisions were made bilaterally through the muscle layer above the ovaries through which the uterine horns were retrieved. The ovaries were removed by cauterization below the oviduct, and the incisions closed using Vicryl sutures (Ethicon, Blue Ash, OH, USA). Topical carprofen was applied, and the skin incision was closed using wound clips (Reflex 7; CellPoint Scientific, Gaithersburg, MD, USA). The mice were left to recover on a heating pad. Sham-treated animals were processed in the same way, except that the ovaries were not removed. Mice were killed 7 h postsurgery, and their pituitaries and serum were collected for analyses.
Primary pituitary cell culture
Adult (≥8-wk-old) mice were killed by CO2 asphyxiation, and their pituitaries were collected in M199 medium containing 10% fetal bovine serum (FBS). A single-cell suspension was prepared as described previously (35). Cells were seeded at a density of 4 × 105/well in 48-well plates. All treatment conditions were performed in duplicate. For viral transduction, adenoviruses expressing enhanced green fluorescent protein (eGFP) or Cre-IRES-eGFP (Baylor College of Medicine Vector Development Laboratory, Houston, TX. USA) were added 24 h after plating at a multiplicity of infection of 60. After 24 h, virus-containing medium was removed and replaced with medium containing 2% FBS with or without 1 nM activin A (R&D Systems, Minneapolis, MN, USA). After 24 h incubation, cells from duplicate wells were harvested using 0.25% trypsin, and pooled. RNA and DNA were extracted using the Qiagen Allprep DNA/RNA kit. RNA was eluted in 25 μl RNase-free water and reverse transcribed. The resulting cDNA was analyzed by qPCR.
FACS
For FACS, pituitaries were dissociated as above, and the resulting cell suspensions were passed through a 40-μm nylon mesh to eliminate cell clumps. Yellow fluorescent protein (YFP)+ and YFP− cells were isolated on a FACSAria cell sorter (BD Biosciences, San Jose, CA, USA) at the flow cytomery core facility of the McGill University Life Sciences Complex. Approximately 5000–10000 YFP+ cells (3–5% of all sorted cells) were routinely obtained from each pituitary in these experiments. RNA and genomic DNA were isolated from sorted cells as described above for mixed pituitary cultures.
Cloning and expression vector construction
The primers used in the “primer walk” experiment are listed in Supplemental Table S1. For expression vector construction, the full-length and truncated Smad3 transcripts were amplified by PCR from S2/3cKO pituitary cDNA, using a sense primer 120 bp upstream of the canonical translation start site in exon 1 and an antisense primer immediately after the STOP codon in exon 9 (see Supplemental Table S1). The resulting fragments were digested with HindIII and BamHI (engineered onto the 5′ end of the primers) and ligated into the same sites in pcDNA3.0. To generate epitope-tagged constructs, the same strategy was used, except that the antisense primer replaced the stop codon with a ClaI restriction site. The resulting fragments were digested with HindIII and ClaI and ligated inframe upstream of a 3X-HA tag in a previously modified pcDNA3.0 vector. Constructs were verified by sequencing (Genome, Montreal, QC, Canada). The −846/+1 mFshb-luc reporter, as well as the SMAD4 and FOXL2 expression vectors were described previously (15, 20).
Cell lines culture, transfections, reporter assays, and Western blotting
LβT2 cells (a gift from Dr. Pamela Mellon, University of California, San Diego, CA, USA) were seeded at a density of 125,000 cells/well in 48-well plates 1 d before transfection. HeLa cells were cultured as described previously (35). Cells were transfected using Lipofectamine 2000 (Invitrogen), and reporter assays were performed as described previously (18, 20, 23, 36). For protein analyses, confluent HeLa cells in 6-well plates were transfected with 2 μg expression vector using Lipofectamine/Plus reagent (Invitrogen) and harvested the next day in radioimmunoprecipitation assay (RIPA) lysis buffer before Western blot analysis (13). Antibodies used were mouse anti-HA (Sigma H9658, St. Louis, MO, USA), mouse anti-β-actin (Sigma A2228) and goat anti-mouse IgG-horseradish peroxidase conjugate secondary antibody (Bio-Rad 170–6515, Hercules, CA, USA).
Statistical analysis
Serum hormones, pituitary and ovarian transcripts, estrous cycle frequency, sperm counts, CL counts, and organ weights were compared using unpaired t tests. Estrous cycle stages and gene expression in sorted cells and reporter experiments were analyzed using 2-way ANOVA followed by Tukey post hoc tests. Ovariectomy experiments were analyzed using 1-way ANOVA followed by Newman-Keuls multiple comparison test. Primary culture experiments were analyzed using 1-way repeated-measures ANOVA followed by Tukey post hoc test. Data were log transformed when variances were not equal between groups. Statistical analyses were performed using Systat 10.2 (Systat Software, Chicago, IL, USA) or GraphPad Prism 5 (GraphPad, San Diego, CA, USA). Values of P < 0.05 were considered statistically significant.
RESULTS
Generation of gonadotrope-specific Smad2/3 double-knockout mice
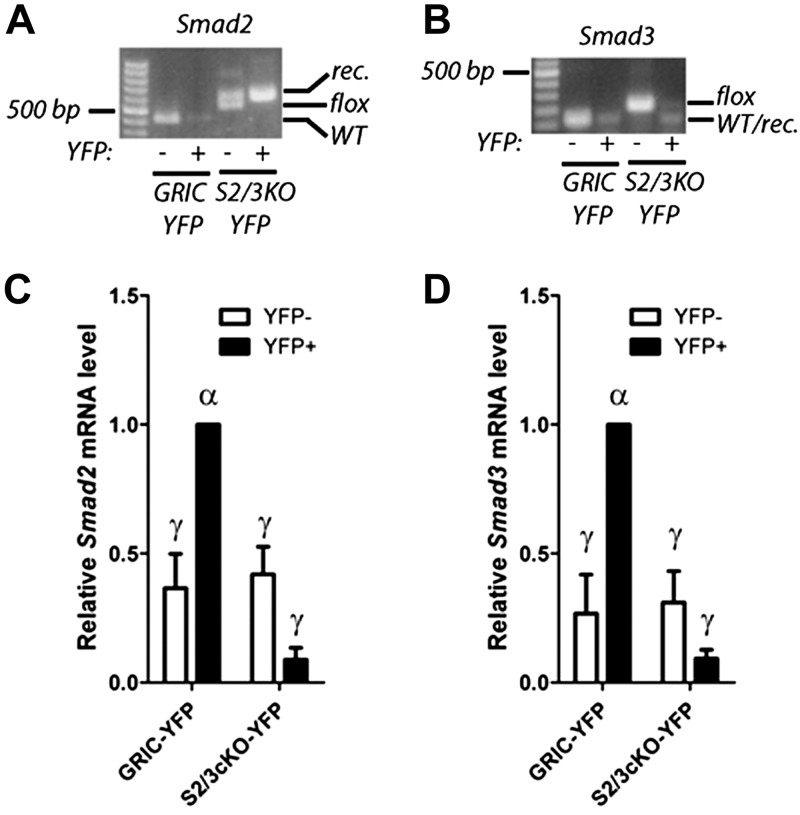
To assess the role of SMAD2/3 signaling in FSH synthesis and fertility in vivo, we generated mice lacking the canonical forms of SMAD2 and 3 in gonadotropes by crossing animals expressing Cre recombinase exclusively in gonadotropes (Gnrhr-IRES-Cre or GnrhrGRIC; ref. 32) with mice carrying conditional (“floxed”) alleles of Smad2 and Smad3 (29, 30). The resulting S2/3cKO double-knockout mice have the genotype Smad2fl/fl;Smad3fl/fl;GnrhrGRIC/+. Control mice were littermates carrying homozygous conditional alleles (Smad2fl/fl;Smad3fl/fl). To assess the deletion efficiency of Smad2/3 in the double-knockout mice, we crossed in the conditional ROSA26eYFP reporter allele (31) on the S2/3cKO background (hereafter S2/3cKO-YFP). In these animals, Cre recombinase expression in gonadotropes triggers both recombination of Smad2/3 and expression of enhanced YFP (eYFP), thus enabling the high efficiency purification of this cell population by FACS (25, 37). PCR analysis of genomic DNA from sorted YFP+ and YFP− cells of S2/3cKO mice indicated essentially complete recombination of the Smad2 and Smad3 loci in gonadotropes (Fig. 1A, B). Consistent with this, qPCR on cDNA prepared from the same cells, using primers directed at the deleted exons, showed a profound loss of Smad2 and Smad3 transcripts in S2/3cKO gonadotropes compared with those of GnrhrGRIC;ROSA26eYFP (GRIC-YFP) controls (Fig. 1C, D). Interestingly, the same analysis indicated higher expression of both Smad2 and Smad3 in gonadotropes (YFP+) compared with other pituitary cell types (YFP−). Furthermore, down-regulation of Smad2 and Smad3 transcripts (by ∼30%) in whole pituitaries from S2/3cKO mice far exceeded the values one would expect if the genes were uniformly expressed across all pituitary cells types, as gonadotropes [and GnrhrGRIC-expressing cells (32, 38)] represent only 5–10% of the total cell population (Supplemental Fig. S1A, B).
Figure 1.
Generation and validation of gonadotrope-specific Smad2/3-knockout mice. A, B) YFP+ and YFP− cells were isolated from GRIC-YFP and S2/3cKO-YFP mice by FACS. Genomic DNA was extracted and analyzed by genotyping PCR for the wild-type (WT), floxed (flox), and recombined (rec.) alleles of Smad2 (A) and Smad3 (B). Data are representative from 1 of 3 sorting experiments. C, D) cDNA was prepared from total RNA from sorted cells and analyzed by qPCR using primers overlapping the deleted exons in Smad2 (C: forward primer in exon 10; reverse primers in exon 11) and Smad3 (D: forward primer in exon 3; reverse primer in exon 4). Smad2 and Smad3 transcript levels were normalized to the levels of the housekeeping gene Rpl19. Data represent means ± se from 3 independent sorting experiments assayed in triplicate. Bars with different symbols differ significantly.
In addition to S2/3cKO mice, we also generated single-gene-knockout mice lacking either Smad2 or Smad3 in gonadotropes. As these mice showed no abnormalities in any of the experiments performed (data not shown), we focus here on the analysis of the double-knockout model.
Puberty and reproductive function in S2/3cKO females
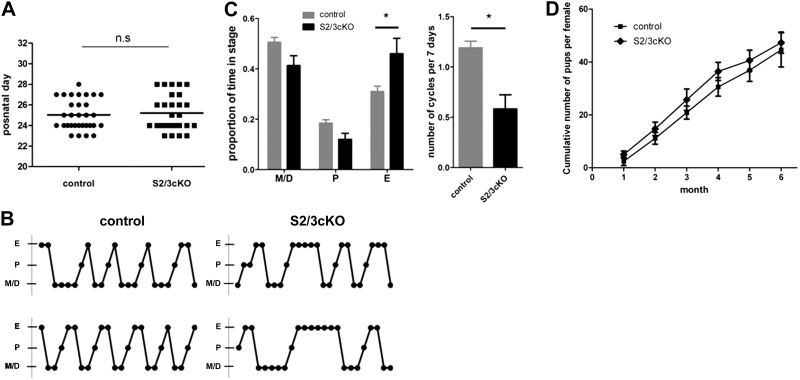
To evaluate the effect of gonadotrope-specific loss of SMAD2/3 on hypothalamic-pituitary-gonadal (HPG) axis activity, we monitored reproductive maturation in S2/3cKO and control females. First, we assessed the day of vaginal opening (v.o.), an estrogen-dependent external marker of puberty onset in mice (33). S2/3cKO mice and control littermates showed a comparable onset of v.o. (Fig. 2A). Subsequently, we examined estrous cyclicity by daily vaginal smears in a cohort of 7 S2/3cKO and 7 control females over a period of 3 wk. Mice of both genotypes exhibited cyclic variation in vaginal cytology, including several 4- to 5-d cycles, as is typical in mice (33). However, 5 of 7 S2/3cKO females showed 1 or more prolonged (4 consecutive days or more) periods of estrus, whereas such events were never observed in control littermates (Fig. 2B). As a result, S2/3cKO females had a significant reduction in estrous cycle frequency and an increase in the proportion of time spent in estrus (Fig. 2C).
Figure 2.
Largely intact reproductive maturation and normal fertility in S2/3cKO females. A) Day of v.o. in S2/3cKO and control mice. Each dot represents an individual mouse. Horizontal line represents the group mean. B) Representative estrous cyclicity profile from 2 control (left) and 2 S2/3cKO (right) mice. M/D, metestrus/diestrus; P, proestrus; E, estrus. C) Proportion of time spent in each cycle stage (left) and estrus cycle frequency (right) for control and S2/3cKO mice (n=7/genotype). D) Cumulative number of pups delivered per female over the course of a 6 mo breeding trial, calculated at the end of each month. Data represent means ± se of 7 mice/genotype. *P ≤ 0.05.
Next, we monitored fertility in the same cohort of animals by pairing them individually with wild-type C57BL/6J male mice for a period of 6 mo. Unexpectedly, S2/3cKO females showed normal fertility. They produced litters of similar sizes and at a comparable frequency to control females, which was reflected in the cumulative number of pups delivered over the period of the breeding trial (Fig. 2D). To gain a more comprehensive view of HPG axis activity, we examined the reproductive organs of an additional cohort of 10-wk-old females. The uteri and ovaries of S2/3cKO females were comparable in morphology and weight to those of control littermates (Supplemental Fig. S1C, D, and data not shown). Ovaries of S2/3cKO mice were histologically normal, with follicles at all stages of development. Moreover, quantitative analyses revealed similar numbers of CL in S2/3cKO and control mice (Supplemental Fig. S1E). Ovarian expression of the FSH-responsive genes cyclin D2 (Ccnd2), cytochrome P450; subfamily A; polypeptide 1 (aromatase) (Cyp19a1), and luteinizing hormone receptor (Lhr) (39) did not differ between genotypes (Supplemental Fig. S1F). Collectively, the data indicate that S2/3cKO females had increased variability in estrous cyclicity but otherwise exhibited normal reproductive function.
Reduced testes weights and sperm production in S2/3cKO males
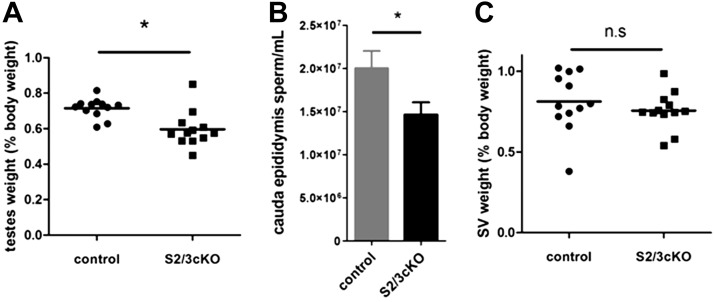
Although male mice do not require FSH for fertility, Fshb-knockout males do exhibit gonadal dysfunction, most notably small testes and oligospermia (1). Furthermore, as the GnrhrGRIC allele is active in male germ cells (32), deletion of the SMAD proteins therein might affect sperm function or survival (40, 41). We therefore examined the reproductive organs of S2/3cKO males. Double knockouts had significantly decreased testes weights relative to controls (Fig. 3A). This was accompanied by a reduction in epididymal sperm counts (Fig. 3B). Testicular histology was largely unremarkable, although some seminiferous tubules appeared smaller in diameter in the testes of S2/3cKO males compared with controls (Supplemental Fig. S1G). Seminal vesicle weight, a marker of circulating testosterone levels, was comparable between S2/3cKO male and control littermates (Fig. 3C). Consistent with their minor reproductive organ anomalies, S2/3cKO males exhibited normal fertility (data not shown).
Figure 3.
Small reductions in testes weights and epididymal sperm counts in S2/3cKO males. A) Testes weight (sum of both testes), expressed as a percentage of body weight, in 10-wk-old control and S2/3cKO male mice. Each point represents an individual mouse. Horizontal line represents the group mean. B) Caudal epididymal sperm count (per milliliter of homogenization buffer) in 10-wk-old control and S2/3cKO male mice. C) Wet seminal vesicle (SV) weight, normalized to body weight in the same animals as in A. n.s., not significant. *P ≤ 0.05.
Normal pituitary Fshb expression and FSH synthesis in S2/3cKO mice
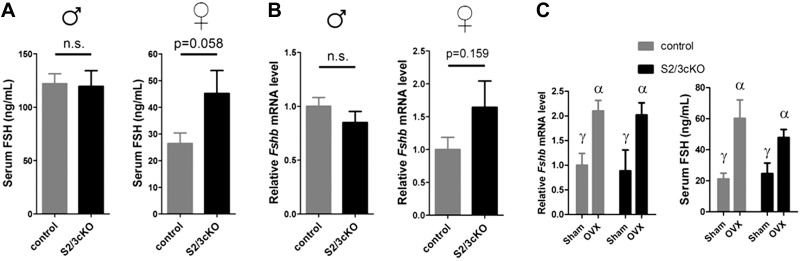
In light of their unexpectedly mild reproductive abnormalities and normal fertility, we next asked whether FSH synthesis was impaired in S2/3cKO mice, as we predicted a priori. Surprisingly, serum FSH levels were equivalent in S2/3cKO mice and control littermates of both sexes (Fig. 4A). Indeed, in metestrus/diestrus female S2/3cKO mice, there was even a trend for increased circulating FSH (71% increase; P=0.058; Fig. 4A). Consistent with the serum FSH values, pituitary Fshb transcript levels, analyzed in the same animals, were not significantly different between the genotypes; although, again, there was a nonsignificant increase in S2/3cKO females (64% increase; P=0.159; Fig. 4B). Similarly, circulating LH levels were normal in these 10-wk-old S2/3cKO mice (Supplemental Fig. S2A). However, we observed significantly higher serum LH, but not FSH, in older S2/3cKO than control females (metestrus/diestrus) retired from the breeding trials (Supplemental Fig. S2B, C).
Figure 4.
Normal FSH synthesis and Fshb expression in S2/3cKO mice. A) Circulating FSH levels in 10-wk-old control and S2/3cKO males (left) and metestrus/diestrus females (right). Data represent means +se of 12 mice/group. B) Relative pituitary Fshb mRNA levels,(with controls set to 1) assayed by qPCR in the same animals as in A. C) Adult metestrus/diestrus control and S2/3cKO females received ovariectomy (OVX) or sham surgery (sham). After 7 h, mice were killed, and their serum and pituitaries were collected. Serum FSH (left) and pituitary Fshb mRNA levels (right) were analyzed in the same animals. Data represent means ± se of 6–7 mice/group. Bars with different symbols differ significantly.
Next, we probed the ability of S2/3cKO females to up-regulate FSH synthesis following the removal of gonad-derived hormone negative feedback. In female rodents, an acute phase of increased FSH synthesis occurs within a few hours after ovariectomy. Because this increase is gonadotropin-releasing hormone (GnRH) independent, it is assumed to reflect increased activin-driven FSH production following the loss of ovarian inhibin feedback (42). Therefore, we ovariectomized adult mestestrus/diestrus females and analyzed serum FSH and pituitary Fshb transcript levels after 7 h. In both control and S2/3cKO mice, there was a significant increase in circulating FSH levels and pituitary Fshb expression in ovariectomized mice compared with sham-operated littermates (Fig. 4C). Collectively, these results suggest that activin-dependent Fshb expression and FSH synthesis are unimpaired in S2/3cKO mice.
Impaired Fshb transcription on acute ablation of Smad2/3 in primary pituitary cells
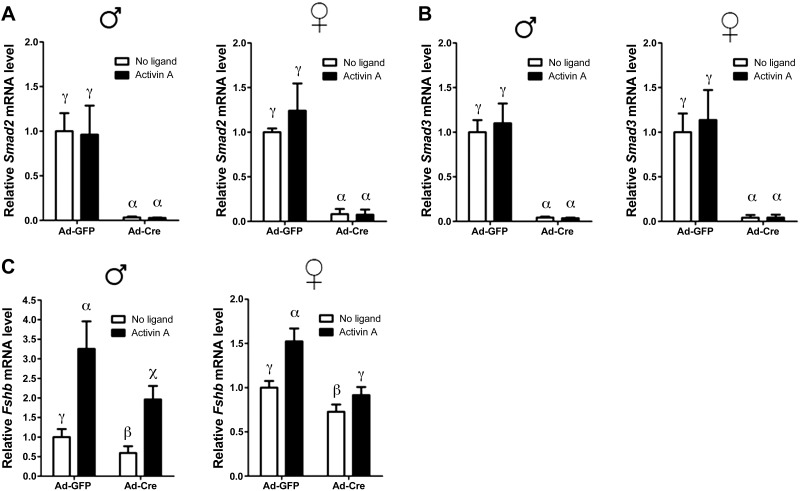
Given the unexpectedly intact FSH secretion in S2/3cKO mice, we revisited the roles of SMAD2 and SMAD3 in activin-stimulated Fshb transcription. Most of the evidence implicating these proteins in Fshb transcription regulation comes from overexpression and knockdown studies in immortalized gonadotrope-like and heterologous cell lines (13–18). To assess whether SMAD2/3 are similarly important in primary gonadotropes, we prepared pituitary cultures from mice homozygous for the Smad2/3 conditional alleles (Smad2fl/fl;Smad3fl/fl) and infected these cells with adenovirus expressing Cre recombinase (Ad-Cre) or control adenovirus expressing green fluorescent protein (Ad-GFP) to induce recombination of Smad2/3 ex vivo. This procedure was highly efficient, as Smad2 and Smad3 mRNA levels were depleted by >95% after transduction with Ad-Cre (Fig. 5A, B). Basal Fshb mRNA levels, which depend on autocrine/paracrine activin (or activin-like) signaling (42, 43), were reduced in Ad-Cre-infected male or female cultures (Fig. 5C). Furthermore, exogenous activin A stimulated Fshb expression in Ad-Cre-transduced cultures but to a lesser extent than in Ad-GFP-infected cell (Fig. 5C). This difference was especially striking in cultures prepared from female mice. Deletion of Smad2 or Smad3 alone had similar, but generally milder, effects on basal and activin A-stimulated Fshb mRNA levels (Supplemental Fig. S3A, B). Together, these data suggest that activin regulation of Fshb expression is at least partially SMAD2/3-dependent in cultured pituitary cells.
Figure 5.
Partial dependence of basal and activin-stimulated Fshb expression on full-length SMAD2/3 in primary pituitary cultures. A, B) Primary pituitary cultures were prepared from Smad2fl/fl;Smad3fl/fl male (left) and female mice (right), and infected for 24 h with Ad-Cre or Ad-GFP. Cells were stimulated for 24 h with 1 nM activin A, or left untreated (no ligand) before RNA extraction. Smad2 (A) and Smad3 (B) mRNA levels were assessed by qPCR. C) Expression of Fshb in the same samples as in A and B. In all panels, data represent means ± se of 5 independent experiments measured in triplicate (n=5). Bars with different symbols differ significantly.
Retention of a transcript encoding a functional, truncated SMAD3 protein in S2/3cKO gonadotropes
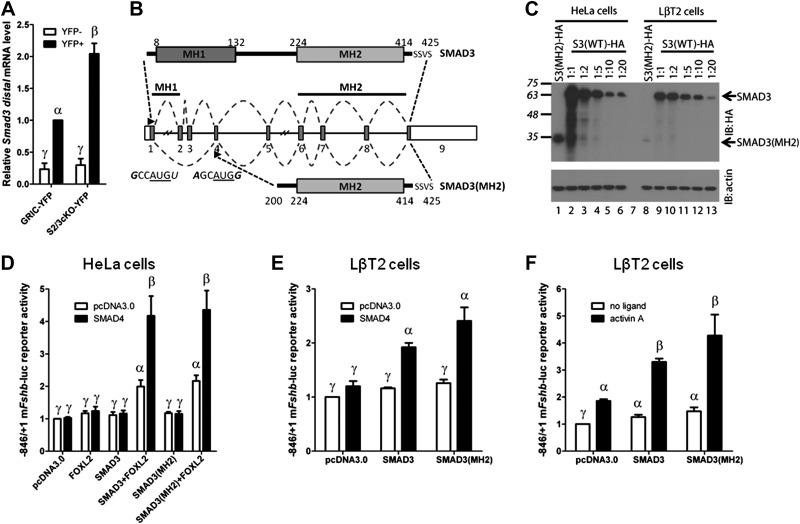
The recombined Smad2 allele used in our study produces a truncated protein (30). However, this protein cannot be C-terminally phosphorylated and does not have activity in functional assays (44). We therefore questioned whether, in contrast, functional Smad3 transcripts might be retained in gonadotropes of S2/3cKO animals. Indeed, qPCR analysis of YFP+ and YFP− cells sorted from the pituitaries of control (GRIC-YFP) and S2/3cKO-YFP animals indicated a 2-fold up-regulation of Smad3 mRNA containing distal exons (exons 8 and 9) in S2/3cKO gonadotropes (Fig. 6A). This contrasted with a robust loss of Smad3 transcripts containing the deleted exons (exons 2 and 3; see Fig. 1D). We further characterized retained Smad3 transcripts in S2/3cKO gonadotropes using a primer walk PCR strategy on cDNA obtained from FACS-sorted gonadotropes from control or S2/3cKO animals with an antisense primer in the terminal exon 9 and sense primers in exons 1 through 8. A single PCR product, of a smaller size than the full-length transcript (data not shown), was amplified from S2/3cKO (but not control) gonadotropes using the sense primer in exon 1. This transcript contained exon 1 spliced to exon 4 and then proceeded normally to include all the remaining exons of Smad3 (Fig. 6B).
Figure 6.
Retention of a truncated Smad3 transcript coding for a functional protein in S2/3cKO mice. A) Distal Smad3 mRNA levels (forward primer in exon 8; reverse primer in exon 9), analyzed by qPCR in YFP+ and YFP− cells from the same experiment as in Fig. 1. Data represent means ± se of 3 independent sorting experiments measured in triplicate (n=3). B) Top: schematic representation of the wild-type SMAD3 protein. Amino acids are numbered. SSVS sequence at the C terminus contains the serine residues phosphorylated by the type I receptor. Middle: Smad3 gene structure with numbered exons. Splicing of the wild-type and recombined Smad3 alleles is depicted with dashed lines. Arrowheads indicate the start of translation in exon 1, and the putative alternative translation initiation site in exon 4. The nucleotide sequence surrounding the AUG sequence (underlined) of both sites is indicated. Bases that conform to the consensus Kozak sequence, −3RCCAUGG+4, at positions −3 and +4, are indicated in italics. Bottom: Representation of the truncated protein, SMAD3(MH2), arising from the recombined Smad3 allele. C) Western blot analysis of whole-cell protein lysates from HeLa (lanes 1–6) or LβT2 (lanes 8–13) cells transfected with 2 μg expression vector encoding C-terminally tagged 3X-HA tagged truncated SMAD3 (SMAD3-MH2; lane 1 and 8) or differing amounts of wild-type SMAD3-HA expression vector (lanes 2–6 and 9–13: 1:1–2 μg; 1:2-1 μg; 1:5-0.4 μg; 1:10-0.2 μg; 1:20-0.1 μg). The blot was probed with anti-HA (IB: HA, top) and anti-β-actin (IB: actin, bottom) antibodies. Arrows indicate the relative migration of full-length SMAD3 and SMAD3-MH2 products. D) HeLa cells were transfected with 225 ng/well of the −846/+1 murine Fshb-luc reporter, with 25 ng of FOXL2 and/or SMAD4 expression vectors, together with 25 ng of SMAD3 or 5 times more (125 ng) SMAD3(MH2) expression vectors. Whole-cell lysates were subjected to luciferase assays. The amount of transfected vectors was balanced across all conditions with pcDNA3.0. E) LβT2 cells were transfected with 225 ng/well of the −846/+1 murine Fshb-luc reporter, with 25 ng of SMAD4 expression vector, together with 25 ng of SMAD3 or 20 times more (500 ng) SMAD3(MH2) expression vector as indicated. The amount of transfected vectors was balanced across all conditions with pcDNA3.0. Whole-cell lysates were subjected to luciferase assays. F) LβT2 cells were transfected with 225 ng/well of the −846/+1 murine Fshb-luc reporter, along with 25 ng SMAD3 or 20 times more (500 ng) SMAD3(MH2) expression vector. The amount of transfected vectors was balanced across all conditions with pcDNA3.0. Cells were stimulated with 1 nM activin A for 24 h, or left untreated, before assaying luciferase activity. Data represent means ± se of 7 (D) or 3 (E, F) independent experiments performed in triplicate. Bars with different symbols differ significantly.
Translation of the SMAD3 protein normally initiates in exon 1, and the relevant translational start site (TSS) is retained in the truncated transcript. Removal of exons 2–3 introduces a frameshift in exon 4, thus preventing the production of a functional protein from the canonical TSS. However, as the TSS in exon 1 does not conform to a consensus Kozak sequence (45), we considered the possibility that an alternative TSS, downstream of exon 3, might exist and be utilized in the novel Smad3 transcript. We identified 2 potential AUG start codons in exon 4, one of which occurred in the context of a consensus Kozak sequence. This raised the possibility that the knockout transcript might encode a protein containing the entirety of the Mad homology 2 (MH2) domain in the event of leaky ribosomal scanning or translation reinitation (Fig. 6B).
To assess this possibility, we generated expression vectors for the full-length and truncated transcript (both starting upstream of the canonical TSS in exon 1) fused to a 3X-HA epitope tag at the C terminus and transfected them into HeLa and LβT2 (immortalized gonadotrope-like) cells. Western blot analysis of cells transfected with the wild-type SMAD3 expression vector revealed a protein product with a molecular mass of 55–60 kDa, consistent with the predicted size of full-length 3X-HA-tagged SMAD3 (Fig. 6C, lanes 2–6 and 9–13). In cells transfected with the truncated SMAD3 expression vector, the full-length protein was not observed; however, a novel, lower abundance band, appeared at ∼30 kDa (Fig. 6C, lanes 1 and 8). The size of this product was consistent with translation initiation in exon 4. Because this protein is predicted to retain the SMAD3 MH2 domain, but lack all of the Mad homology 1 (MH1) domain and most of the linker region, it is hereafter referred to as SMAD3(MH2). Titration experiments indicated that a SMAD3:SMAD3(MH2) expression vector ratio of 1:5 in HeLa cells and 1:20 in LβT2 cells yielded comparable protein expression levels (Fig. 6C, compare lane 4 with lane 1, and lane 13 with lane 8, respectively). It should be noted that SMAD3(MH2) corresponds to a naturally occurring variant previously described in murine pituitary and gonadotrope-like cells (46). This protein is phosphorylated and accumulates in the nucleus in response to activin A.
Using promoter-reporter assays, we assessed whether SMAD3(MH2) retains functional activity at the murine Fshb promoter. To directly compare the activity of SMAD3(MH2) and wild-type SMAD3, we transfected the amount of expression vector required to produce comparable levels of the 2 proteins, determined by our titration experiments described above (Fig. 6C). We previously reported that ectopic expression of FOXL2 and SMAD3 stimulates murine Fshb promoter activity in heterologous cells and that this effect is potentiated by SMAD4 (23). We reproduced these results here in HeLa cells using a −846/+1 murine Fshb-luciferase reporter (Fig. 6D). SMAD3(MH2) synergistically activated the Fshb promoter with FOXL2 and SMAD4 to the same extent as wild-type SMAD3 (Fig. 6D). To extend these analyses to a homologous system, we next employed LβT2 cells, which express endogenous FOXL2. Both wild-type SMAD3 and SMAD3(MH2) synergistically activated the −846/+1 murine Fshb-luc reporter with SMAD4 (Fig. 6E) and activin A (Fig. 6F). SMAD3(MH2) tended to show greater activity than wild-type in these assays, although this was not statistically significant. Collectively, these data indicate that the recombined Smad3 allele encodes a truncated transcript and protein capable of activating Fshb transcription in cooperation with SMAD4 and FOXL2.
DISCUSSION
SMAD2 and SMAD3 are the canonical activin-induced signaling molecules, and both were previously implicated in activin-regulated Fshb transcription. We were therefore surprised to observe quantitatively normal FSH synthesis and fertility in gonadotrope-specific Smad2/3 conditional knockout mice. Although the most parsimonious explanation for these results might be incomplete or insufficient recombination of the floxed alleles, at least 3 lines of evidence argue against this possibility. First, we observe robust (>90%) suppression of full-length Smad2 and Smad3 transcripts in genetically labeled gonadotropes of S2/3cKO mice. Second, the GnrhrGRIC allele has demonstrated specificity and efficiency in other models (25, 32, 38). Third, we further decreased Smad2/3 gene dosage by globally deleting 1 allele each of Smad2 and Smad3 such that only 1 floxed allele per gene required recombination in gonadotropes (Smad2fl/−;Smad3fl/−;GnrhrGRIC/+) but failed to observe additional phenotypes (data not shown). In light of these observations, we consider 3 alternative explanations for the absence of FSH deficiency in S2/3cKO mice: compensation by a residual but truncated form of SMAD3, SMAD3(MH2); compensation by activin-dependent but SMAD2/3-independent signaling; or compensation by activin-independent signaling. We consider each of these possibilities in turn, with greatest emphasis on the first.
Our data clearly rule out necessary roles for SMAD3 DNA-binding activity and SMAD2 for quantitatively normal FSH synthesis in vivo. This result might have been anticipated for SMAD2 based on previous in vitro manipulations of SMAD2/3 levels (by knockdown or overexpression), which indicated a quantitatively more important role for SMAD3 than SMAD2 in Fshb transcriptional regulation (13, 16, 17, 23, 36). Furthermore, FOXL2, which is required for FSH synthesis and fertility in vivo, interacts more strongly with SMAD3 than SMAD2 (36, 47). In contrast with in vivo observations, but perhaps more in line with prior cell line data, we observed significant impairments of basal and activin A-stimulated Fshb expression following acute ablation of full-length Smad2/3 in primary pituitary cells. Two, nonmutually exclusive, possibilities may explain these apparently discrepant results. First, the short time-frame of primary culture experiments may not have enabled the development of compensatory mechanisms similar to those established in vivo. Second, gonadotropes in dissociated cultures may rely more on ligands signaling through SMAD2/3 to maintain Fshb expression than do gonadotropes in the context of the intact gland and/or animal. Consistent with the first possibility, basal and activin A-stimulated Fshb mRNA expression were normal in pituitary cultures of female S2/2cKO mice (data not shown).
These in vivo and in vitro discrepancies aside, the most parsimonious explanation for the absence of FSH deficiency in S2/3cKO mice was the failure of our mouse model to ablate all of SMAD3 function. That is, the recombined Smad3 allele produces a transcript that encodes a truncated but still functional SMAD3 protein: SMAD3(MH2). This protein is likely generated via translation reinitiation or leaky ribosomal scanning from the novel mRNA transcribed in these mice (45). The translation start site in exon 1, from which full-length SMAD3 is ordinarily derived, does not conform to a consensus Kozak sequence, with a thymine (uracil in the mRNA) rather than a guanine at the + 4 position (with the adenine of the ATG/AUG denoted as+1). In the context of the mRNA lacking exons 2 and 3, translation appears to initiate at a consensus Kozak sequence in exon 4. Translation from this site is not unprecedented as an alternative Smad3 transcript initiating in the third intron (generating a novel exon 3a) was previously described in gonadotrope-like LβT2 cells (46). Although we were unable to confirm the presence of this particular transcript in control or S2/3cKO gonadotropes, the resulting protein would be indistinguishable from SMAD3(MH2) described here. As indicated above, the authors of the previous study showed that this truncated protein is phosphorylated and accumulates in the nucleus on activin A stimulation. In contrast to what we report here, they suggested that the truncated protein acts as a dominant-negative when coexpressed with wild-type SMAD3. However, its independent actions were not assessed in their experiments and we propose that their results might be alternatively explained by hypomorphic (rather than dominant-negative) activity, as we observed in our reporter assays. That is, SMAD3(MH2) is expressed at lower levels than wild-type SMAD3 when equivalent amounts of expression vector are employed. We had to titrate the amount of wild-type vector to achieve equivalent expression of the 2 proteins to demonstrate their similar activities in transcriptional assays.
Based on previous observations, we predict that SMAD3(MH2) fulfills SMAD3 functions necessary for proper Fshb transcription. Indeed, we previously reported that either SMAD3 or SMAD4 (but not both) must bind DNA to stimulate murine Fshb transcription, suggesting that SMAD3 DNA-binding activity is dispensable for FSH synthesis (23). Furthermore, SMAD3 physically interacts with SMAD4 and FOXL2 via its MH2 domain, which is preserved in the truncated protein described here. Consistent with these observations, SMAD3(MH2) activates the murine Fshb promoter in cooperation with SMAD4 and FOXL2 in in vitro assays. Supporting the important functionality of the SMAD3 MH2 domain, Smad3Δexon8-knockout mice have reproductive anomalies and pituitary Fshb deficiency. It is important to note, however, that the reduction in Fshb expression in Smad3Δexon8 KO mice is quantitatively modest (around 30%; ref. 26), and females actually have elevated serum FSH levels (28). However, these observations may be confounded by ovarian abnormalities (27, 28) and/or partial compensation by SMAD2 in these mice.
In our in vitro experiments, transfection of the same amount of truncated and full-length Smad3 expression vectors produced notably less SMAD3(MH2) than SMAD3 protein. This result is expected for proteins generated through leaky ribosomal scanning or translation reinitiation (45). Nevertheless, when expressed at the same protein level, SMAD3(MH2) was functionally equivalent to wild-type SMAD3. The implications of these observations for the potential of SMAD3(MH2) to compensate for the loss of full-length SMAD3 in vivo are hard to predict for at least 2 reasons. First, it is unclear what threshold level of SMAD3 activity (expression) is required to sustain quantitatively normal FSH synthesis in vivo. Second, we observed that transcripts containing distal exons were upregulated ∼2-fold in S2/3cKO compared with control gonadotropes. Therefore, it is possible that the amount of SMAD3(MH2) protein in mutant gonadotropes is closer to wild-type levels than suggested by our in vitro overexpression studies. Unfortunately, due to the paucity of gonadotropes (we typically isolate 5,000–7,000 cells per pituitary), we are unable to obtain sufficient numbers of purified cells from GRIC-YFP and S2/3cKO-YFP mice for protein analysis. Therefore, establishing the necessity of SMADsin pituitary FSH synthesis will require cell-specific removal of all protein function, likely with a novel conditional Smad3 allele.
Our observation that the recombined Smad3 allele encodes a functional protein may also clarify discrepancies in the phenotypes of existing Smad3-knockout mouse lines. Indeed, global deletion of different Smad3 exons has produced divergent phenotypes, with the only common observation being smaller body size (27, 44, 48, 49). Smad3Δexon2-knockout embryos produce an identical transcript (exon 1 splicing to exon 4) as the one reported here, presumably resulting in the production of the SMAD3(MH2) protein. Interestingly, this is apparently the only Smad3-null strain that develops fully penetrant colorectal cancer (49, 50). Unique phenotypes reported in other Smad3-knockout lines include mild forelimb malformation in Smad3Δexon1 mice (48) and immune system dysfunction and aortic aneurysms in Smad3Δexon8 mice (44, 51). The only allele that disrupts the coding sequence for the MH2 domain is Smad3Δexon8. In light of our results, it is perhaps not surprising that this is the only Smad3-deficient strain in which reproductive defects and pituitary Fshb deficiency have been reported (27, 28). However, Smad3Δexon8 mice also produce a truncated SMAD3 protein, comprising the MH1 domain, which can act as a dominant-negative in some contexts (44). Thus, as mentioned above, a novel conditional Smad3 allele, which completely removes protein function and lacks dominant-negative activity, is needed to assess the role of SMAD3 not only in gonadotropes, but in all cell types. Indeed, such a mouse model will enable, if not necessitate, repetition of investigations using other modified Smad3 alleles.
Although it seems likely that the retention of SMAD3(MH2) explains, at least in part, normal FSH levels in S2/3cKO mice, other mechanisms of compensation may exist. Other receptor-regulated SMADs (R-SMADs), in particular SMAD8, can activate Fshb transcription in vitro (52–54). Further, activins can signal through noncanonical R-SMADs in some contexts (55), although the extent to which this also occurs in gonadotropes is unknown. In addition, it was proposed that activins may regulate Fshb transcription via a SMAD-independent mechanism involving the kinases transforming growth factor β-activated kinase 1 (TAK1) and p38 (56). However, a subsequent study showed that the small molecule TAK1 inhibitor 5Z-7-oxozeaenol nonspecifically blocks activin receptor-like kinase family member 4 (ALK4) activity. This and other studies also failed to confirm a role for p38 in this system (14, 57, 58). Therefore, in vitro studies do not, at present, demonstrate SMAD-independent mechanism of FSH regulation by activins. This question could be definitively resolved, however, by disrupting all SMAD-dependent signaling by selectively ablating the common partner Smad4 in gonadotropes. Indeed, preliminary data along these lines from our laboratory appear to confirm a necessary role for SMAD signaling in Fshb expression in vivo (59).
Signaling by hormones other than activins may regulate Fshb transcription and FSH synthesis in vivo, perhaps bypassing a requirement for SMAD2/3. For example, bone morphogenetic proteins (BMPs) regulate Fshb transcription and FSH synthesis in primary pituitary cells and gonadotrope-like cell lines (53, 54, 57, 60, 61). However, their contributions to FSH regulation in vivo remain to be determined. Another obvious candidate is GnRH, a well-established and potent stimulator FSH synthesis (3, 62). Indeed, mice that lack GnRH or the GnRH receptor exhibit profound FSH deficiency (63, 64). However, because GnRH also regulates the expression of follistatin (65, 66), an activin antagonist, FSH phenotypes in GnRH-deficient mice may be attributable, at least in part, to dysregulation of activin or activin-like signaling. The relative roles of activins and GnRH in FSH regulation in vivo have not been clearly established; however, it is possible that GnRH might assume a more important and compensatory role in the absence of signaling via SMAD2/3. Finally, compensatory regulation by steroids may help sustain normal FSH production. In rodents, on the morning of estrus, a selective rise in FSH synthesis and secretion drives ovarian follicle recruitment and maturation (67). Although this FSH surge is thought to be driven by increased activin signaling in the face of lower inhibin levels (68, 69), it is also blocked by progesterone and glucocorticoid receptor antagonists (70–73). Thus, it is possible that the steroid milieu may enable appropriate FSH production in the absence of activin signaling.
In summary, our results demonstrate, for the first time, that Fshb transcription and FSH synthesis can occur independently of SMAD3 DNA-binding activity and SMAD2 in vivo. Further, they reveal that deletion of exons 2 and 3 of Smad3 results in the production of a novel Smad3 transcript, which encodes a functional protein. This latter observation has important implications for investigations (past, present, and future) of SMAD3 function using existing Smad3-knockout mouse lines.
Supplementary Material
Acknowledgments
The authors acknowledge Xiang Zhou for help with the ovariectomy procedures and Ken McDonald for assistance with FACS.
J.F. was supported by a Doctoral Research Award from the Canadian Institutes of Health Research (CIHR). This work was funded by CIHR operating grant MOP-89991 (to D.J.B). D.J.B. is a Senior Research Scholar of the Fonds de la Recherche en Santé de Québec. U.B. is funded by DFG BO1743/2. The University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core is supported by the U.S. National Institutes of Health National Institute of Child Health and Human Development (Specialized Cooperative Centers Program in Reproduction Research) grant U54-HD28934.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- Ad-Cre
- adenovirus expressing Cre recombinase
- Ad-GFP
- adenovirus expressing green fluorescent protein
- cKO
- conditional knockout
- eGFP
- enhanced green fluorescent protein
- eYFP
- enhanced yellow fluorescent protein
- FACS
- fluorescence-activated cell sorting
- FOXL2
- forkhead box L2
- FSH
- follicle-stimulating hormone
- Fshb
- follicle-stimulating hormone β subunit
- GFP
- green fluorescent protein
- GnRH
- gonadotropin-releasing hormone
- Gnrhr
- gonadotropin-releasing hormone receptor
- HPG
- hypothalamic-pituitary-gonadal
- LH
- luteinizing hormone
- MH1
- Mad homology 1
- MH2
- Mad homology 2
- qPCR
- quantitative polymerase chain reaction
- R-SMAD
- receptor-regulated SMAD
- S2/3cKO
- Smad2/3 conditional knockout
- SMAD2/3/4
- mammalian homolog of Drosophila mothers against decapentaplegic; family member 2/3/4
- TSS
- translation start site
- YFP
- yellow fluorescent protein
- v.o
- vaginal opening
REFERENCES
- 1. Kumar T. R., Wang Y., Lu N., Matzuk M. M. (1997) Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat. Genet. 15, 201–204 [DOI] [PubMed] [Google Scholar]
- 2. Themmen A. P. N., Huhtaniemi I. T. (2000) Mutations of gonadotropins and gonadotropin receptors: elucidating the physiology and pathophysiology of pituitary-gonadal function. Endocr. Rev. 21, 551–583 [DOI] [PubMed] [Google Scholar]
- 3. Bernard D. J., Fortin J., Wang Y., Lamba P. (2010) Mechanisms of FSH synthesis: what we know, what we don't, and why you should care. Fertil. Steril. 93, 2465–2485 [DOI] [PubMed] [Google Scholar]
- 4. Ling N., Ying S. Y., Ueno N., Esch F., Denoroy L., Guillemin R. (1985) Isolation and partial characterization of a Mr 32,000 protein with inhibin activity from porcine follicular fluid. Proc. Natl. Acad. Sci. U. S. A. 82, 7217–7221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ling N., Ying S. Y., Ueno N., Shimasaki S., Esch F., Hotta M., Guillemin R. (1986) Pituitary FSH is released by a heterodimer of the beta-subunits from the two forms of inhibin. Nature 321, 779–782 [DOI] [PubMed] [Google Scholar]
- 6. Vale W., Rivier C., Hsueh A., Campen C., Meunier H., Bicsak T., Vaughan J., Corrigan A., Bardin W., Sawchenko P., (1988) Chemical and biological characterization of the inhibin family of protein hormones. Recent. Prog. Horm. Res. 44, 1–34 [DOI] [PubMed] [Google Scholar]
- 7. Vale W., Rivier J., Vaughan J., McClintock R., Corrigan A., Woo W., Karr D., Spiess J. (1986) Purification and characterization of an FSH releasing protein from porcine ovarian follicular fluid. Nature 321, 776–779 [DOI] [PubMed] [Google Scholar]
- 8. Ying S. Y. (1988) Inhibins, activins, and follistatins: gonadal proteins modulating the secretion of follicle-stimulating hormone. Endocr. Rev. 9, 267–293 [DOI] [PubMed] [Google Scholar]
- 9. Thackray V. G., Mellon P. L., Coss D. (2009) Hormones in synergy: regulation of the pituitary gonadotropin genes. Mol. Cell. Endocrinol. 314, 192–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bilezikjian L. M., Justice N. J., Blackler A. N., Wiater E., Vale W. W. (2012) Cell-type specific modulation of pituitary cells by activin, inhibin and follistatin. Mol. Cell. Endocrinol. 359, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pangas S. A., Woodruff T. K. (2000) Activin signal transduction pathways. Trends Endocrinol. Metab. 11, 309–314 [DOI] [PubMed] [Google Scholar]
- 12. Shi Y., Massague J. (2003) Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 13. Bernard D. J. (2004) Both SMAD2 and SMAD3 mediate activin-stimulated expression of the follicle-stimulating hormone beta subunit in mouse gonadotrope cells. Mol. Endocrinol. 18, 606–623 [DOI] [PubMed] [Google Scholar]
- 14. Dupont J., McNeilly J., Vaiman A., Canepa S., Combarnous Y., Taragnat C. (2003) Activin signaling pathways in ovine pituitary and LbetaT2 gonadotrope cells. Biol. Reprod. 68, 1877–1887 [DOI] [PubMed] [Google Scholar]
- 15. Lamba P., Santos M. M., Philips D. P., Bernard D. J. (2006) Acute regulation of murine follicle-stimulating hormone beta subunit transcription by activin A. J. Mol. Endocrinol. 36, 201–220 [DOI] [PubMed] [Google Scholar]
- 16. Suszko M. I., Balkin D. M., Chen Y., Woodruff T. K. (2005) Smad3 mediates activin-induced transcription of follicle-stimulating hormone beta-subunit gene. Mol. Endocrinol. 19, 1849–1858 [DOI] [PubMed] [Google Scholar]
- 17. Suszko M. I., Lo D. J., Suh H., Camper S. A., Woodruff T. K. (2003) Regulation of the rat follicle-stimulating hormone beta-subunit promoter by activin. Mol. Endocrinol. 17, 318–332 [DOI] [PubMed] [Google Scholar]
- 18. Wang Y., Fortin J., Lamba P., Bonomi M., Persani L., Roberson M. S., Bernard D. J. (2008) Activator protein-1 and smad proteins synergistically regulate human follicle-stimulating hormone beta-promoter activity. Endocrinology 149, 5577–5591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y., Libasci V., Bernard D. J. (2010) Activin A induction of FSHbeta subunit transcription requires SMAD4 in immortalized gonadotropes. J. Mol. Endocrinol. 44, 349–362 [DOI] [PubMed] [Google Scholar]
- 20. Lamba P., Fortin J., Tran S., Wang Y., Bernard D. J. (2009) A novel role for the forkhead transcription factor FOXL2 in activin A-regulated follicle-stimulating hormone beta subunit transcription. Mol. Endocrinol. 23, 1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lamba P., Khivansara V., D'Alessio A. C., Santos M. M., Bernard D. J. (2008) Paired-like homeodomain transcription factors 1 and 2 regulate follicle-stimulating hormone beta-subunit transcription through a conserved cis-element. Endocrinology 149, 3095–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suszko M. I., Antenos M., Balkin D. M., Woodruff T. K. (2008) Smad3 and Pitx2 cooperate in stimulation of FSHbeta gene transcription. Mol. Cell. Endocrinol. 281, 27–36 [DOI] [PubMed] [Google Scholar]
- 23. Tran S., Lamba P., Wang Y., Bernard D. J. (2011) SMADs and FOXL2 synergistically regulate murine FSHbeta transcription via a conserved proximal promoter element. Mol. Endocrinol. 25, 1170–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Justice N. J., Blount A. L., Pelosi E., Schlessinger D., Vale W., Bilezikjian L. M. (2011) Impaired FSHbeta expression in the pituitaries of Foxl2 mutant animals. Mol. Endocrinol. 25, 1404–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tran S., Zhou X., Lafleur C., Calderon M. J., Ellsworth B. S., Kimmins S., Boehm U., Treier M., Boerboom D., Bernard D. J. (2013) Impaired fertility and FSH synthesis in gonadotrope-specific Foxl2 knockout mice. Mol. Endocrinol. 27, 407–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Coss D., Thackray V. G., Deng C. X., Mellon P. L. (2005) Activin regulates luteinizing hormone beta-subunit gene expression through Smad-binding and homeobox elements. Mol. Endocrinol. 19, 2610–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tomic D., Brodie S. G., Deng C., Hickey R. J., Babus J. K., Malkas L. H., Flaws J. A. (2002) Smad 3 may regulate follicular growth in the mouse ovary. Biol. Reprod. 66, 917–923 [DOI] [PubMed] [Google Scholar]
- 28. Tomic D., Miller K. P., Kenny H. A., Woodruff T. K., Hoyer P., Flaws J. A. (2004) Ovarian follicle development requires Smad3. Mol. Endocrinol. 18, 2224–2240 [DOI] [PubMed] [Google Scholar]
- 29. Li Q., Pangas S. A., Jorgez C. J., Graff J. M., Weinstein M., Matzuk M. M. (2008) Redundant roles of SMAD2 and SMAD3 in ovarian granulosa cells in vivo. Mol. Cell. Biol. 28, 7001–7011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y., Festing M. H., Hester M., Thompson J. C., Weinstein M. (2004) Generation of novel conditional and hypomorphic alleles of the Smad2 gene. Genesis 40, 118–123 [DOI] [PubMed] [Google Scholar]
- 31. Srinivas S., Watanabe T., Lin C. S., William C. M., Tanabe Y., Jessell T. M., Costantini F. (2001) Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wen S., Schwarz J. R., Niculescu D., Dinu C., Bauer C. K., Hirdes W., Boehm U. (2008) Functional characterization of genetically labeled gonadotropes. Endocrinology 149, 2701–2711 [DOI] [PubMed] [Google Scholar]
- 33. Caligioni C. S. (2009) Assessing reproductive status/stages in mice. Curr. Protoc. Neurosci. Appendix 4, Appendix 4I [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schmittgen T. D., Livak K. J. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 35. Fortin J., Kumar V., Zhou X., Wang Y., Auwerx J., Schoonjans K., Boehm U., Boerboom D., Bernard D. J. (2013) NR5A2 regulates Lhb and Fshb transcription in gonadotrope-like cells in vitro, but is dispensable for gonadotropin synthesis and fertility in vivo. PLoS One 8, e59058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lamba P., Wang Y., Tran S., Ouspenskaia T., Libasci V., Hebert T. E., Miller G. J., Bernard D. J. (2010) Activin A regulates porcine follicle-stimulating hormone beta-subunit transcription via cooperative actions of SMADs and FOXL2. Endocrinology 151, 5456–5467 [DOI] [PubMed] [Google Scholar]
- 37. Hoivik E. A., Bjanesoy T. E., Mai O., Okamoto S., Minokoshi Y., Shima Y., Morohashi K., Boehm U., Bakke M. (2011) DNA methylation of intronic enhancers directs tissue-specific expression of steroidogenic factor 1/adrenal 4 binding protein (SF-1/Ad4BP). Endocrinology 152, 2100–2112 [DOI] [PubMed] [Google Scholar]
- 38. Wen S., Ai W., Alim Z., Boehm U. (2010) Embryonic gonadotropin-releasing hormone signaling is necessary for maturation of the male reproductive axis. Proc. Natl. Acad. Sci. U. S. A. 107, 16372–16377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Itman C., Wong C., Hunyadi B., Ernst M., Jans D. A., Loveland K. L. (2011) Smad3 dosage determines androgen responsiveness and sets the pace of postnatal testis development. Endocrinology 152, 2076–2089 [DOI] [PubMed] [Google Scholar]
- 40. Xu J., Beyer A. R., Walker W. H., McGee E. A. (2003) Developmental and stage-specific expression of Smad2 and Smad3 in rat testis. J. Androl. 24, 192–200 [DOI] [PubMed] [Google Scholar]
- 41. Dalkin A. C., Knight C. D., Shupnik M. A., Haisenleder D. J., Aloi J., Kirk S. E., Yasin M., Marshall J. C. (1993) Ovariectomy and inhibin immunoneutralization acutely increase follicle-stimulating hormone-beta messenger ribonucleic acid concentrations: evidence for a nontranscriptional mechanism. Endocrinology 132, 1297–1304 [DOI] [PubMed] [Google Scholar]
- 42. Corrigan A. Z., Bilezikjian L. M., Carroll R. S., Bald L. N., Schmelzer C. H., Fendly B. M., Mason A. J., Chin W. W., Schwall R. H., Vale W. (1991) Evidence for an autocrine role of activin B within rat anterior pituitary cultures. Endocrinology 128, 1682–1684 [DOI] [PubMed] [Google Scholar]
- 43. Rejon C. A., Ho C. C., Wang Y., Zhou X., Bernard D. J., Hebert T. E. (2013) Cycloheximide inhibits follicle-stimulating hormone beta subunit transcription by blocking de novo synthesis of the labile activin type II receptor in gonadotrope cells. Cell Signal. 25, 1403–1412 [DOI] [PubMed] [Google Scholar]
- 44. Yang X., Letterio J. J., Lechleider R. J., Chen L., Hayman R., Gu H., Roberts A. B., Deng C. (1999) Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. EMBO J. 18, 1280–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene 234, 187–208 [DOI] [PubMed] [Google Scholar]
- 46. Kim S. Y., Zhu J., Woodruff T. K. (2011) A truncated, activin-induced Smad3 isoform acts as a transcriptional repressor of FSHbeta expression in mouse pituitary. Mol. Cell. Endocrinol. 342, 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blount A. L., Schmidt K., Justice N. J., Vale W. W., Fischer W. H., Bilezikjian L. M. (2009) FoxL2 and Smad3 coordinately regulate follistatin gene transcription. J. Biol. Chem. 284, 7631–7645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Datto M. B., Frederick J. P., Pan L., Borton A. J., Zhuang Y., Wang X. F. (1999) Targeted disruption of Smad3 reveals an essential role in transforming growth factor beta-mediated signal transduction. Mol. Cell. Biol. 19, 2495–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhu Y., Richardson J. A., Parada L. F., Graff J. M. (1998) Smad3 mutant mice develop metastatic colorectal cancer Cell 94, 703–714 [DOI] [PubMed] [Google Scholar]
- 50. Maggio-Price L., Treuting P., Zeng W., Tsang M., Bielefeldt-Ohmann H., Iritani B. M. (2006) Helicobacter infection is required for inflammation and colon cancer in SMAD3-deficient mice. Cancer Res. 66, 828–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ye P., Chen W., Wu J., Huang X., Li J., Wang S., Liu Z., Wang G., Yang X., Zhang P., Lv Q., Xia J. (2013) GM-CSF contributes to aortic aneurysms resulting from SMAD3 deficiency. J. Clin. Invest. 123, 2317–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ho C. C., Bernard D. J. (2010) Bone morphogenetic protein 2 acts via inhibitor of DNA binding proteins to synergistically regulate follicle-stimulating hormone beta transcription with activin A. Endocrinology 151, 3445–3453 [DOI] [PubMed] [Google Scholar]
- 53. Ho C. C., Bernard D. J. (2009) Bone morphogenetic protein 2 signals via BMPR1A to regulate murine follicle-stimulating hormone beta subunit transcription. Biol. Reprod. 81, 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee K. B., Khivansara V., Santos M. M., Lamba P., Yuen T., Sealfon S. C., Bernard D. J. (2007) Bone morphogenetic protein 2 and activin A synergistically stimulate follicle-stimulating hormone beta subunit transcription. J. Mol. Endocrinol. 38, 315–330 [DOI] [PubMed] [Google Scholar]
- 55. Besson-Fournier C., Latour C., Kautz L., Bertrand J., Ganz T., Roth M. P., Coppin H. (2012) Induction of activin B by inflammatory stimuli up-regulates expression of the iron-regulatory peptide hepcidin through Smad1/5/8 signaling. Blood 120, 431–439 [DOI] [PubMed] [Google Scholar]
- 56. Safwat N., Ninomiya-Tsuji J., Gore A. J., Miller W. L. (2005) Transforming growth factor beta-activated kinase 1 is a key mediator of ovine follicle-stimulating hormone beta-subunit expression. Endocrinology 146, 4814–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nicol L., Faure M. O., McNeilly J. R., Fontaine J., Taragnat C., McNeilly A. S. (2008) Bone morphogenetic protein-4 interacts with activin and GnRH to modulate gonadotrophin secretion in LbetaT2 gonadotrophs. J. Endocrinol. 196, 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang Y., Bernard D. J. (2012) Activin A induction of murine and ovine follicle-stimulating hormone beta transcription is SMAD-dependent and TAK1 (MAP3K7)/p38 MAPK-independent in gonadotrope-like cells. Cell. Signal. 24, 1632–1640 [DOI] [PubMed] [Google Scholar]
- 59. Fortin J., Boehm U., Deng C., Bernard D. J. (2013) SMAD signaling in gonadotropes is critical for normal FSH synthesis and fertility in mice. 95th Annual Meeting of the Endocrine Society. Endocr. Rev. 34, OR04–06 [Google Scholar]
- 60. Huang H. J., Wu J. C., Su P., Zhirnov O., Miller W. L. (2001) A novel role for bone morphogenetic proteins in the synthesis of follicle-stimulating hormone. Endocrinology 142, 2275–2283 [DOI] [PubMed] [Google Scholar]
- 61. Otsuka F., Shimasaki S. (2002) A novel function of bone morphogenetic protein-15 in the pituitary: selective synthesis and secretion of FSH by gonadotropes. Endocrinology 143, 4938–4941 [DOI] [PubMed] [Google Scholar]
- 62. Marshall J. C., Dalkin A. C., Haisenleder D. J., Griffin M. L., Kelch R. P. (1993) GnRH pulses–the regulators of human reproduction. Trans. Am. Clin. Climatol. Assoc. 104, 31–46 [PMC free article] [PubMed] [Google Scholar]
- 63. Cattanach B. M., Iddon C. A., Charlton H. M., Chiappa S. A., Fink G. (1977) Gonadotrophin-releasing hormone deficiency in a mutant mouse with hypogonadism. Nature 269, 338–340 [DOI] [PubMed] [Google Scholar]
- 64. Wu S., Wilson M. D., Busby E. R., Isaac E. R., Sherwood N. M. (2010) Disruption of the single copy gonadotropin-releasing hormone receptor in mice by gene trap: severe reduction of reproductive organs and functions in developing and adult mice. Endocrinology 151, 1142–1152 [DOI] [PubMed] [Google Scholar]
- 65. Besecke L. M., Guendner M. J., Schneyer A. L., Bauer-Dantoin A. C., Jameson J. L., Weiss J. (1996) Gonadotropin-releasing hormone regulates follicle-stimulating hormone-beta gene expression through an activin/follistatin autocrine or paracrine loop. Endocrinology 137, 3667–3673 [DOI] [PubMed] [Google Scholar]
- 66. Kirk S. E., Dalkin A. C., Yasin M., Haisenleder D. J., Marshall J. C. (1994) Gonadotropin-releasing hormone pulse frequency regulates expression of pituitary follistatin messenger ribonucleic acid: a mechanism for differential gonadotrope function. Endocrinology 135, 876–880 [DOI] [PubMed] [Google Scholar]
- 67. Hoak D. C., Schwartz N. B. (1980) Blockade of recruitment of ovarian follicles by suppression of the secondary surge of follicle-stimulating hormone with porcine follicular field. Proc. Natl. Acad. Sci. U. S. A. 77, 4953–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bilezikjian L. M., Blount A. L., Leal A. M., Donaldson C. J., Fischer W. H., Vale W. W. (2004) Autocrine/paracrine regulation of pituitary function by activin, inhibin and follistatin. Mol. Cell. Endocrinol. 225, 29–36 [DOI] [PubMed] [Google Scholar]
- 69. Woodruff T. K., Besecke L. M., Groome N., Draper L. B., Schwartz N. B., Weiss J. (1996) Inhibin A and inhibin B are inversely correlated to follicle-stimulating hormone, yet are discordant during the follicular phase of the rat estrous cycle, and inhibin A is expressed in a sexually dimorphic manner. Endocrinology 137, 5463–5467 [DOI] [PubMed] [Google Scholar]
- 70. Knox K. L., Ringstrom S. J., Schwartz N. B. (1993) RU486 blocks the effects of inhibin antiserum or luteinizing hormone on the secondary follicle-stimulating hormone surge. Endocrinology 133, 277–283 [DOI] [PubMed] [Google Scholar]
- 71. Knox K. L., Ringstrom S. J., Szabo M., Perlyn C. A., Sutandi S., Schwartz N. B. (1996) RU486 on an estrogen background blocks the rise in serum follicle-stimulating hormone induced by antiserum to inhibin or ovariectomy. Endocrinology 137, 1226–1232 [DOI] [PubMed] [Google Scholar]
- 72. Knox K. L., Schwartz N. B. (1992) RU486 blocks the secondary surge of follicle-stimulating hormone in the rat without blocking the drop in serum inhibin. Biol. Reprod 46, 220–225 [DOI] [PubMed] [Google Scholar]
- 73. Szabo M., Knox K. L., Ringstrom S. J., Perlyn C. A., Sutandi S., Schwartz N. B. (1996) Mechanism of the inhibitory action of RU486 on the secondary follicle-stimulating hormone surge. Endocrinology 137, 85–89 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.