Abstract
Purpose
FBXW7 is a tumor suppressor gene responsible for the degradation of several proto-oncogenes. Preclinical data suggest that FBXW7 mutations sensitize cells to mTOR inhibitors. Clinicopathologic characteristics of cancer patients with FBXW7 mutations and their responses to mTOR inhibitors remain unknown.
Methods
Using multiplex gene panels we evaluated how the FBXW7 mutation affected the cancer phenotype of patients referred to a phase I clinic starting January 2012. Whenever possible patients positive for FBXW7 mutation were treated with regimens containing an mTOR inhibitors and their outcomes were reviewed.
Results
FBXW7 mutations were detected in 17 of 418 patients (4.0%). Among tumor types with more than 10 patients tested, FBXW7 mutations occurred in colorectal cancer (7/49; 14.3%), squamous cell cancer of head and neck (2/18; 11.1%), liver (1/13; 7.7%), and ovarian cancers (1/40; 2.5%). No one clinical, pathological or demographic feature was characteristic of the FBXW7-mutated patient population. The mutation occurred in isolation in only 2/17 (12%) patients, and KRAS was frequently found as a concomitant mutation, especially in patients with colorectal cancer (6/7; 86%). Ten patients were treated on a protocol containing an mTOR inhibitor, with a median time to treatment failure of 2.8 months (range, 1.3–6.8). One patient with liver cancer (fibrolamellar subtype) continues to have a prolonged stable disease for 6.8+ months.
Conclusion
In patients with advanced cancers, somatic mutations in FBXW7 usually occur with other simultaneous molecular aberrations, which can contribute to limited therapeutic efficacy of mTOR inhibitors.
Introduction
The identification of molecular aberrations that are predictive of response to targeted therapy has been the focus of intensive research. Preclinical data from numerous cancer cell lines and mice models have correlated specific genetic mutations with susceptibility to agents inhibiting the pathway putatively activated in the mutated state. [1], [2]. Indeed, major therapeutic advances have recently been made in oncology tailoring treatment to molecular characteristics of some tumors.[3]–[7] Additionally, the strategy of matching druggable genetic abnormalities with targeted agents has demonstrated efficacy in umbrella protocols. [8], [9] However, much remains unknown regarding the efficacy of novel targeted agents and how genetic alterations can be translated to the clinic, and current preclinical models are incomplete. [10].
Extensive comprehensive molecular profiling is commercially available for cancer patients and some results suggest potential treatment options based exclusively on the mutations found in tested tumors. Establishing a correlation between the preclinical activity of targeted agents with clinical data is essential to optimize this approach.
FBXW7 is a tumor suppressor gene that is mutated in various human tumors. [11] This gene encodes a F-box protein responsible for ubiquitination and turnover of several oncoproteins and its loss of function has been associated with genetic instability and tumor growth. [12], [13] mTOR is one of the substrates of FBXW7-mediated protein degradation, and loss of function of FBXW7 increases the levels of total and activated mTOR. [14] Preclinical data have suggested that inactivating mutations of FBXW7 could predict sensitivity to the mTOR inhibitor rapamycin,. [14], [15]; however, their clinical utility remains unknown.
Therefore, we investigated the FBXW7 mutational status and clinical and demographic characteristics of patients with advanced cancer referred to our Phase I Clinical Trials Program and the outcomes of such patients treated with agents targeting the mTOR pathway.
Patients and Methods
Patients
We reviewed the electronic medical records of all patients with advanced solid tumors tested for FBXW7 mutations referred to the Department of Investigational Cancer Therapeutics (Phase I Clinical Trials Program) at The University of Texas MD Anderson Cancer Center starting in January 2012. Patients who tested positive for FBXW7 mutations were included in further analyses. Patients with colorectal cancer who tested negative for FBXW7 mutations were included as controls for the colorectal cancer subgroup. This study and all associated treatments were conducted in accordance with the guidelines of the MD Anderson Institutional Review Board (IRB). This study was part of an umbrella protocol approved by MD Anderson IRB. The need for written informed consent was waived due to the retrospective nature of the study.
Tissue Samples and Mutation Analysis
FBXW7 mutations were investigated in archival formalin-fixed, paraffin-embedded tissue blocks or material from fine needle aspiration biopsies obtained from diagnostic and/or therapeutic procedures. All histologies were centrally reviewed at MD Anderson. FBXW7 mutation analysis was performed in different Clinical Laboratory Improvement Amendment-certified laboratories as part of a gene panel analysis. These included 182 genes in targeted next-generation sequencing Foundation One platform (Foundation Medicine, Cambridge, MA), 46 genes in Ion Torrent next-generation sequencing (Baylor’s Cancer Genetics Laboratory, Houston, TX) and 53 genes in Sequenom Mass ARRAY platform (Knight Diagnostics,Portland, OR). Information about mutations in genes other than FBXW7 discovered in these multiplex panels was also registered.
Treatment and Evaluation
Patients presenting with FBXW7 mutations were enrolled, whenever possible, in clinical trials containing inhibitors of the mTOR pathway, particularly protocols testing rapalogs, thought to be primarily anti-mTORC1 agents. Treatment continued until disease progression, withdrawal of consent by the patient, clinical judgment deeming the necessity of removing a patient from a clinical trial, or development of unacceptable toxicity or death.
Clinical assessments were performed as specified in each protocol, typically before the initiation of therapy and then, at a minimum, at the beginning of each new treatment cycle. Treatment response was assessed using computed tomography scans, magnetic resonance imaging and/or positron emission tomography scans at baseline before treatment initiation, and then every 2 cycles (6–8 weeks). All radiographs were read in the Department of Radiology at MD Anderson and reviewed in the Department of Investigational Cancer Therapeutics tumor measurement clinic. Responses were categorized using RECIST on the basis of specific protocol requirements [16], [17], and were reported as best response, defined as the maximum shrinkage of tumor or stabilization of the disease during all the assessments obtained.
Statistical Analysis
Patient characteristics, including demographics, tumor type, FBXW7 mutation status and associated genetic abnormalities were summarized using frequency distributions and percentages. Fisher’s exact test was used to assess the association among categorical variables and FBXW7 mutation status. Time to treatment failure (TTF) was defined as the interval from the start of therapy to treatment discontinuation for any reason, including disease progression, treatment toxicity, patient preference, physician judgment, or death. The Wilcoxon signed rank test assessed TTF differences within patients. All tests were 2-sided, and P<0.05 was considered statistically significant. All statistical analyses were carried out using S+ sofware, ver 8.2 (TIBCO Software Inc, Houston, TX).
Results
Patient Characteristics
The tumors from 418 patients with advanced cancer were assessed for FBXW7 mutations using various multigene panels, and in 17 (4.0%) a mutation in FBXW7 was identified. Of these 17 patients, 9 (52%) were male and 7 (41%) had colorectal cancer. The median number of prior therapies before initial evaluation in the Phase I Clinic was 2 (0–9). Patient characteristics are summarized in Tables 1 and 2. The following tumor types had more than 10 patients tested and no FBXW7 mutation detected: adenoid cystic (n = 14), breast cancer (n = 37), gastroesophageal (n = 29), lung (n = 15), kidney (n = 10) and soft tissue sarcoma (n = 61). We tested a mixed of primary and metastatic tumors and of the 17 positive samples 12 (71%) were derived from primary and 5 (29%) from metastatic lesions. The prevalence of FBXW7 mutation was similar in samples from primary and metastatic tumors.
Table 1. Baseline characteristics of FBXW7 mutation-positive patients and prevalence of associated mutations.
| Category | Subcategory | Patients (n = 17) |
| Age (years) Median (range) | 60 (16–74) | |
| Gender (%) | Male | 9 (53) |
| Female | 8 (47) | |
| Ethnicity (%) | White | 13 (76) |
| African American | 2 (12) | |
| Hispanic | 2 (12) | |
| Tumor type [n/patients tested: (%)] | Colorectal | 7/49 (14.3) |
| Head and Neck (squamous) | 2/18 (11.1) | |
| Bladder | 1/8 (12.5) | |
| Cervix | 1/10 (10) | |
| Endometrial | 1/7 (14.3) | |
| Liver | 1/13 (7.7) | |
| Ovarian | 1/40 (2.5) | |
| Mesothelioma | 1/4 (25) | |
| Pancreatic | 1/2 (50) | |
| Teratoma | 1/1 (100) | |
| FBXW7 mutation (%) | Inactivating | 13 (76) |
| Unknown function | 4 (24) | |
| KRAS mutation (%) | Positive | 6 (35) |
| Negative | 11 (65) | |
| TP53 mutation (%) | Positive | 10 (59) |
| Negative | 7 (41) | |
| PI3KCA mutation (%) | Positive | 3 (18) |
| Negative | 14 (82) | |
| APC mutation (%) | Positive | 3 (18) |
| Negative | 14 (82) |
Table 2. Characteristics of 18 patients with FBXW7 mutation-positive tumors.
| Patient No. | Tumor type | Histology | FBXW7 mutation | Type of aberration | Concomitant mutations | mTOR therapy | Best response | TTF (mos)a |
| 1 | Bladder | Undifferentiated carcinoma | W244* | Inactivating | TP53 | Sirolimus, HCQ | SD | 2.7 |
| 2 | Cervix | Squamous | W244* | Inactivating | CBL, MLH1, PRKDC | – | ||
| 3 | Colorectal | Adenocarcinoma | R465C | Inactivating | KRAS, APC, SMAD4 | – | ||
| 4 | Colorectal | Adenocarcinoma | R278* | Inactivating | KRAS, SMAD4 | – | ||
| 5 | Colorectal | Adenocarcinoma | R505C | Inactivating | KRAS, TP53 | Everolimus, pazopanib | SD | 3 |
| 6 | Colorectal | Adenocarcinoma | R479Q | Inactivating | KRAS, TP53 | – | ||
| 7 | Colorectal | Adenocarcinoma | G499Vfs*25 | Unknown | KRAS | Sirolimus,HCQ | SD | 4.4 |
| 8 | Colorectal | Adenocarcinoma | R505C | Inactivating | KRAS, TP53, PIK3CA | Everolimus, anakinra | PD | 1.4 |
| 9 | Colorectal | Adenocarcinoma | R222* | Inactivating | TP53, APC, HER-2 | Temsirolimus, bevacizumab, cetuximab | SD | 3.6 |
| 10 | Endometrial | Clear cell | R465C | Inactivating | TP53, PIK3R1, MAP3K1 | – | ||
| 11 | Liver | Fibrolamellar HCC | E192A | Unknown | None | Sirolimus, vorinostat | SD | 6.8+ |
| 12 | Head and Neck | Squamous | S282* | Inactivating | TP53, CDKN2A | Temsirolimus, bevacizumab, valproic acid | SD | 2.2 |
| 13 | Head and Neck | Squamous | R479Q | Inactivating | MLL2 | – | ||
| 14 | Pleura | Mesothelioma | R658* | Unknown | TP53, NF2 | Sirolimus, lapatinib | PD | 1.3 |
| 15 | Ovarian | Serous | R465H | Inactivating | TP53, PIK3CA | Everolimus, Anastrozole | PD | 3.3 |
| 16 | Pancreatic | Neuroendocrine | E113D | Unknown | None | – | ||
| 17 | Teratoma | Adenocarcinoma | 726+1 G>A splice | Inactivating | TP53, ARID1A | Temsirolimus, bevacizumab, carboplatin | SD | 2.6 |
Abbreviations- HCQ: hydroxichloroquine; SD: stable disease, PD: progressive disease; PR: partial response; TTF: time to treatment failure, HCC: hepatocellular carcinoma a+ denotes not progressing at the time of the analysis.
Concomitant Mutations
Genetic abnormalities analyzed using different multiplex gene assays revealed isolated FBXW7 mutations in 2 of 17 (12%) patients. The most frequent concomitant genetic abnormality was TP53 mutation in 10 (59%) patients. Other frequently occurring abnormalities were KRAS, PIK3CA and APC mutations (Table 1 and 2).
Comparison of Clinical and Mutational Characteristics
Because 7 out of 17 (41%) patients with FBXW7 mutations had colorectal cancer, we compared the clinical, pathological and mutational characteristics between the colorectal cancer population and the non-colorectal cancer patients with mutation (Table 3). In addition, 41 patients with colorectal cancer without FBXW7 mutation were used as a control group. Of the 10 patients with cancers other than colorectal and FBXW7 mutations, none (0%) had simultaneous KRAS mutations or APC mutations compared to 6/7 (86%) patients with KRAS mutations and 3/7 (43%) with APC mutations in the colorectal cancer group (p = 0.0006 and p = 0.022, respectively Table 3). No statistical differences were observed in sex, ethnicity, age and type of metastasis between these groups.
Table 3. Comparison of characteristics of subgroups of FBXW7-positive patients and a control group of CRC patients tested negative for a FBXW7 mutation.
| Characteristic | Non-CRC FBXW7 pos (N = 10) (%) | CRC FBXW7 pos (N = 7) (%) | CRC FBXW7 neg (N = 41) (%) |
| Female | 6 (60) | 2 (29) | 23 (56) |
| White | 8 (80) | 5 (71) | 27 (66) |
| Median Age | 59 (15–74) | 57 (36–67) | 52 (23–74) |
| >3 metastatic sites | 1 (10) | 1 (14) | 6 (15) |
| Liver metastasis | 3 (30) | 5 (71) | 33 (80) |
| Lung metastasis | 5 (50) | 6 (86) | 32 (78) |
| KRAS mutation | 0 (0)a | 6 (86)a | 33 (80) |
| TP53 mutation | 6 (60) | 4 (57) | 17 (41) |
| APC mutation | 0 (0)b | 3/5 (60)b | 16/24 (67) |
| PIK3CA mutation | 1 (10) | 1 (14) | 8 (20) |
p = 0.0006;
p = 0.022 (Fisher exact test).
CRC: colorectal cancer.
Treatment of Patients with mTOR Inhibitors
Of the 17 patients with FBXW7 mutations, 10 (59%) were enrolled in phase I protocols including mTOR inhibitors combined with other agents (Table 2). All of these patients were evaluable for response. The doses of mTOR inhibitors ranged from 50 to 100% of the maximum tolerated dose or the highest expected dose when the dose escalation was ongoing at the time of analysis. All doses were considered to be active based on prior experience with the drugs. There was no partial or complete response and 7 (70%) had stable disease (SD) as their best response, including 2 (20%) patients with prolonged SD lasting more than 16 weeks (Figure 1).
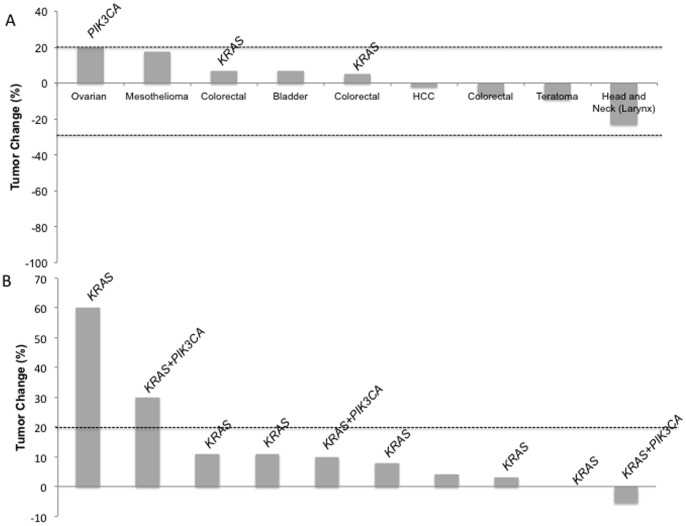
Figure 1. Waterfall plot of patients with measurable disease by RECIST treated with mTOR inhibitors.
(A) Responses of patients who were positive for FBXW7 mutations. (B) Responses of colorectal cancer patients negative for FBXW7 mutations. Concomitant KRAS and PIK3CA mutations are indicated.
The median TTF for the cohort of 10 patients with FBXW7 mutations treated with mTOR inhibitors was 2.8 months (1.3–6.8), including 1 patient still being treated at the time of this analysis. All of these patients had received prior therapies and a trend for an worse TTF on mTOR inhibitor protocols was detected when compared to the treatment immediately before referral to the Phase I Clinic (TTF = 5.7 months [1.3–18] from their prior therapy versus 2.8 months [1.3–6.8] from an mTOR-based treatment, p = 0.055). Of note, the patient with the highest TTF while on an mTOR-based regimen had a tumor that did not harbor KRAS mutations. Due to the small sample size, we could not detect a difference in the median TTF on mTOR therapy between patients with a FBXW7 mutation, with or without concomitant KRAS mutations (median TTF = 3.0 and 2.7 months, respectively, p = 0.99).
Comparison of Outcomes of Colorectal Patients Treated with mTOR Inhibitors
A total of 4 patients with colorectal cancer and a FBXW7 mutation were treated with an mTOR inhibitor and 3 patients (75%) had SD as their best response. Of the 41 patients with colorectal cancer without a FBXW7 mutation, 12 (29%) were enrolled on a protocol with an mTOR inhibitor and 4 (33%) had SD (Figure 1). The median TTF of FBXW7-positive colorectal cancer patients was 3.3 months (1.4–4.4) on mTOR inhibitors versus 1.8 months (0.6–4.9) for FBXW7-negative patients with colorectal cancer (p = 0.27).
Discussion
FBXW7 mutations were found in 17 of 418 (4.0%) patients referred to our phase I unit with various advanced tumor types. Although colorectal cancer was not the most prevalent cancer among the patients, it comprised the greatest number of patients testing positive for FBXW7 mutations. No particular demographic or pathological feature was characteristic of the FBXW7-mutated population in our sample, but we found that the mutation rarely occurs in isolation, especially in colorectal cancer. Our series revealed an overall low activity of mTOR inhibitors in patients with FBXW7 mutations, contrasting with previous pre-clinical data.
Previous studies revealed FBXW7 mutations in approximately 6% of cancer patients. A substantial variation in the frequency of mutation among tumor types was also described, with the highest mutation rate in cholangiocarcinoma (30%), followed by gastrointestinal tumors (including colorectal cancers), and endometrial and prostate cancers, in the range of 4–15%.[18]–[21].
As of May 2013, the Catalogue of Somatic Mutations in Cancer (COSMIC) database reported an overall frequency of the FBXW7 mutation in 4% of cancer patients, identical to our data. [22] The highest frequencies were in biliary tumors (22%), endometrial (11%), urinary (8%), and colorectal cancers (7%). Our study demonstrated the highest frequency of the FBXW7 mutation in colorectal cancer (14.3%). We also detected this mutation in endometrial (1/7 patients) and bladder cancer (1/8 patients), but none of the 3 patients tested with cholangiocarcinoma harbored a FBXW7 mutation. Of note, our data describe a FBXW7 mutation in liver cancer, specifically a fibrolamellar variant, and also in rare tumors, such as mesothelioma and teratoma. In COSMIC database tested samples derived from primary tumors, while in our study samples from metastatic tumors were also tested.
FBXW7 mutations as single molecular abnormalities were rare in our study, which might reflect the difficulty of targeting this mutation with agents blocking single pathways. Indeed, prevalence of concomitant mutations in TP53 (59%), KRAS (35%) and PIK3CA (18%) in our patients with FBXW7 mutations was higher than previously reported with technologies preceding multiplex genomic technologies, which had limited detection capabilities. A prior study described the following prevalence of mutations on the same genes: TP53, 37%, KRAS, 18% and PIK3CA 10%, respectively. [8].
Occurrence of concomitant KRAS mutations was more frequently seen in colorectal cancer patients with FBXW7 mutations in comparison to non-colorectal group. The analysis of a control group of colorectal patients who tested negative for a FBXW7 mutation showed that this association was, in fact, due to the high prevalence of KRAS mutations in our colorectal cancer population and, therefore, we cannot conclude about any possible association between these mutations.
We described a similar previously reported [18] pattern of the FBXW7 mutation available in the COSMIC database. The vast majority of the mutations detected in our study were single nucleotide changes, predominantly missense substitutions. Of these, the most common mutations were found in two mutational hotspots in the Arg465 and Arg479 codons. Interestingly, our series of patients with more advanced and refractory solid tumors had a higher frequency of nonsense substitutions than previously described. [18] Since the functional consequences of both types of substitutions are not well compared in the literature it is hard to speculate about the possible implications of this finding.
Considering that most of these mutations are within the WD40 domain responsible for the recognition of substrates by FBXW7 or result in a stop of the translation process prior to inclusion in this domain, FBXW7 mutations are expected to inactivate the translated protein. [11] Most FBXW7 mutations are heterozygous, but because the protein dimerizes, how they affect protein functionality varies. [23] Mutations that result in retention of the dimerization domain may have a dominant negative effect that is more deleterious than mutations resulting in allele deletions or premature stop codons. [11] Hence, similar to other druggable oncogenic aberration, it is anticipated that the functional consequences of FBXW7 mutations upon substrates are quite variable, and responses to targeted therapies might also be heterogeneous. [24], [25].
When we evaluated the response to mTOR inhibitors in a subset of patients with tumors harboring FBXW7 mutations, variable responses were demonstrated. Previous studies showed that tumor cell lines with FBXW7 gene mutations are sensitive to rapamycin [14], suggesting a potential rationale for treating such tumors with mTOR inhibitors. Overall, however, we found only limited activity in phase I trials using mTOR inhibitors in this population. The TTF for the overall population was 2.8 months, which was worse than the TTF on the therapies used immediately prior to enrollment on phase I protocols with mTOR inhibitors. The absence of tumor responses with mTOR inhibitors also indicates a lack of activity of mTOR inhibitors for FBXW7 positive patients. It is important to note that our data is limited to mTORC1 inhibitors, which have some concerns related to re-activation of mTOR pathway by reversal of a feed-back loop. [26].
Of interest, one patient with a refractory fibrolamellar hepatocellular carcinoma demonstrated prolonged SD for 6.8 months on sirolimus based combination and is still on treatment at the time of this report. This patient was the only one among those treated with mTOR inhibitors who had FBXW7 mutation without other simultaneous molecular abnormalities. It is plausible that similar to other malignancies simultaneous molecular aberrations can lead to activation of other molecular pathways, which can lead to therapeutic resistance. [24], [27] Specifically for mTOR inhibitors, it has been shown that KRAS mutation is a mechanism of resistance even in the presence of sensitizing mutations, such as the PIK3CA mutations. [24], [28], [29] Therefore, absence of simultaneous mutations along with better understanding of functional consequences of specific mutation types can be crucial for patient selection for mTOR inhibitors.
One study suggested that rapalogs can delay FBXW7-induced tumorigenesis [30], but it is not clear whether these drugs produce a cytoreductive effect. Obtaining disease stabilization as best response in our study points to a direction where mTOR inhibitors are unable to produce tumoral reductions of FBXW7 positive tumors. In addition, the absence of responses in our population combined with a short period of disease control suggests again that pathways other than PI3K/AKT/mTOR confer resistance to mTOR-based therapies. In contrast to preclinical models were FBXW7 are studied as an isolated event, we showed that the predictive information generated with these models do not translate necessarily to the in vivo context. There are many possible reasons (including association of concomitant mutations, stromal effects among others) that preclude the validity of the preclinical data for treatment-based decisions. These models are essentially hypothesis generating and should encourage the development of clinical protocols to test the potential findings that they generate.
The limitations of this study are its retrospective nature, small sample size, and inclusion of protocol treatments that were not exclusively mTOR inhibitors. Additionally, a possible selection bias and absence of randomization to study drugs characterizes this study as exploratory in nature. However, to our knowledge, this is the first series reporting the clinical characteristics and treatment outcomes of an FBXW7-mutated cancer patient population. Additional series are needed to better understand therapeutic alternatives that can be used to target this mutation, by exploring use of alternatives to mTOR-1 inhibition and correlating FBXW7 mutations with mTOR pathway activation in vivo. That is a strong possibility given that several oncogenes are regulated by FBXW7, including MYC, Notch, JUN and cyclin E. Thus, targeting different pathways in addition to the mTOR pathway might be necessary to effectively kill cancer cells in tumors driven by FBXW7 alterations. [11].
In conclusion, we demonstrated that FBXW7 mutations are found in several types of solid tumors. In addition, the diverse types of FBXW7 mutations are likely related to various functional effects that the FBXW7 mutation has on protein functionality. Also, the frequent concomitance of other oncogene mutations provides challenges to targeting tumors harboring FBXW7 abnormalities. Most patients with FBXW7 mutations had limited benefit from mTOR based therapies; however, studying mTOR inhibitors in cancers lacking simultaneous molecular abnormalities as well as describing functional consequences of specific FBXW7 mutation subtypes warrants further investigation.
Acknowledgments
The authors acknowledge Joann Aaron, MA, in the Department of Investigational Cancer Therapeutics for editorial support.
Funding Statement
The authors have no support or funding to report.
References
- 1. Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, et al. (2012) Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483: 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tran B, Dancey JE, Kamel-Reid S, McPherson JD, Bedard PL, et al. (2012) Cancer genomics: technology, discovery, and translation. J Clin Oncol 30: 647–660. [DOI] [PubMed] [Google Scholar]
- 3. Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, et al. (2001) Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med 344: 1031–1037. [DOI] [PubMed] [Google Scholar]
- 4. Demetri GD, von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, et al. (2002) Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med 347: 472–480. [DOI] [PubMed] [Google Scholar]
- 5. Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, et al. (2010) Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 363: 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shaw AT, Yeap BY, Solomon BJ, Riely GJ, Gainor J, et al. (2011) Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol 12: 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Falchook GS, Lewis KD, Infante JR, Gordon MS, Vogelzang NJ, et al. (2012) Activity of the oral MEK inhibitor trametinib in patients with advanced melanoma: a phase 1 dose-escalation trial. Lancet Oncol 13: 782–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tsimberidou AM, Iskander NG, Hong DS, Wheler JJ, Falchook GS, et al. (2012) Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res 18: 6373–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Janku F, Tsimberidou AM, Garrido-Laguna I, Wang X, Luthra R, et al. (2011) PIK3CA mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. Mol Cancer Ther 10: 558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, et al. (2013) Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A 110: 3507–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Welcker M, Clurman BE (2008) FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer 8: 83–93. [DOI] [PubMed] [Google Scholar]
- 12. Wang Z, Inuzuka H, Zhong J, Wan L, Fukushima H, et al. (2012) Tumor suppressor functions of FBW7 in cancer development and progression. FEBS Lett 586: 1409–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, et al. (2004) Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature 432: 775–779. [DOI] [PubMed] [Google Scholar]
- 14. Mao JH, Kim IJ, Wu D, Climent J, Kang HC, et al. (2008) FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science 321: 1499–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Liu Y, Lu J, Zhang P, Wang Y, et al.. (2013) Rapamycin inhibits FBXW7 loss-induced epithelial-mesenchymal transition and cancer stem cell-like characteristics in colorectal cancer cells. Biochem Biophys Res Commun. [DOI] [PMC free article] [PubMed]
- 16. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, et al. (2009) New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45: 228–247. [DOI] [PubMed] [Google Scholar]
- 17. Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, et al. (2000) New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 92: 205–216. [DOI] [PubMed] [Google Scholar]
- 18. Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, et al. (2007) FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res 67: 9006–9012. [DOI] [PubMed] [Google Scholar]
- 19. Lee JW, Soung YH, Kim HJ, Park WS, Nam SW, et al. (2006) Mutational analysis of the hCDC4 gene in gastric carcinomas. Eur J Cancer 42: 2369–2373. [DOI] [PubMed] [Google Scholar]
- 20. Hubalek MM, Widschwendter A, Erdel M, Gschwendtner A, Fiegl HM, et al. (2004) Cyclin E dysregulation and chromosomal instability in endometrial cancer. Oncogene 23: 4187–4192. [DOI] [PubMed] [Google Scholar]
- 21. Koh MS, Ittmann M, Kadmon D, Thompson TC, Leach FS (2006) CDC4 gene expression as potential biomarker for targeted therapy in prostate cancer. Cancer Biol Ther 5: 78–83. [DOI] [PubMed] [Google Scholar]
- 22.The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Wellcome Trust Sanger Institute. [DOI] [PMC free article] [PubMed]
- 23. Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, et al. (2007) The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med 204: 1825–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, et al. (2013) PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res 73: 276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gazdar AF (2009) Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 28 Suppl 1S24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, et al. (2006) mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res 66: 1500–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nazarian R, Shi H, Wang Q, Kong X, Koya RC, et al. (2010) Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468: 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Di Nicolantonio F, Arena S, Tabernero J, Grosso S, Molinari F, et al. (2010) Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J Clin Invest 120: 2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ihle NT, Lemos R Jr, Wipf P, Yacoub A, Mitchell C, et al. (2009) Mutations in the phosphatidylinositol-3-kinase pathway predict for antitumor activity of the inhibitor PX-866 whereas oncogenic Ras is a dominant predictor for resistance. Cancer Res 69: 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y, Huang Y, Wang Z, Huang Y, Li X, et al. (2013) Temporal mTOR inhibition protects Fbxw7-deficient mice from radiation-induced tumor development. Aging (Albany NY) 5: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]