Abstract
Background:
About 50% of non-small cell lung cancer (NSCLC) patients develop distant metastases following pulmonary resection. Currently, there are no reliable factors allowing for individual selection of high-risk patients for adjuvant systemic therapies.
Methods:
We assessed by quantitative reverse transcription PCR microRNA (miRNA) expression in 273 stage I–IIIA NSCLC samples. Expression of 677 miRNAs was evaluated in fresh-frozen tumour samples in the training cohort of 50 squamous cell carcinoma (SCC) patients who underwent curative surgery. Of those, 20 patients developed distant metastases, and 30 were free of recurrence for >4 years. In the second step, miRNAs with highest predictive value for distant relapse were re-evaluated in formalin-fixed paraffin-embedded material in an independent group of 134 stage I–IIIA SCC patients. Additionally, the same miRNAs were investigated in 89 lung adenocarcinoma (AC) patients and in normal lung parenchyma (NLP).
Results:
In the training cohort of SCC, six miRNAs were differently expressed in the non-recurrent vs recurrent groups and correlated with distant recurrence-free survival, however none reached the level of significance after correction for multiple testing. Of these six miRNAs, miR-662, -192 and -192* were confirmed as prognostic in the independent SCC cohort. Expression of miR-128, -10b, -502-3p and -192 differed between SCC and AC, and miR-128 and -192 – between NLP and NSCLC.
Conclusions:
We identified three new miRNAs predictive of distant relapse in operable SCC. Future miRNA studies should account for differences between NSCLC subtypes.
Keywords: microRNA, quantitative reverse transcription polymerase chain reaction (qRT–PCR), non-small cell lung cancer (NSCLC), squamous cell lung cancer (SCC), lung adenocarcinoma (AC), prognostic marker
Non-small cell lung cancer (NSCLC) is the primary cause of cancer-related deaths (Siegel et al, 2013). Only around 20% of cases are diagnosed at the early stage, allowing for effective pulmonary resection (Chee et al, 2008). Additionally, ∼50% of those who undergo surgery will subsequently relapse, mainly at distant sites (Winton et al, 2005). Adjuvant chemotherapy was shown to improve the 5-year survival by around 5% (Pignon et al, 2008), therefore selection of patients who may benefit from this approach is essential.
In the past three decades, several molecular abnormalities have been investigated to select high-risk subsets among early stage NSCLC patients. One-gene aberrations, for example, mutations of TP53 or K-ras gene, are not predictive for disease outcomes (Huncharek et al, 2000; Aviel-Ronen et al, 2006). Similarly, the prognostic role of protein markers, such as ERCC1 or RRM1, remains controversial (Bepler et al, 2011; Friboulet et al, 2013). More recently, new gene-expression markers associated with increased risk of NSCLC recurrence were postulated, very few have been reliably validated though (Zhu et al, 2010; Xie et al, 2011; Kratz et al, 2012), and none in a prospective manner.
MicroRNAs (miRNAs) are small non-coding RNAs that have a key role in cell biology, as they control expression of more than 30% of human genes (Calin and Croce, 2006). These molecules execute gene silencing through binding to target sequences on mRNA, whereby they block translation. They are commonly encoded in the introns of protein-coding genes or as independent genes (Ying and Lin, 2009). Recently, expression of miRNAs in NSCLC was also shown to correlate with prognosis (Takamizawa et al, 2004; Yu et al, 2008; Landi et al, 2010). Notably, these molecules are stable in formalin-fixed paraffin-embedded (FFPE) samples, which may facilitate their clinical use. Finally, miRNAs are relatively robust; for example, in contrast to mRNA expression, their expression measurements are independent of the storing time of archival samples (Hall et al, 2012).
In a recent study, the prognostic value of several miRNAs was demonstrated in NSCLC and validated in a sizeable cohort of lung adenocarcinoma (AC) patients (Lu et al, 2012). Another study addressing prognostic significance of miRNA expression did not account for NSCLC subtypes in the validation phase (Yu et al, 2008). Owing to different expression of several miRNAs or mRNAs in particular histological types of NSCLC, prognostic analyses including more than one subtype of NSCLC are questionable (Landi et al, 2010; Skrzypski et al, 2013). In the current study, we investigated whether miRNA expression may predict disease dissemination in operable squamous cell lung carcinoma (SCC), which in many countries still constitutes the major histological subtype of NSCLC. To this end, nearly 700 miRNAs were screened in the training cohort, and the miRNAs with the highest predictive value for distant metastases were subsequently assessed in the independent cohort of patients, in accordance with REMARK guidelines for prognostic markers development (McShane et al, 2005). In addition, we compared miRNA expression in SCC and AC patients, and in normal lung parenchyma (NLP) from NSCLC patients and non-lung cancer individuals.
Materials and Methods
Patients
The study was approved by the Ethics Committee of the Medical University of Gdańsk. The study subjects were NSCLC patients who underwent curative pulmonary resection between 2001 and 2012 and subsequently either developed distant metastases (‘cases') or survived at least 4 years without any type of recurrence (‘controls'), with median follow-up of 5.8 years; range, 4.1–10.1. Patients who developed isolated local recurrence, for example, limited to the bronchial stump, hilar or mediastinal lymph nodes, were excluded. All patients underwent pathologically confirmed complete anatomical resection of the primary tumour and adequate mediastinal lymph node dissection. None of the patients received preoperative or postoperative chemotherapy, or adjuvant radiotherapy.
Training cohort: fresh-frozen tumour tissue samples
Expression of 677 miRNAs was analysed in 50 stage I–IIIA (non-N2) SCC patients (Table 1). Study group included 50 patients, 20 of whom subsequently developed distant metastases and 30 who were free of any type of recurrence for the median follow-up of 6.3 years (range 4.6–9.4). In all cases, fresh-frozen samples of tumour tissue were obtained during pulmonary resection. Major clinical characteristics in relapsed and not-relapsed cohorts did not differ significantly (Table 1).
Table 1. Patient characteristics.
| Variable | Category | Total | Relapsed | Relapse-free | P-value |
|---|---|---|---|---|---|
|
Training set – SCC | |||||
| |
|
n=50 (100%) |
n=20 (100%) |
n=30 (100%) |
|
| Sex | Male | 39 (78) | 17 (85) | 22 (73) | 0.53 |
| |
Female |
11 (22) |
3 (15) |
8 (27) |
|
| Stage | IA | 6 (12) | 0 (0) | 6 (20) | 0.08 |
| IB | 25 (50) | 11 (55) | 14 (47) | ||
| IIA | 0 (0) | 0 (0) | 0 (0) | ||
| IIB | 17 (34) | 9 (45) | 8 (27) | ||
| |
IIIA |
2 (4) |
0 (0) |
2 (7) |
|
| Grade | G1 | 4 (8) | 2 (10) | 2 (7) | 0.65 |
| G2 | 34 (68) | 12 (60) | 22 (73) | ||
| |
G3 |
12 (24) |
6 (30) |
6 (20) |
|
| Mean age (range) |
|
67 (48–77) |
63 (48–75) |
68 (49–77) |
0.38 |
|
Confirmatory set – SCC | |||||
| |
|
n=134 (100%) |
n=54 (100%) |
n=80 (100%) |
|
| Sex | Male | 101 (75) | 39 (72) | 62 (77) | 0.55 |
| |
Female |
33 (25) |
15 (28) |
18 (23) |
|
| Stage | IA | 29 (22) | 5 (9) | 24 (30) | 0.034 |
| IB | 65 (48) | 25 (46) | 40 (50) | ||
| IIA | 4 (3) | 1 (2) | 3 (4) | ||
| IIB | 31 (23) | 20 (37) | 11 (14) | ||
| |
IIIA |
5 (4) |
3 (6) |
2 (2) |
|
| Grade | G1 | 5 (4) | 2 (4) | 3 (4) | 0.52 |
| G2 | 78 (58) | 29 (54) | 49 (61) | ||
| |
G3 |
51 (38) |
23 (42) |
28 (35) |
|
| Mean age (range) |
|
64 (39–81) |
62 (39–79) |
64 (47–81) |
0.033 |
|
AC | |||||
| |
|
n=89 (100%) |
n=35 (100%) |
n=54 (100%) |
|
| Sex | Male | 57 (64) | 24 (69) | 33 (61) | 0.62 |
| |
Female |
32 (36) |
11 (31) |
21 (39) |
|
| Stage | IA | 28 (31) | 7 (20) | 21 (39) | 0.093 |
| IB | 34 (38) | 12 (34) | 22 (41) | ||
| IIA | 3 (3) | 2 (6) | 1 (2) | ||
| IIB | 23 (26) | 13 (37) | 10 (18) | ||
| |
IIIA |
1 (1) |
1 (3) |
0 (0) |
|
| Predominant histological type | Acinar | 34 (38) | 18 (51) | 16 (30) | 0.31 |
| Solid | 30 (34) | 10 (29) | 20 (37) | ||
| Papillary | 7 (8) | 3 (8) | 4 (7) | ||
| Micro-papillary | 3 (3) | 1 (3) | 2 (4) | ||
| Lepidic | 8 (9) | 1 (3) | 7 (13) | ||
| |
Mucinous |
7 (8) |
2 (6) |
5 (9) |
|
| Mean age (range) | 61 (45–81) | 61 (45–81) | 61 (46–75) | 0.46 | |
Abbreviations: AC=adenocarcinoma; SCC=squamous cell carcinoma.
For one RNA isolation, 20 slices of 5 μm were cut into the lysis buffer (Qiagen, Valencia, CA, USA) from the fresh-frozen tumour blocks in criostat, so that the first and the last slice were set aside for haematoxylin and eosin staining to assess the viable tumour content. Only samples from blocks that contained at least 50% of viable tumour tissue on both sections were used for RNA isolation.
Total RNA containing miRNA was isolated using miRNeasy Mini Kit (Qiagen). The RNA concentration was measured with NanoDrop and quality was analysed with RNA Lab Chip Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). Reverse transcription was carried out with 750 ng of RNA with TaqMan MiRNA RT kit (Applied Biosystems, Valencia, CA, USA) and pools A or B of stem-loop primers (Megaplex Primer Pools, Human Pools Set v2.0, Applied Biosystems), in accordance with the manufacturer's recommendations. Each primer pool included primers for amplifying 378 different miRNAs. The resultant cDNA was quantified with qRT–PCR reactions that contained polymerase with 5′ nuclease activity, target specific primers and fluorescent TaqMan probes in microfluidic cards (TaqMan Array Human miRNA A+B Cards Set v2.0, Applied Biosystems) in HT 7900 cycler (Applied Biosystems), with reaction conditions in accordance with the manufacturer's recommendations. Raw expression results (Ct values) were obtained through SDS.2.1 software (Applied Biosystems).
Expression of miRNA in SCC confirmatory cohort and in lung AC: FFPE tumour tissue samples
The miRNAs that best predicted for distant metastases in the training set were subsequently assessed in an independent cohort of 134 stage I–IIIA (non-N2) SCC patients. In addition, expression of these miRNAs was investigated in 89 stage I-IIIA (non-N2) AC patients. These groups included patients who developed distant metastases after surgery (SCC: 41%, AC: 42%) or were free of relapse for the median follow-up of 5.6 years (range, 4.1–10.0) and 6.2 years (range, 4.3–10.1) for SCC and AC, respectively (Table 1). No significant differences were found between the groups with and without metastases, except for stage and age in the SCC group (Table 1).
In all patients, paraffin blocks containing primary tumour were reviewed and one slice from each block was obtained for haematoxylin and eosin staining. The block with the highest percentage of cancer tissue was chosen for molecular analysis. To further decrease the content of non-carcinomatous tissue, admixture of normal lung, chondroid tissue, lymph nodes and necrotic tissue were removed by macrodissection.
RNA was isolated from paraffin-embedded blocks that after macrodissection contained at least 80% of viable tumour tissue. Four slices of 20 μm each were cut for total RNA isolation. To avoid cross-contamination, blades used for macrodissection and for slice cutting were changed after each case.
Total RNA containing miRNA was isolated from FFPE tumour tissue with RecoverAll Kit (Ambion, Valencia, CA, USA). The concentration of RNA was assessed in NanoDrop. Reverse transcription was carried out with 300 ng of total RNA with TaqMan MiRNA RT kit (Applied Biosystems) and pools A and B of stem-loop primers (Megaplex Primer Pools, Human Pools Set v3.0, Applied Biosystems), in accordance with the manufacturer's recommendations. The obtained cDNA was quantified by qRT–PCR in triplicates with the use of miRNAs specific primers and fluorescent TaqMan probes, and polymerase with 5′ nuclease activity in customised microfluidic cards (Custom TaqMan Array Microfluidic Cards, Applied Biosystems) in HT 7900 cycler (Applied Biosystems), with reaction conditions in accordance with the manufacturer's recommendations (Applied Biosystems). Raw expression results (Ct values) were obtained through SDS.2.1 software (Applied Biosystems).
miRNA expression in matched specimens of normal lung from lung cancer patients and from non-lung cancer individuals
Twenty samples of pathologically verified uninvolved lung parenchyma in close proximity to tumour were acquired from lung cancer patients whose tumoral tissue was subjected to miRNA expression analyses. In addition, samples of NLP were obtained from 17 individuals who underwent thoracotomy for pneumothorax (5 women and 12 men, age range 19–70 years). Of those, 10 patients never smoked and the remaining 7 were active moderate smokers (4–20 pack-years). One patient suffered from Marfan's syndrome. Six patients had previous episodes of pneumothorax and the remaining 11 (including the Marfan's syndrome patient) were treated for pneumothorax for the first time. None of these patients had a current or previous history of lung cancer or any pulmonary disease other than pneumothorax.
Pathological assessment
In all cases, histological type of cancer was re-evaluated using haematoxylin and eosin staining independently by two pathologists (PC and WB), in accordance with the WHO criteria. The discordant or dubious cases were further diagnosed with immunohistochemistry, namely p63 and CK5/6 positive staining was considered indicative of SCC, whereas TTF-1 and CK7 of AC. Cases with equivocal diagnosis, for example, with co-expression of p63 and TTF-1, were excluded from the study.
SCC grade was determined according to the 2004 classification of lung cancer (Travis et al, 2004). The subtypes of AC were determined using the new lung AC classification (Travis et al, 2011), considering the predominant morphological component of AC.
Owing to the retrospective character of this study, we used originally determined tumour stage, according to the 6th edition of the TNM classification (Sobin and Wittekind, 2002).
Statistical analysis
miRNAs with no amplification signal (Ct⩾40) in at least 80% of samples were excluded from the training cohort data set and were not subjected to further analysis. The individual Ct values for target miRNAs were normalised against the mean of the Ct values of U6 RNA and RNU48 and miRNA expression was obtained with the 2−(ΔCt) method (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008). In the training cohort, the miRNA expression was correlated with distant metastases-free survival (DMFS) using univariate Cox regression model to identify miRNAs predictive of distant relapse. To delineate miRNAs with highest fold change between ‘recurrent' and ‘non-recurrent' groups, that is, potentially constituting the most robust markers, their expression was also compared between the groups with Student's t-test. miRNAs significant in both analyses at P<0.05 (unadjusted) were deemed potentially relevant, and their expression was subsequently analysed in the confirmatory cohort. In the confirmatory phase, miRNA expression was normalised against the mean of the Ct values of U6 RNA and RNU48 with ΔCt method and was correlated with DMFS using Cox regression model (ΔCt method). P-values were adjusted for multiple hypotheses testing using Benjamini–Hochberg algorithm in both training and confirmatory cohorts. The clinical relevance of the miRNA expression was further tested in the multivariate models that included three variables: miRNA expression (as −ΔCt), the stage and the histological grade. The negative ΔCt allowed for the intuitive interpretation of resulting HR value, that is, for the cases where the high miRNA expression was associated with the high risk of relapse, the obtained HR value was greater than 1. The stage was projected to a binary low/high status (stage I vs stage II–IIIA) and the grade was included with the values of 1, 2 or 3, respectively. The log-rank analyses were performed for miRNAs in the validation phase with cut-offs corresponding to: (i) 60th percentile of expression, where high miRNA expression correlated with high risk of recurrence or (ii) 40th percentile of expression, where low miRNA expression correlated with high risk of recurrence. The cut-offs (40th/60th percentiles) were applied according to the number of patients in the ‘recurrent' group (around 40%) in SCC confirmatory cohort and in the AC group. The DMFS curves were generated using Kaplan–Meier method. Expression of six miRNAs analysed in the confirmatory cohort was compared between SCC and AC, and between cancer and NLP, using Mann–Whitney test. To assess the applicability of the results from fresh-frozen samples to FFPE samples, the expression of these six miRNAs was also assessed in 47 matched pairs of fresh-frozen and FFPE samples from the same patients and compared with Pearson's test.
Box-plots were drawn using R software. Before plotting, measurements outside median +/− 3IQR were discarded. Box marks the first to third quartile range, whiskers extends to the most extreme data points, but no more than 1.5 times IQR from the box.
Results
Expression of 494 miRNAs (73% of 677 investigated) was detected in at least 20% of samples from the training set. The quality of the RNA from fresh-frozen tissues was in the range of 7.5–9.8 RIN. This was reflected by the narrow ranges of Ct values for RNAs chosen for normalisation: U6 RNA (range 17.1–19.0) and RNU48 (range 20.0–22.3) for frozen tissue samples. In the confirmatory series including FFPE SCC samples, the Ct ranges of expression were wider: U6 RNA: 19.0–28.3 and RNU48: 23.4–29.1, and respectively for AC: U6 RNA: 18.1–27.8 and RNU48: 22.7–29.6. The Ct values for normalisation and target miRNAs were different between fresh-frozen and FFPE samples (P<0.001). Except for miR-662, there was a significant correlation of normalised expression of target miRNAs between fresh-frozen and FFPE samples (correlation coefficients in the range of 0.39–0.63). However, only for miR-10b the coefficient of correlation was above 0.5 (Supplementary Table 1).
Association between miRNA expression and DMFS
To identify in the training cohort potentially prognostic miRNAs, we selected candidate miRNAs, that is, those that met simultaneously two statistical criteria: their expression correlated with DMFS (unadjusted P<0.05 in Cox test) and was different between ‘relapsed' and ‘not relapsed' patients (unadjusted P<0.05 in Student's t-test). These criteria were met by six miRNAs: miR-10b, miR-662, miR-502-3p, miR-192*, miR-192 and miR-128 (Table 2), none of whom, however, reached the level of significance after correction for multiple testing. In the second step, these six miRNAs were formally assessed in an independent cohort of SCC, and subjected to additional analysis in AC patients. The list of correlations of expression of all analysed miRNAs with DMFS (Cox test) and comparison of miRNAs expression between ‘relapsed' and ‘not relapsed' patients (Student's t-test) in the training cohort are shown in the Supplementary Table 2.
Table 2. miRNA deemed as potentially relevant prognostic markers in the training cohort of SCC patients.
| miRNA | Cox P-value | Cox corrected P-value | t-Student P-value | t-Student corrected P-value |
|---|---|---|---|---|
| 10b |
0.0017 |
0.61 |
<0.001 |
0.25 |
| 662 |
0.0026 |
0.61 |
0.025 |
0.99 |
| 502-3p |
0.007 |
0.65 |
0.021 |
0.99 |
| 192* |
0.007 |
0.65 |
0.027 |
0.99 |
| 192 |
0.017 |
0.89 |
0.025 |
0.99 |
| 128 | 0.045 | 0.89 | 0.034 | 0.99 |
Abbreviation: SCC=squamous cell lung carcinoma.
Corrected P-values were computed with Benjamini–Hochberg algorithm.
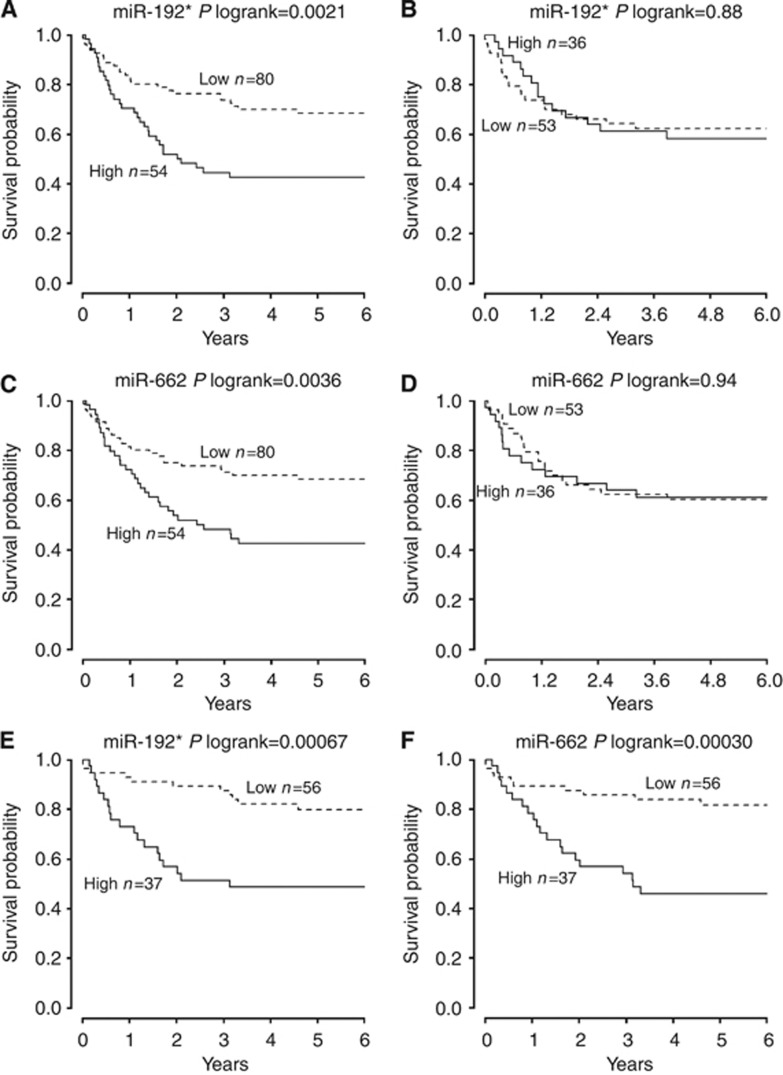
In the SCC confirmatory cohort, expression of miR-662, miR-192 and miR-192* significantly correlated with DMFS (adjusted P<0.05). None of the analysed miRNAs was significant in AC patients (Table 3). DMFS probability curves for significant miRNAs are shown in Figure 1. When the correction for multiple comparisons was applied, miR-662 (P=0.003), miR-192 (P=0.007) and miR-192* (P=0.034) retained their significance in the confirmatory SCC group. Of those, miR-662 (adjusted P<0.001), miR-192 (adjusted P=0.004) and miR-192* (adjusted P=0.009) were also significant in the subset of stage I SCC. In the multivariate analyses including miRNA expression, TNM stage and tumour grade, miR-662 (P<0.001, HR=1.23, 95% CI: 1.09–1.38), miR-192 (P=0.009, HR=1.24, 95% CI: 1.06–1.45), miR-192* (P=0.037, HR=1.12, 95% CI: 1.01–1.24) and stage (P<0.002, HR in the range of 2.41–2.58) were independently correlated to DMFS in the SCC confirmatory group (Table 4).
Table 3. Relationship between miRNA expression and DMFS in the confirmatory cohort of SCC patients, in the exploratory subset of stage I SCC from the confirmatory SCC cohort, and in AC patients.
| miRNA | All SCC P-value | All SCC FDR | All SCC Cox coefficients | Stage I SCC P-value | Stage I SCC FDR | Stage I SCC Cox coefficients | All AC P-value | All AC Cox coefficients |
|---|---|---|---|---|---|---|---|---|
| 662 |
<0.001 |
0.003 |
−0.22 |
<0.001 |
<0.001 |
−0.38 |
0.27 |
−0.102 |
| 192 |
0.002 |
0.007 |
−0.23 |
0.001 |
0.0044 |
−0.30 |
0.87 |
0.0118 |
| 192* |
0.017 |
0.034 |
−0.126 |
0.004 |
0.0088 |
−0.200 |
0.55 |
−0.042 |
| 502-3p |
0.057 |
0.085 |
0.106 |
0.043 |
0.065 |
0.142 |
0.125 |
0.111 |
| 128 |
0.106 |
0.128 |
0.104 |
0.068 |
0.082 |
0.143 |
0.90 |
0.0116 |
| 10b | 0.4 | 0.40 | −0.070 | 0.29 | 0.29 | −0.118 | 0.62 | −0.055 |
Abbreviations: AC=lung adenocarcinoma; DMFS=distant metastasis-free survival; SCC=squamous cell lung carcinoma.
Negative sign of Cox coefficient denotes that high miRNA expression correlates with shorter DMFS. Differences significant after Benjamini–Hochberg correction (FDR<0.05) are marked in bold.
Figure 1.
Distant metastasis-free survival (DMFS) according to expression of selected miRNAs in squamous cell lung cancer (SCC) and adenocarcinoma (AC). (A) DMFS according to the miR-192* expression in stage I–IIIA SCC. (B) DMFS according to miR-192* expression in stage I–IIIA AC. (C) DMFS according to miR-662 expression in stage I–IIIA SCC. (D) DMFS according to miR-662 expression in stage I–IIIA AC. (E) DMFS according to miRNA-192* expression in stage I SCC. (F) DMFS according to miRNA-662 expression in stage I SCC.
Table 4. P-values and HR with 95% CI for prognostic multivariate Cox models including: miRNA expression, stage and grade.
| |
miRNA |
Stage |
Grade |
|||
|---|---|---|---|---|---|---|
| miRNA | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) |
| 662 |
<0.001 |
1.23 (1.09–1.38) |
<0.001 |
2.58 (1.52–4.37) |
0.45 |
1.20 (0.75–1.91) |
| 192 |
0.009 |
1.24 (1.06–1.45) |
0.0011 |
2.41 (1.42–4.09) |
0.56 |
1.14 (0.73–1.78) |
| 192* |
0.037 |
1.12 (1.01–1.24) |
<0.001 |
2.51 (1.48–4,26) |
0.41 |
1.21 (0.77–1.91) |
| 502-3p |
0.123 |
0.91 (0.81–1.03) |
<0.001 |
2.51 (1.48–4.27) |
0.46 |
1.17 (0.77–1.78) |
| 10b |
0.29 |
1.10 (0.93–1.30) |
<0.001 |
2.65 (1.56–4.50) |
0.32 |
1.25 (0.80–1.96) |
| 128 | 0.42 | 0.94 (0.82–1.09) | <0.001 | 2.50 (1.45–4.29) | 0.45 | 1.19 (0.76–1.84) |
Abbreviations: CI=confidence interval; HR=hazard ratio.
Differences significant after Benjamini–Hochberg correction (FDR<0.05) are marked in bold.
Validation of the prognostic relevance of the miRNAs previously reported as prognostic in either NSCLC or SCC
The prognostic significance of previously reported miRNAs was scrutinised in the training cohort where the expression data for 677 miRNAs were available. Among 46 miRNAs correlating with DMFS in the training cohort at the level of P<0.1, 5 were previously reported as prognostic in NSCLC or SCC (Supplementary Table 2, Supplementary Figure 1). MiR-15a (P=0.004), miR-137 (P=0.051) and miR-193a-3p (P=0.079) were upregulated in the ‘recurrent' vs ‘non-recurrent' SCC cohorts, whereas miR-128 (P=0.045) and miR-149 (P=0.057) were underexpressed.
miRNAs expression in SCC vs AC
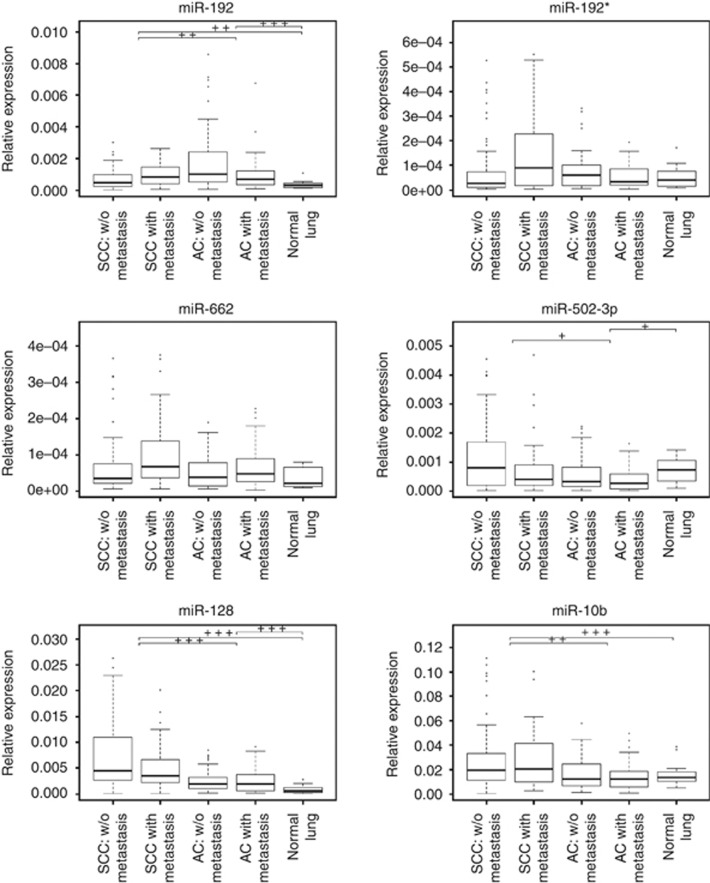
To test for potential differences in the miRNA expression between histopathological subtypes of NSCLC, we compared between SCC and AC the expression of six miRNAs selected for confirmatory analysis in SCC. Of those, expression of miR-128, miR-10b, miR-502-3p and miR-192 was significantly different between SCC and AC (adjusted P<0.05; Figure 2).
Figure 2.
Expression of miRNAs analysed in validation: miR-10b, miR-128, miR-192, miR-502-3p, miR-662 and miR-192* in: NLP from non-lung cancer individuals (‘normal lung'); squamous cell lung carcinoma that did not recur (‘SCC w/o metastasis'); squamous cell lung carcinoma that did recur (‘SCC with metastasis'); from lung adenocarcinoma that did not recur (‘AC w/o metastasis'); from lung adenocarcinoma that did recur (‘AC with metastasis'); *P<0.05, **P<0.001 and ***P<0.0001.
miRNA expression in NSCLC vs NLP
Expression of miR-128 and miR-192 differed significantly between NLP and SCC (adjusted P<0.006), irrespective of relapse status (Table 5, Figure 2). Expression of miR-662 and miR-10b was significantly different between relapsed SCC and NLP (adjusted P<0.03) (Supplementary Table 3). MiR-192 and miR-128 in NLP was differently expressed compared with AC (adjusted P<0.001), irrespective of relapse status (Table 5, Figure 2). Expression of miR-502-3p was significantly different between relapsed AC and NLP (adjusted P=0.011), as well as between AC and NLP (adjusted P=0.048) (Supplementary Table 3). There were no significant differences in expression between NLP from non-lung cancer individuals and NLP adjacent to tumour from lung cancer patients (adjusted P>0.05).
Table 5. Expression fold change and FDR: AC vs SCC, SCC vs NL parenchyma, AC vs NL parenchyma.
| miRNA | Normalised expression fold change AC/SCC | SCC vs AC FDR | Normalised expression fold change SCC/NL | SCC vs NL FDR | Normalised expression fold change AC/NL | AC vs NL FDR |
|---|---|---|---|---|---|---|
| 128 |
0.48 |
<0.001 |
8.04 |
<0.001 |
3.88 |
<0.001 |
| 10b |
0.66 |
0.003 |
3.12 |
<0.001 |
2.04 |
0.063 |
| 192 |
4.00 |
0.003 |
1.19 |
0.005 |
4.75 |
<0.001 |
| 502-3p |
0.55 |
0.016 |
1.69 |
0.865 |
0.94 |
0.048 |
| 662 |
0.54 |
0.3 |
2.02 |
0.053 |
1.09 |
0.326 |
| 192* | 1.24 | 0.52 | 3.77 | 0.481 | 4.66 | 0.2 |
Abbreviations: AC=lung adenocarcinoma; NL=normal lung; SCC=squamous cell lung carcinoma.
Differences significant after Benjamini–Hochberg correction (FDR<0.05) are marked in bold.
Discussion
This is the first report on the prognostic significance of the expression of miR-662, miR-192 and miR-192* in early stage SCC. Notably, the prognostic relevance of these miRNAs was confirmed in a sizeable independent cohort of patients. Our data also suggest that these miRNAs are most likely not prognostic in lung AC. The study was devised in two steps: (i) relevant miRNAs were identified by screening of the expression of 677 miRNAs in the training cohort, and (ii) potentially relevant miRNAs were subsequently assessed in the independent cohort of SCC, and additionally determined in AC patients and in NLP. A number of stringent methodological requirements were met to verify the study hypothesis. First, we assumed that the loco-regional recurrences in the bronchial stump, hilar or mediastinal lymph nodes and the chest wall most often occur stochastically, as a result of incomplete tumour removal at submicroscopic level. Therefore, all cases with isolated loco-regional recurrences of NSCLC were excluded from the analyses on the premise that they likely occur irrespective of the primary tumour potential for distant spread. Consequently, miRNA expression was compared between the two groups of patients who either developed distant metastases or were free of recurrence after long-term follow-up. This approach has the potential to provide the clearest experimental setting for development of markers of increased risk of distant recurrence. To our knowledge, none of the previously reported studies on prognostic gene signatures mandated the exclusion of cases with the isolated local recurrences (Raponi et al, 2006; Guo et al, 2008; Raponi et al, 2009; Zhu et al, 2010; Lu et al, 2012).
Second, as 85% of NSCLC recurrences become evident within 3 years after resection (Pepek et al, 2011), the minimum follow-up of 4 years was required for all patients considered as ‘controls' to ensure a reasonable probability that they were truly free of cancer dissemination. Relatively short follow-up of patients might have confounded the analyses in several previous studies. For example, Yu et al (2008) reported the censoring rate of 66%. Indeed, short postoperative observation renders a non-relapsed group inadequate for prognostic analyses, as a patient without an evident relapse after, for example, 12 months is still at a relatively high risk of developing metastases in due course. Further, it was required that the patients had not been administered induction or adjuvant chemotherapy, as we reasoned that its use might obfuscate the actual risk of tumour dissemination.
In this ‘proof of concept' study, we identified and confirmed the prognostic significance of three new miRNAs in operable SCC. Owing to the differences in miRNA expression between the fresh-frozen and FFPE samples, we did not attempt to determine the optimal cut-offs for both materials. The hazard ratios for the prognostic miRNAs were lower than those for the stage; however, the prognostic effect of each miRNA was independent of other factors including stage. The prognostic value of miRNAs in NSCLC may likely be strengthened by their pooling into a multi-gene classifier. However, we intentionally forwent here the development of multi-gene prognostic models, as their parameters (cut-offs, coefficients weighted for each miRNA in risk score equation etc.) tend to be unstable if developed in cohorts including fewer than 250 cases (Michiels et al, 2005; van Vliet et al, 2008). Moreover, a formal validation of a multi-gene model necessitates a collection of numerous samples, preferably from many centres. This project is currently under way.
A relatively small training cohort (n=50) is an apparent limitation of our study, as it allows for reliable detection of only a strong HR, in the range of >4, with a power of 0.8 in the case of a single hypothesis. Indeed, to detect a modest prognostic effect of miRNAs (e.g., HR=2), more than 600 patients are needed in a discovery set for high throughput scenario (>100 miRNAs tested). Thus, the results of the training phase in our study allowed only for ranking miRNAs according to their potential prognostic relevance, which was subsequently verified in an independent sizeable cohort of patients. Also, the morphological heterogeneity of NSCLC, even among its major histological subtypes, is being increasingly recognised. Warth et al (2012) showed that almost 93% of ACs displayed at least two histological patterns according to the novel IASLC/ATS/ERS AC classification (Travis et al, 2011). Molecular heterogeneity of SCC was recently reported as a part of the cancer mutational atlas (Network CGAR, 2012). Also, four mRNA expression-based classes of SCC were previously reported, where ‘primitive' subtype conferred poorer prognosis in reference to ‘classical', ‘secretory' and ‘basal' SCC subtypes (Wilkerson et al, 2010). The prognostic role of those molecular features has not yet been validated and therefore they were not included in the current analysis.
None of the prognostically relevant miRNAs from our series has been previously reported in SCC specific analyses. However, high expression of miR-192* was recently found to correlate with shorter relapse-free survival in the training cohort of 357 NSCLC patients, but not in separate analyses including AC (n=189) or SCC (n=106) (Lu et al, 2012). In gastric cancer, miR-192 was shown to be overexpressed in vivo and exerted cell growth and migration-promoting effects in vitro (Jin et al, 2011). High expression of miR-192 was also found in pancreatic cancer stem cells (Jung et al, 2011). This miRNA is encoded on chromosome 11 in the vicinity of a gene for CDC42 binding protein kinase gamma (DMPK-like) that has been implicated as a marker of aggressive course of NSCLC. In the study by Lu et al (2012), the low expression of miR-662 correlated with shorter survival in AC, but not in SCC and in the entire population of NSCLC. In the current study, however, the high expression of miR-662 strongly correlated with short DMFS in SCC in both training and confirmatory cohorts. MiR-662 is encoded on chromosome 16 in proximity of mesothelin (MSLN). Overexpression of mesothelin was reported in breast cancer metastases (Wu et al, 2008) and was found to promote anchorage-independent growth of breast cancer cells (Uehara et al, 2008). In paediatric cancers, the expression of 63% of miRNAs located inside coding genes was significantly correlated with that of their host genes (Wei et al, 2009). Further research is warranted to define the prognostic role of the genes potentially co-expressed with the reported miRNAs.
The computationally predicted mRNA targets of miRNAs associated with an elevated metastatic potential of SCC were scrutinised in an attempt to find their functional correlates. The overexpression of miR-662, miR-192* or miR-192 is expected to result in the downregulation of their mRNA targets. Along these lines, overexpression of miR-662 may provide an alternative mechanism of downregulation of a tumour suppressor gene GDF10 that belongs to TGF-β family ligands. Intriguingly, miR-662 also targets the NLRC5 that was recently identified as a transactivation factor for MHC I complexes, crucial for adaptive immune response against viruses and malignancy (Meissner et al, 2010).
Among the top targets for miR-192* is RAB21, a GTP-binding proteins that regulates intracellular trafficking of integrins, a process fundamental for cell adhesion and motility. Another target potentially downregulated by miR-192* is TSHZ3, a newly described candidate tumour suppressor gene for breast and prostate cancers (Yamamoto et al, 2011). MiR-192 and miR-192* simultaneously target 13 mRNA, including zinc finger proteins ZEB2 and integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51).
We confirmed a potential prognostic role of some previously reported miRNAs. For example, the adverse effect of the high expression of miR-15a, reported by Raponi et al (2009), miR-137, reported by Yu et al (2008), and miR-193-3p, reported by Lu et al (2012) was also significant or borderline in our training cohort (Cox analysis; P=0.004, P=0.051 and P=0.079, respectively). Notably, Lu et al (2012) reported an inverse correlation between expression of miR-15a and the risk of progression. The prognostic impact of miR-128 and miR-149 reported by these authors was confirmed in the current study, yet the overlap with other prognostic miRNAs from previous studies in NSCLC or SCC was limited. These discrepancies may stem from the differences in methodology used by different research groups and, in particular, normalisation procedures. In the current study, we used the mean of the expression of U6 and RNU48.
Importantly, miRNAs predictive of the dissemination of early stage SCC did not predict for distant recurrence in AC. It may be therefore envisaged along the lines of previous reports (Landi et al, 2010; Lu et al, 2012) that SCC and AC employ different molecular pathways during progression. Moreover, the expression of two miRNAs prognostic for SCC (miR-662 and miR-192*) was significantly different between SCC and AC. This finding additionally underscores the need for separate investigations of the prognostic effect of quantitative markers in these two major NSCLC subtypes. The differential expression of miR-128 and miR-502-3p between SCC and AC is consistent with the results of a previous study (Lu et al, 2012), whereas the differences in expression of miR-10b and miR-192 were not previously reported. Finally, expression of two prognostic miRNAs (miR-192 and miR-662) was significantly different in SCC compared with NLP. Such differences render these miRNAs particularly suited as constituents of clinically useful prognostic signatures.
In conclusion, we identified and confirmed three novel miRNAs that are predictive of dissemination in operable SCC and may constitute the components of multi miRNAs signatures. If corroborated in further studies, these findings may be relevant to clinical practice, as they could facilitate identification of SCC patients for adjuvant therapies.
Acknowledgments
We would like to thank Dr David Gandara for reviewing the manuscript and for valuable comments. The study was supported by the Polish Ministry of Science grant no. N N403 210139.
Jacek Jassem and Marcin Skrzypski disclose the option agreement with Novartis Pharma A.G. regarding the prognostic test in non-small cell lung cancer based on microRNA expression. The remaining authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Aviel-Ronen S, Blackhall FH, Shepherd FA, Tsao MS. K-ras mutations in non-small-cell lung carcinoma: a review. Clin Lung Cancer. 2006;8 (1:30–38. doi: 10.3816/CLC.2006.n.030. [DOI] [PubMed] [Google Scholar]
- Bepler G, Olaussen KA, Vataire AL, Soria JC, Zheng Z, Dunant A, Pignon JP, Schell MJ, Fouret P, Pirker R, Filipits M, Brambilla E. ERCC1 and RRM1 in the international adjuvant lung trial by automated quantitative in situ analysis. Am J Pathol. 2011;178 (1:69–78. doi: 10.1016/j.ajpath.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66 (15:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- Chee KG, Nguyen DV, Brown M, Gandara DR, Wun T, Lara PN. Positron emission tomography and improved survival in patients with lung cancer: the Will Rogers phenomenon revisited. Arch Intern Med. 2008;168 (14:1541–1549. doi: 10.1001/archinte.168.14.1541. [DOI] [PubMed] [Google Scholar]
- Friboulet L, Olaussen KA, Pignon JP, Shepherd FA, Tsao MS, Graziano S, Kratzke R, Douillard JY, Seymour L, Pirker R, Filipits M, André F, Solary E, Ponsonnailles F, Robin A, Stoclin A, Dorvault N, Commo F, Adam J, Vanhecke E, Saulnier P, Thomale J, Le Chevalier T, Dunant A, Rousseau V, Le Teuff G, Brambilla E, Soria JC. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. N Engl J Med. 2013;368 (12:1101–1110. doi: 10.1056/NEJMoa1214271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo N, Wan Y, Tosun K, Lin H, Msiska Z, Flynn D, Remick S, Vallyathan V, Dowlati A, Shi X, Castranova V, Beer D, Qian Y. Confirmation of gene expression-based prediction of survival in non-small cell lung cancer. Clin Cancer Res. 2008;14 (24:8213–8220. doi: 10.1158/1078-0432.CCR-08-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JS, Taylor J, Valentine HR, Irlam JJ, Eustace A, Hoskin PJ, Miller CJ, West CM. Enhanced stability of microRNA expression facilitates classification of FFPE tumour samples exhibiting near total mRNA degradation. Br J Cancer. 2012;107 (4:684–694. doi: 10.1038/bjc.2012.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huncharek M, Kupelnick B, Geschwind JF, Caubet JF. Prognostic significance of p53 mutations in non-small cell lung cancer: a meta-analysis of 829 cases from eight published studies. Cancer Lett. 2000;153 (1–2:219–226. doi: 10.1016/s0304-3835(00)00381-5. [DOI] [PubMed] [Google Scholar]
- Jin Z, Selaru FM, Cheng Y, Kan T, Agarwal R, Mori Y, Olaru AV, Yang J, David S, Hamilton JP, Abraham JM, Harmon J, Duncan M, Montgomery EA, Meltzer SJ. MicroRNA-192 and -215 are upregulated in human gastric cancer in vivo and suppress ALCAM expression in vitro. Oncogene. 2011;30 (13:1577–1585. doi: 10.1038/onc.2010.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung DE, Wen J, Oh T, Song SY. Differentially expressed microRNAs in pancreatic cancer stem cells. Pancreas. 2011;40 (8:1180–1187. doi: 10.1097/MPA.0b013e318221b33e. [DOI] [PubMed] [Google Scholar]
- Kratz JR, He J, Van Den Eeden SK, Zhu ZH, Gao W, Pham PT, Mulvihill MS, Ziaei F, Zhang H, Su B, Zhi X, Quesenberry CP, Habel LA, Deng Q, Wang Z, Zhou J, Li H, Huang MC, Yeh CC, Segal MR, Ray MR, Jones KD, Raz DJ, Xu Z, Jahan TM, Berryman D, He B, Mann MJ, Jablons DM. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet. 2012;379 (9818:823–832. doi: 10.1016/S0140-6736(11)61941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi M, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen A, Rubagotti M, Goldstein A, Linnoila I, Marincola F, Tucker M, Bertazzi P, Pesatori A, Caporaso N, McShane L, Wang E. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16 (2:430–441. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25 (4:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu Y, Govindan R, Wang L, Liu PY, Goodgame B, Wen W, Sezhiyan A, Pfeifer J, Li YF, Hua X, Wang Y, Yang P, You M. MicroRNA profiling and prediction of recurrence/relapse-free survival in stage I lung cancer. Carcinogenesis. 2012;33 (5:1046–1054. doi: 10.1093/carcin/bgs100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics REporting recommendations for tumour MARKer prognostic studies (REMARK) Br J Cancer. 2005;93 (4:387–391. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner TB, Li A, Biswas A, Lee KH, Liu YJ, Bayir E, Iliopoulos D, van den Elsen PJ, Kobayashi KS. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci USA. 2010;107 (31:13794–13799. doi: 10.1073/pnas.1008684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet. 2005;365 (9458:488–492. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- Network CGAR Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489 (7417:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepek JM, Chino JP, Marks LB, Damico TA, Yoo DS, Onaitis MW, Ready NE, Hubbs JL, Boyd J, Kelsey CR. How well does the new lung cancer staging system predict for local/regional recurrence after surgery?: a comparison of the TNM 6 and 7 systems. J Thorac Oncol. 2011;489 (7417:519–525. doi: 10.1097/JTO.0b013e31821038c0. [DOI] [PubMed] [Google Scholar]
- Pignon J, Tribodet H, Scagliotti G, Douillard J, Shepherd F, Stephens R, Dunant A, Torri V, Rosell R, Seymour L, Spiro S, Rolland E, Fossati R, Aubert D, Ding K, Waller D, Le Chevalier T, Group LC. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26 (21:3552–3559. doi: 10.1200/JCO.2007.13.9030. [DOI] [PubMed] [Google Scholar]
- Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, Beer D. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69 (14:5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- Raponi M, Zhang Y, Yu J, Chen G, Lee G, Taylor JM, Macdonald J, Thomas D, Moskaluk C, Wang Y, Beer DG. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66 (15:7466–7472. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- Schmittgen T, Livak K. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3 (6:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63 (1:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Skrzypski M, Dziadziuszko R, Jassem E, Szymanowska-Narloch A, Gulida G, Rzepko R, Biernat W, Taron M, Jelitto-Górska M, Marjański T, Rzyman W, Rosell R, Jassem J. Main histologic types of non-small-cell lung cancer differ in expression of prognosis-related genes. Clin Lung Cancer. 2013;14 (6:666–673.e2. doi: 10.1016/j.cllc.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Sobin LH, Wittekind C.2002TNM Classification of Malignant Tumours6th edn.John Wiley & Sons: Hoboken, NJ, USA [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64 (11:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Müller-Hermelink HK, Harris CC.(eds) (2004World Health Organization Classification of Tumors. Pathology and Genetics. Tumours of the Lung, Pleura, Thymus and Heart IARC Press: Lyon, France [Google Scholar]
- Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ, Van Schil PE, Garg K, Austin JH, Asamura H, Rusch VW, Hirsch FR, Scagliotti G, Mitsudomi T, Huber RM, Ishikawa Y, Jett J, Sanchez-Cespedes M, Sculier JP, Takahashi T, Tsuboi M, Vansteenkiste J, Wistuba I, Yang PC, Aberle D, Brambilla C, Flieder D, Franklin W, Gazdar A, Gould M, Hasleton P, Henderson D, Johnson B, Johnson D, Kerr K, Kuriyama K, Lee JS, Miller VA, Petersen I, Roggli V, Rosell R, Saijo N, Thunnissen E, Tsao M, Yankelewitz D. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6 (2:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara N, Matsuoka Y, Tsubura A. Mesothelin promotes anchorage-independent growth and prevents anoikis via extracellular signal-regulated kinase signaling pathway in human breast cancer cells. Mol Cancer Res. 2008;6 (2:186–193. doi: 10.1158/1541-7786.MCR-07-0254. [DOI] [PubMed] [Google Scholar]
- van Vliet MH, Reyal F, Horlings HM, van de Vijver MJ, Reinders MJ, Wessels LF. Pooling breast cancer datasets has a synergetic effect on classification performance and improves signature stability. BMC Genomics. 2008;9:375. doi: 10.1186/1471-2164-9-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth A, Muley T, Meister M, Stenzinger A, Thomas M, Schirmacher P, Schnabel PA, Budczies J, Hoffmann H, Weichert W. The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage-independent predictor of survival. J Clin Oncol. 2012;30 (13:1438–1446. doi: 10.1200/JCO.2011.37.2185. [DOI] [PubMed] [Google Scholar]
- Wei JS, Johansson P, Chen QR, Song YK, Durinck S, Wen X, Cheuk AT, Smith MA, Houghton P, Morton C, Khan J. microRNA profiling identifies cancer-specific and prognostic signatures in pediatric malignancies. Clin Cancer Res. 2009;15 (17:5560–5568. doi: 10.1158/1078-0432.CCR-08-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson MD, Yin X, Hoadley KA, Liu Y, Hayward MC, Cabanski CR, Muldrew K, Miller CR, Randell SH, Socinski MA, Parsons AM, Funkhouser WK, Lee CB, Roberts PJ, Thorne L, Bernard PS, Perou CM, Hayes DN. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res. 2010;16 (19:4864–4875. doi: 10.1158/1078-0432.CCR-10-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winton T, Livingston R, Johnson D, Rigas J, Johnston M, Butts C, Cormier Y, Goss G, Inculet R, Vallieres E, Fry W, Bethune D, Ayoub J, Ding K, Seymour L, Graham B, Tsao MS, Gandara D, Kesler K, Demmy T, Shepherd F, National Cancer Institute of Canada Clinical Trials Group, National Cancer Institute of the United States Intergroup JBR.10 Trial Investigators Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352 (25:2589–2597. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- Wu JM, Fackler MJ, Halushka MK, Molavi DW, Taylor ME, Teo WW, Griffin C, Fetting J, Davidson NE, De Marzo AM, Hicks JL, Chitale D, Ladanyi M, Sukumar S, Argani P. Heterogeneity of breast cancer metastases: comparison of therapeutic target expression and promoter methylation between primary tumors and their multifocal metastases. Clin Cancer Res. 2008;14 (7:1938–1946. doi: 10.1158/1078-0432.CCR-07-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Xiao G, Coombes KR, Behrens C, Solis LM, Raso G, Girard L, Erickson HS, Roth J, Heymach JV, Moran C, Danenberg K, Minna JD, Wistuba II. Robust gene expression signature from formalin-fixed paraffin-embedded samples predicts prognosis of non-small-cell lung cancer patients. Clin Cancer Res. 2011;17 (17:5705–5714. doi: 10.1158/1078-0432.CCR-11-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Cid E, Bru S, Yamamoto F. Rare and frequent promoter methylation, respectively, of TSHZ2 and 3 genes that are both downregulated in expression in breast and prostate cancers. PLoS One. 2011;6 (3:e17149. doi: 10.1371/journal.pone.0017149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying SY, Lin SL. Intron-mediated RNA interference and microRNA biogenesis. Methods Mol Biol. 2009;487:387–413. doi: 10.1007/978-1-60327-547-7_19. [DOI] [PubMed] [Google Scholar]
- Yu S, Chen H, Chang G, Chen C, Chen H, Singh S, Cheng C, Yu C, Lee Y, Chen H, Su T, Chiang C, Li H, Hong Q, Su H, Chen C, Chen W, Liu C, Chan W, Li K, Chen J, Yang P. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13 (1:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Zhu C, Ding K, Strumpf D, Weir B, Meyerson M, Pennell N, Thomas R, Naoki K, Ladd-Acosta C, Liu N, Pintilie M, Der S, Seymour L, Jurisica I, Shepherd F, Tsao M. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28 (29:4417–4424. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.