Abstract
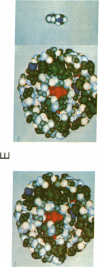
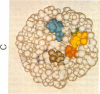
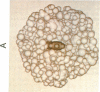
A descriptive medium for the presentation of protein structure has been developed and used to evaluate the structure of the active site of bovine trypsin (EC 3.4.21.4). This technique, involving advanced computer graphics technology, permits the facile display of a representation of the molecular surface of proteins of known structure and employs color to code the structural or chemical features of this surface. Benzamidine derivatives were inserted into the benzamidine-binding site of trypsin and the binary inhibitor-trypsin complex was evaluated by using the computer-generated structure. On the basis of qualitative assessments of the contribution of electrostatic and hydrophobic forces to the binding energy associated with complex formation, we made predictions concerning the effects of interaction of benzamidine substituents and amino acid side chains upon the binding energy associated with inhibitor-protein binding. The computer display of the molecular surfaces of the binary complex of substituted benzamidines and trypsin permitted unique insight into the identity and chemical properties of the atoms that participate at the interface of the molecular surfaces of the inhibitor and the protein. The computer-generated molecular surface display can potentially be combined with quantitative definition of the physical forces involved in the interaction of molecular surfaces. This technology should facilitate the study of the structure-activity relationship of substrates, inhibitors, and drugs that bind to proteins of known three-dimensional structure.
Keywords: protein structure, enzyme-inhibitor complex, drug-protein interaction, structure-activity relationships
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anfinsen C. B., Scheraga H. A. Experimental and theoretical aspects of protein folding. Adv Protein Chem. 1975;29:205–300. doi: 10.1016/s0065-3233(08)60413-1. [DOI] [PubMed] [Google Scholar]
- Bing D. H., Cory M., Fenton J. W., 2nd Exo-site affinity labeling of human thrombins. Similar labeling on the A chain and B chain/fragments of clotting alpha- and nonclotting gamma/beta-thrombins. J Biol Chem. 1977 Nov 25;252(22):8027–8034. [PubMed] [Google Scholar]
- Bode W., Schwager P. The refined crystal structure of bovine beta-trypsin at 1.8 A resolution. II. Crystallographic refinement, calcium binding site, benzamidine binding site and active site at pH 7.0. J Mol Biol. 1975 Nov 15;98(4):693–717. doi: 10.1016/s0022-2836(75)80005-2. [DOI] [PubMed] [Google Scholar]
- Feldmann R. J. The design of computing systems for molecular modeling. Annu Rev Biophys Bioeng. 1976;5:477–510. doi: 10.1146/annurev.bb.05.060176.002401. [DOI] [PubMed] [Google Scholar]
- Furie B., Schechter A. N., Sachs D. H., Anfinsen C. B. An immunological approach to the conformational equilibrium of staphylococcal nuclease. J Mol Biol. 1975 Mar 15;92(4):497–506. doi: 10.1016/0022-2836(75)90305-8. [DOI] [PubMed] [Google Scholar]
- Greer J., Bush B. L. Macromolecular shape and surface maps by solvent exclusion. Proc Natl Acad Sci U S A. 1978 Jan;75(1):303–307. doi: 10.1073/pnas.75.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansch C., Grieco C., Silipo C., Vittoria A. Quantitative structure-activity relationship of chymotrypsin-ligand interactions. J Med Chem. 1977 Nov;20(11):1420–1435. doi: 10.1021/jm00221a013. [DOI] [PubMed] [Google Scholar]
- Huber R., Kukla D., Bode W., Schwager P., Bartels K., Deisenhofer J., Steigemann W. Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor. II. Crystallographic refinement at 1.9 A resolution. J Mol Biol. 1974 Oct 15;89(1):73–101. doi: 10.1016/0022-2836(74)90163-6. [DOI] [PubMed] [Google Scholar]
- Krieger M., Kay L. M., Stroud R. M. Structure and specific binding of trypsin: comparison of inhibited derivatives and a model for substrate binding. J Mol Biol. 1974 Feb 25;83(2):209–230. doi: 10.1016/0022-2836(74)90388-x. [DOI] [PubMed] [Google Scholar]
- Krieger M., Koeppe R. E., 2nd, Stroud R. M. pH dependence of tritium exchange with the C-2 protons of the histidines in bovine trypsin. Biochemistry. 1976 Aug 10;15(16):3458–3464. doi: 10.1021/bi00661a010. [DOI] [PubMed] [Google Scholar]
- Matthews D. A., Alden R. A., Birktoft J. J., Freer T., Kraut J. Re-examination of the charge relay system in subtilisin comparison with other serine proteases. J Biol Chem. 1977 Dec 25;252(24):8875–8883. [PubMed] [Google Scholar]
- Richards F. M. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]