Abstract
Bacteriophage lambda integrates into the chromosome of its Escherichia coli host by means of a site-specific recombination between a locus on the phage chromosome (phage att site) and a locus on the bacterial chromosome (bacterial att site). The nucleotide sequence of four lambda att sites altered in site-specific recombination has been determined. The int-dependent deletions that generated these att sites have one end point within the phage att site and extend either to the left or to the right. As a result of the new internucleotide bond created by deletion formation, these phage have alterations in the 15-base-pair common core region. The new DNA sequences brought to the att sites by the deletions, designated delta for regions to the left and delta' for regions to the right, do not share any discernible homology with their analogous counterparts in the phage att site arms, P and P', respectively, or with the bacterial att site arms, B and B', respectively. The finding of alterations in the 15-base-pair common core region necessitates a reinterpretation of the genetic properties of these att sites in site-specific recombination. The structure of these sites in relation to their genetic properties can be viewed as being consistent with a model in which the only specificity elements in int-dependent site-specific recombination are the common core region, O, and the phage arms, P and P'.
Full text
PDF

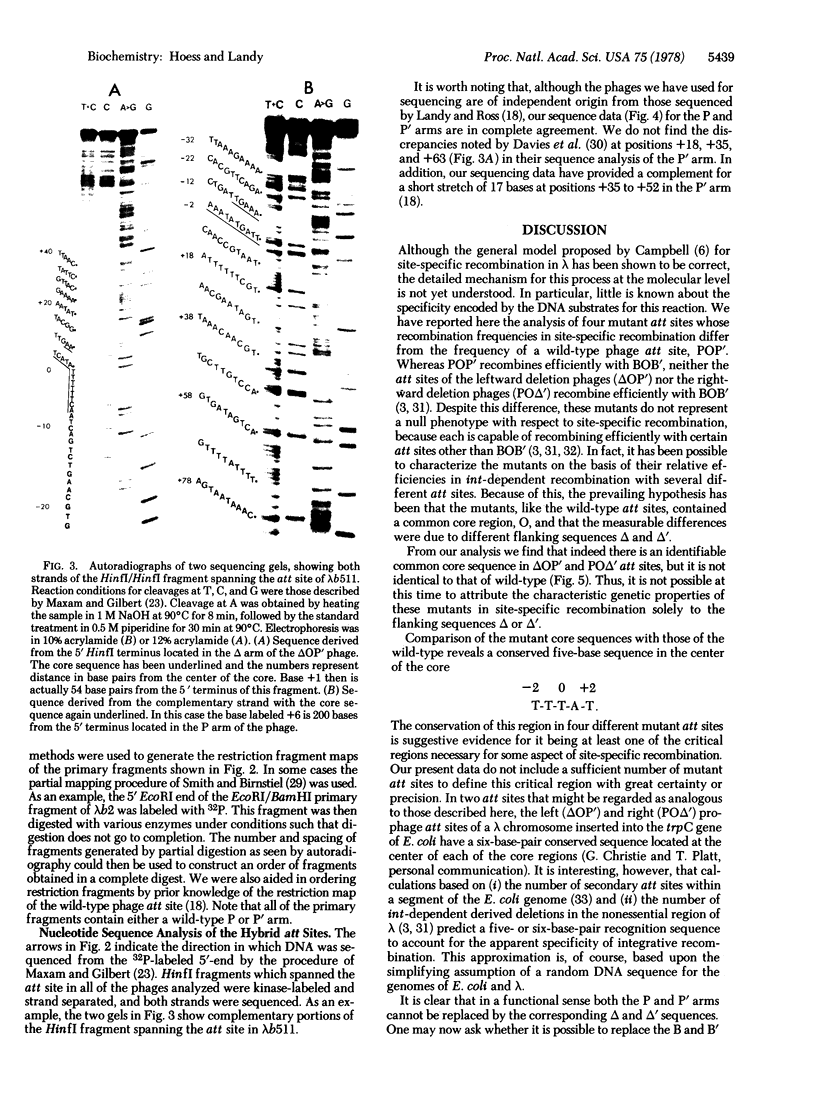


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boulter J., Lee N. Isolation of specialized transducing bacteriophage lambda carrying genes of the L-arabinose operon of Escherichia coli B/r. J Bacteriol. 1975 Sep;123(3):1043–1054. doi: 10.1128/jb.123.3.1043-1054.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R. W., Schreier P. H., Buchel D. E. Nucleotide sequence of the attachment site of coliphage lambda. Nature. 1977 Dec 22;270(5639):757–760. doi: 10.1038/270757a0. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Parkinson J. S. Deletion mutants of bacteriophage lambda. 3. Physical structure of att-phi. J Mol Biol. 1971 Mar 14;56(2):403–423. doi: 10.1016/0022-2836(71)90473-6. [DOI] [PubMed] [Google Scholar]
- Egan J., Landy A. Structural analysis of the tRNA1Tyr gene of Escherichia coli. A 178 base pair sequence that is repeated 3.14 times. J Biol Chem. 1978 May 25;253(10):3607–3622. [PubMed] [Google Scholar]
- Fischer-Fantuzzi L. Integration of lambda and lambda-b2 genomes in nonimmune host bacteria carrying a lambda cryptic prophage. Virology. 1967 May;32(1):18–32. doi: 10.1016/0042-6822(67)90248-6. [DOI] [PubMed] [Google Scholar]
- Gingery R., Echols H. Mutants of bacteriophage lambda unable to integrate into the host chromosome. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1507–1514. doi: 10.1073/pnas.58.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M. E., Yarmolinsky M. B. Integration-negative mutants of bacteriophage lambda. J Mol Biol. 1968 Feb 14;31(3):487–505. doi: 10.1016/0022-2836(68)90423-3. [DOI] [PubMed] [Google Scholar]
- Guarneros G., Echols H. New mutants of bacteriophage lambda with a specific defect in excision from the host chromosome. J Mol Biol. 1970 Feb 14;47(3):565–574. doi: 10.1016/0022-2836(70)90323-2. [DOI] [PubMed] [Google Scholar]
- Guerrini F. On the asymmetry of lambda integration sites. J Mol Biol. 1969 Dec 28;46(3):523–542. doi: 10.1016/0022-2836(69)90194-6. [DOI] [PubMed] [Google Scholar]
- Haggerty D. M., Scheif R. F. Location in bacteriophage lamdba DNA of cleavage sites of the site-specific endonuclease from Bacillus amyloliquefaciens H. J Virol. 1976 May;18(2):659–663. doi: 10.1128/jvi.18.2.659-663.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KELLENBERGER G., ZICHICHI M. L., WEIGLE J. A mutation affecting the DNA content of bacteriophage lambda and its lysogenizing properties. J Mol Biol. 1961 Aug;3:399–408. doi: 10.1016/s0022-2836(61)80053-3. [DOI] [PubMed] [Google Scholar]
- Kaiser A. D., Masuda T. Evidence for a prophage excision gene in lambda. J Mol Biol. 1970 Feb 14;47(3):557–564. doi: 10.1016/0022-2836(70)90322-0. [DOI] [PubMed] [Google Scholar]
- Landy A., Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977 Sep 16;197(4309):1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini J. C., Weisberg R., Landy A. The isolation of restriction fragments containing the primary and secondary (galT) bacterial att sites of phage lambda. Virology. 1977 Dec;83(2):254–270. doi: 10.1016/0042-6822(77)90170-2. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash H. A. Integration and excision of bacteriophage lambda. Curr Top Microbiol Immunol. 1977;78:171–199. doi: 10.1007/978-3-642-66800-5_6. [DOI] [PubMed] [Google Scholar]
- Parkinson J. S. Deletion mutants of bacteriophage lambda. II. Genetic properties of att-defective mutants. J Mol Biol. 1971 Mar 14;56(2):385–401. doi: 10.1016/0022-2836(71)90472-4. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Robinson L. H., Landy A. HindII, HindIII, and HpaI restriction fragment maps of bacteriophage lambda DNA. Gene. 1977 Sep;2(1):1–31. doi: 10.1016/0378-1119(77)90019-1. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. III. The components of the secondary attachment sites. J Mol Biol. 1975 Apr 25;93(4):415–429. doi: 10.1016/0022-2836(75)90237-5. [DOI] [PubMed] [Google Scholar]
- Shulman M. J., Mizuuchi K., Gottesman M. M. New att mutants of phage lambda. Virology. 1976 Jul 1;72(1):13–22. doi: 10.1016/0042-6822(76)90307-x. [DOI] [PubMed] [Google Scholar]
- Shulman M., Gottesman M. Attachment site mutants of bacteriophage lambda. J Mol Biol. 1973 Dec 25;81(4):461–482. doi: 10.1016/0022-2836(73)90517-2. [DOI] [PubMed] [Google Scholar]
- Signer E. R., Weil J., Kimball P. C. Recombination in bacteriophage lambda. 3. Studies on the nature of the prophage attachment region. J Mol Biol. 1969 Dec 28;46(3):543–563. doi: 10.1016/0022-2836(69)90195-8. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Reeder R. H., Carroll D., Brown D. D., Deutch A., Higashinakagawa T., Dawid I. B. Amplified ribosomal DNA from Xenopus laevis has heterogeneous spacer lengths. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2823–2827. doi: 10.1073/pnas.71.7.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zissler J. Integration-negative (int) mutants of phage lambda. Virology. 1967 Jan;31(1):189–189. doi: 10.1016/0042-6822(67)90030-x. [DOI] [PubMed] [Google Scholar]





