Abstract
Background:
The amplification of epidermal growth factor receptor (EGFR) in triple negative breast carcinomas (TNBC) suggests its potential therapeutic application, as for HER-2, using standardised methods of measurement. In this regard, we aimed to compare several methods for evaluating EGFR amplification along with potential mutations for suitability in clinical practice.
Methods:
Tissue sections of 138 TNBCs were used (1) to compare EGFR amplification and expression by silver in situ hybridisation (SISH) to qPCR and immunohistochemistry (IHC) and (2) to search for EGFR mutations, along with Kras, PI3K, Braf and HER-2 mutations and echinoderm microtubule associated protein like 4-anaplastic lymphoma kinase (EML4-ALK) translocation.
Results:
(1) Amplification of EGFR was observed in well-characterised TNBCs (up to 92%); (2) qPCR correlated with SISH with 94% specificity and 75.6% sensitivity; (3) IHC correlated with SISH with 97% sensitivity and 78% specificity; (4) no EGFR, Kras mutations or EML4-ALK translocations were found, but PI3K and Braf mutations were observed in 26% of cases; and (5) small, acentric circular extrachromosomal DNA similar to ‘double minutes' in glioblastomas was observed in 18% of SISH sections.
Conclusions:
SISH and IHC are methods that are suitable in clinical practice to screen for EGFR amplification and overexpression, which are frequently observed in TNBC. Patients with TNBC are potential candidates for EGFR-targeted therapy combined with PI3K and Braf inhibitors.
Keywords: triple negative breast cancer, EGFR amplification mutation, PI3K, Braf
Triple negative breast carcinomas (TNBC), defined as tumours lacking oestrogen and progesterone receptors and HER-2 gene amplification, account for 10–20% of all breast carcinomas in Asian and Western populations (Thike et al, 2010; De Ruijter et al, 2011). TNBC are usually of high histoprognostic grade and are associated with distinctive, metastatic patterns, particularly with high rates of brain metastases, shorter time-to-recurrence and earlier mortality (Thike et al, 2010). TNBC also commonly express basal markers such as cytokeratins 5/6 and epidermal growth factor receptor (EGFR) (Nielsen et al, 2004; Charpin et al, 2009a; El Guerrab et al, 2011), and are therefore generally accepted as clinical surrogates for basal-like breast cancers (BLBC), although not all basal-like breast carcinomas are triple negative (Corkery et al, 2009). The TNBC phenotype is also acknowledged as an aggressive breast carcinoma molecular subtype with an early age of cancer onset and a high probability of metastases at presentation. Sustained, complete remission in TNBC is rare and additional treatments directed at appropriate molecular targets, such as poly (ADP-ribose) polymerase inhibitors (Rios and Puhalla, 2011), are urgently needed.
Likewise, patients with EGFR-amplified and EGFR-over-expressing TNBC (30–52%) and BLBC (60%) (Reis-Filho, Tutt, 2008) should benefit from EGFR-targeted therapy, as well as from anti-HER-2 therapies. Ongoing clinical trials are exploring the roles of monoclonal antibodies against EGFR, such as cetuximab in TNBC. However, the use of anti-EGFR drugs in TNBC management is still controversial and only a few preliminary studies have been reported to date. The failure of EGFR-targeted therapy in TNBC trials is likely due to an insufficient knowledge of the various types of molecular dysfunction as a result of abnormal EGFR mediation and from inadequate selection of patients for this specific treatment (O'Shaughnessy et al, 2011; Carey et al, 2012). Particularly, because of discrepant data in literature reports, as for HER-2 targeted treatments, reliable and standardised methods of the measurement of EGFR amplification and overexpression that are suitable for the current clinical and pathological practices are also required to properly identify those patients with TNBC, EGFR amplification and EGFR overexpression (Nakajima et al, 2012). Although a recent study (Nakajima et al, 2012) has documented comparative methods (chromogen ISH and immunohistochemistry (IHC) procedures) for EGFR evaluation within TNBC, no extensive study that has focused on this particular point has been reported for TNBC.
In these regards, we (i) first correlated different procedures, including IHC, silver in situ-hybridisation (SISH), and qPCR, to evaluate EGFR overexpression and EGFR amplification in TNBC (as extensively documented for HER-2 in the literature), and (ii) searched for EGFR mutations in TNBC, which are well acknowledged in non-small cell lung carcinoma (NSCLC) (Lynch et al, 2004; Paez et al, 2004). In TNBC, EGFR mutations rarely occur (Bhargava et al, 2005; Reis-Filho et al, 2006; Carey et al, 2010; Jacot et al, 2011) and often account for silent mutations (Generali et al, 2007). However, this point is controversial in TNBC. Most studies have shown that no EGFR-activating mutations occur in TNBC (Nakajima et al, 2012), but one series of 653 Asian patients with TNBC was recently reported to have EGFR mutations in 11.4% of cases (70 out of 653) that were independent of EGFR expression (Teng et al, 2011). This particular point is crucial for a patient's response to anti-EGFR therapies. Our study was also designed to evaluate EGFR mutations in TNBC of non-Asian patients.
In TNBC, such as in NSCLC and colorectal carcinomas (Siena et al, 2009), EGFR deregulation and mutations of downstream pathways, in particular Kras, Braf, PI3K, PTEN, or ALK, have been recently reported (Martin et al, 2012) and might be responsible for the impaired efficacy of EGFR-targeted drugs. Therefore, we studied mutations in the signalling pathways downstream of EGFR, particularly of PI3K, Kras and Braf, in parallel to mutations involved in response to a specific therapy, such as the ALK translocation that is currently observed in NSCLC but is exceptionally found in breast carcinomas (BC) (Lin et al, 2009).
Materials and Methods
Materials
Our series included 161 cases that were distributed into two different tumour sets. The first set (n=47) consisted of current, large sections. The second set (n=114) consisted of tissue microarray (TMA) sections from 915 consecutive, ductal breast carcinomas that were distributed individually as cores (four cores per case) measuring 0.6 mm in diameter.
This study was designed according to the current recommendations of the French National Committee of Cancer INCa (Institut National du Cancer) and the 15189 ISO European recommendations for legal archival procedures in the Pathology Departments and Certification. According to the French law, approval from an ethical committee was not required for our study. IHC and in situ hybridisation tissue arrays were performed from archived, paraffin-embedded and formalin-fixed tissue samples that remained in blocks after current diagnosis in pathology labs. Informed consents from patients who were included in the study for IHC, in situ hybridisation, and analysis of gene alterations were systematically obtained before surgical intervention and tissue sampling.
Tumour sample formalin fixation was controlled (24 h for biopsies and lumpectomy less than 3 cm; and 48 h for larger surgical specimens at room temperature) along with paraffin embedding with a constant control, melted-paraffin temperature (60 °C). Sections were obtained using automated devices calibrated to obtain four micron-thick tissue sections 24 h before immunodetection processing.
Current large sections
Large current sections were also evaluated because they included more tissue that was suitable for PCR and mutation, which was in contrast to the small TMA cores of 0.6 mm in diameter. The tumours that were selected (n=47) for the study included the following: (i) TNBC (n=24 out of 47) without any estrogene receptors (ER)- or progesterone receptors (PR)-positive immune expression and without HER-2 amplification, which was evidenced by a negative SISH test, or (ii) ‘TNBC-like' (n=23 out of 47), which had similar unfavourable histoprognostic criteria and were HER-2-negative (SISH test) but expressed low ER levels (<20%) in positive cells and negative PR levels (n=23 out of 47). This ‘TNBC-like' group was included in the study to show that depending on the cut-off of the percentage of ER-positive cells (0 to 20%), the rate of EGFR amplification in TNBC could considerably vary, as shown by the variability in the literature data. The tumour pTN stages were pT1 and pT2 (mean size 18 mm), and node-positive carcinomas accounted for 38% at the time of diagnosis (n=14 out of 38).
TMA sections
TMA sections were constructed as previously described (Charpin et al, 2009a, 2009b) and included 915 breast carcinomas of any type that were collected from 2000 to 2005. The samples included 114 TNBC that were completely ER- or PR-negative and lacked HER-2 amplification (SISH test). Tumours were not otherwise specified ductal carcinomas.
Tumours measuring from 7 mm to 53 mm (pT1: n=398; pT2: n=488; pT3: n=29) and exhibiting the usual distribution of Ellis and Elston grade (grade 1: 21% grade 2: 61% grade 3: 17%) were included, and 667 were node negative.
Methods
IHC
Automated procedures were assessed (Charpin et al, 2009a, 2009b) on a Ventana Benchmark XT device, using E-view detection Ventana kits for DAB staining and the following specific monoclonal mouse or rabbit antibodies: monoclonal anti-savage EGFR clone 3C6 (Ventana Roche, Meylan Grenoble, France); clone B6 for detection of the E746- A750 exon 19 deletion of the EGFR gene (Cell Signaling Technology, St. Quentin, France); clone 43B2 for L858R, for the detection of mutated EGFR exon 21 (Cell Signaling Technology); anti-ALK clone 5A4 (Abcam, Paris, France) for the detection of the fusion transcript echinoderm microtubule associated protein like 4-anaplastic lymphoma kinase (EML4-ALK); clone SP1 anti-ER (Ventana Roche); clone 1E2 anti PR (Ventana Roche); and clone 4B5 anti-HER-2 (Ventana Roche). The slides were counterstained with hematoxylin and bluing reagent.
The positive controls for mutated or amplified non-EGFR consisted of paraffin sections of breast and qPCR-amplified colonic carcinomas, whereas the controls for mutated EGFR and translocated ALK consisted of paraffin sections of mutated EGFR (exon 19 deletion and exon 21 mutation) or FISH EML4-ALK-translocated lung carcinomas.
In situ hybridisation (ISH)
The ISH procedures included silver ISH (SISH Ventana Roche, INFORM) for the detection of the EGFR gene and chromosome seven amplification and fluorescent ISH for ALK translocation (DAKO Histology FISH accessory kit, ABBOTT DNA ALK probe, Trappes, France). The positive controls for savage EGFR consisted of the EGFR-SISH xenograft control slides that were provided with the Ventana Roche kits. Normal labelling of stromal cells with two EGFR silver dots in normal cells also served as a positive control for tissue quality control for EGFR and chromosome seven. In some cases, a polysomia was identified that contained more than 2N chromosomes in diploid, normal cells and when more than two spots were observed for chromosome seven within tumour cells. The mean number of EGFR spots, which reflected the number of EGFR copies was evaluated in 100 tumour cells (HPF x 60 Zeiss Axiophot). Amplified tumours were those with a ratio of the mean number of EGFR copies or silver dots versus the mean number of chromosome 7⩾2.
In addition, a qualitative pattern of silver spot distribution within EGFR-amplified tumour cells was recorded as follows: (i) dense clusters of silver dots within the nucleus, (ii) loose nuclear distribution of variable amounts of distinct, silver, small or tiny dots and (iii) a straight or curved linear arrangement of silver spots within the cytoplasm but close to the nuclear membrane that produced the ‘double minute' pattern that was observed in glioblastomas (Vogt et al, 2004; Gibaud et al, 2010).
Analysis of gene alterations
DNA extraction: All analyses were performed on formalin-fixed, paraffin-embedded blocks. The first stage consisted in dewaxing by Microclearing (DiaPath, Martingo, Italy) followed by slow-tissue rehydration in successive ethanol soakings from 100 to 70%. The tumour area was then scraped with a lancet blade and collected in tubes containing 40 μl of a mixture of Pronase (Roche, Mannheim, Germany), Tris Plug, SDS and CaCl2. The tubes were then incubated with endo and exopeptidases overnight at 55 °C. DNA was purified by automated extraction on EVO75 (Tecan, Mannedorf, Switzerland) according to the M&N protocol NR (NucleoSpin 96 Blood; NucleoSpin 8 Viruses Binding Strips). Elution was carried out in bovine serum albumin at 500 μg ml−1, and the DNA was stored at −20 °C.
High resolution melting analysis and DNA sequencing: Our laboratory routinely uses qPCR-HRM as a pre-screening tool, which prevents useless sequencing of wild-type DNA. The choice of primers, concentrations, temperature and time of reaction were adapted to each set according to MIQE recommendations (Bustin et al, 2009). The negative predictive value of qPCR-HRM was validated on 300 samples for each set of primers/exon. The analytical chain was accredited according to the Iso-Norm 15 189 (Guzel and Guner, 2009; Yanikkaya-Demirel, 2009), the sample and mix (HRM Master Mix, Roche) distribution were automated and performed in 96-well plates (Evo75, Tecan), and reactions were performed in duplicate. Mutated reference samples (cell lines), wild-type DNA (placenta), dewaxing, extraction, and PCR negative controls were included with the samples to be analysed.
The PCR was then carried out on a LightCycler 480 (Roche) and the samples dissociation profiles were compared with those of reference samples. The samples were then classified as ‘wild type' or ‘not wild type' using the gene-scanning software (Roche). The exponential phase of qPCR-HRM allowed for the quantification of the initial DNA and detection of the presence of PCR inhibitors. Only the samples defined as ‘not wild type' by qPCR-HRM were sequenced by the Sanger method (Sanger et al, 1977) (MixBigDye 5X, Roche) after DNA purification (ExoSap-IT) and automated on an Evo75, (Tecan). The sequences were analysed using a 3500 or 3130 Dx Genetic Analyser (Applied Biosystems, St. Aubin, France). The search for mutations in exon 9 of the gene PI3KCA was performed after removal of a pseudogene that was localised on chromosome 22 and was similar to exons/introns 9–13.
Quantitative qPCR: qPCR was developed following the MIQE recommendations for RT–PCR dosage (Bustin et al, 2009). The copy number was normalised to a gene located in the same chromosome, beta-actin (ACTB, 7p22.1), and a gene located on another chromosome (glyceraldehyde-3-phosphate dehydrogenase or GAPDH, 1 2p13.31). This strategy has been used in previous studies (Lamy et al, 2006; Lamy et al, 2011) and allowed us to distinguish real changes in the copy number variation from aneusomy. The qPCR reaction conditions were adapted to each primer-probe set. Each well contained 18 μl of mix prepared with the Mix 2X LightCycler 480 Honest Master, 10 μM forward and reverse primers, 2.5 μM fluorescent probe and sterile water. Two microliters of sample DNA were added to each well in triplicate.
The PCR plate also contained a genomic DNA standard scale (Roche) containing 12 000, 1200, 120 and 12 gene copies. The standard scale allowed for the conversion of the results obtained in Cq (quantitative cycle) into amplicon copy numbers for each gene by integrating the efficiency of the reaction to each reaction. One DNA that was known to have an amplified EGFR gene was used as a positive control (EGFR/ACTB=12.67±2.31) and human placental DNA was used as a normal control (EGFR/ACTB=0.55±0.06). The theoretical decision threshold of gene amplification for a sample was EGFR/beta-actin (n=2). Below this threshold, the sample was considered ‘wild type,' and above this threshold, the sample was considered ‘amplified.'
Statistical analysis
The associations between qualitative variables was assessed using a χ2-test (or a Fisher exact test when appropriate). A two-sided value of P<0.05 was considered statistically significant.
Results
EGFR overexpression and amplification in TMA (cores of TNBC)
IHC
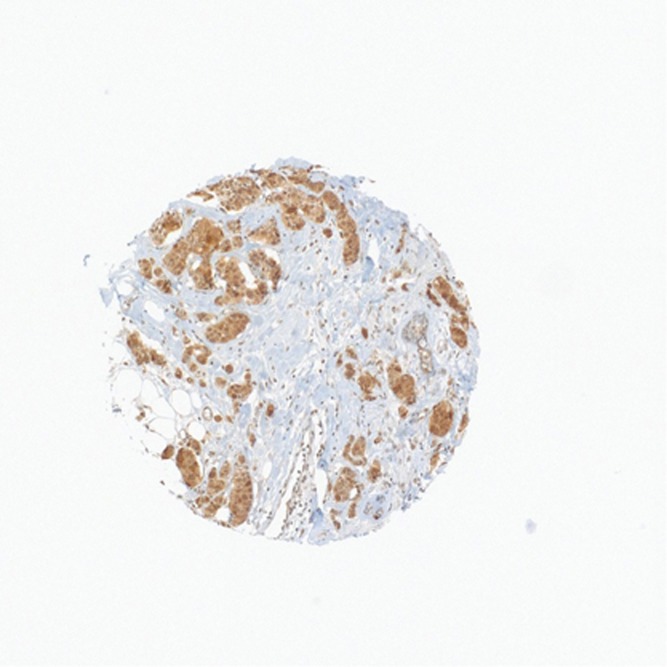
By TMA, 159 out of 915 sporadic breast carcinomas were EGFR-positive (17%) and included a majority of TNBC (n=97) and non-TNBC (n=62). With regards to TNBC (n=114 out of 915), which is strictly defined by a lack of HER-2 amplification (ISH or qPCR) and completely negative ER and PR immunoexpression, 97 out of 114 were anti-EGFR-positive (85%) (Figure 1). In contrast, EGFR-positive, non-TNBC (n=62 out of 801; 6.4%) accounted for 19 TNBC-like (19 out of 170; 11%) and 45 ‘not otherwise specified,' high-grade ductal carcinomas (45 out of 277; 13%).
Figure 1.
Positive anti-EGFR immunohistochemical reaction in TMA spot of ductal triple negative breast carcinoma.
In situ hybridisation
Among the 114 TNBC of the 159 cases, 92% (105 out of 114) were positive for EGFR-SISH, which was similar to the ratio (87%) that was observed in large sections (Table 1).
Table 1. EGFR amplification evaluated by SISH, IHC and QPCR.
| Tumour type | No. of cases | P-value |
|---|---|---|
| Large sections: | ||
| TNBC | 24 | |
| TNBC-like | 23 | |
| Total | 47 | |
| TMA: TNBC |
114 |
|
|
IHC | ||
| Large sections: | ||
| TNBC | 23 (96%) | <0.0001 |
| TNBC-like | 9 (39%) | |
| Total | 32 (67%) | |
| TMA: TNBC |
97 (85%) |
|
|
SISH | ||
| Large sections: | ||
| TNBC | 21 (87%) | <0.0005 |
| TNBC-like | 8 (33%) | |
| Total | 29 (62%) | |
| TMA: TNBC |
105 (92%) |
|
|
QPCR | ||
| Large sections: | ||
| TNBC | 21 (91%) | <0.0103 |
| TNBC-like | 8 (29%) | |
| Total | 29 (64%) | |
| TMA: TNBC |
NT |
|
|
Minute | ||
| Large sections: | ||
| TNBC | 4 (18%) | |
| TNBC-like | 0 | |
| Total | 4 (18%) | |
| TMA: TNBC | 16 (14%) | |
Abbreviations: IHC=immunohistochemistry; SISH=silver in situ hybridisation; TMA=tissue microarray; TNBC=triple negative breast carcinomas. All data are presented as n (%).
EGFR gene amplification/qPCR
The evaluation of EGFR gene amplification could not be assessed by TMA for the cores of the 114 TNBC that were included in our 915 series of sporadic breast carcinomas. The small sizes of the tumour cores measuring 0.6 mm in diameter (and 4 μm-thick sections) were not appropriate for qPCR.
Correlation between SISH/qPCR/IHC in large sections
In current clinical practice, diagnostic ISH is a cost-effective procedure that is presently recommended for HER-2 amplification evaluation and requires a small amount of formalin paraffin-embedded tissue that is appropriate to the expertise of the pathologist. We therefore correlated the SISH to qPCR and IHC data.
qPCR versus SISH: The levels of amplification in large sections varied from 2 to 17 copies. In two tumours, the low density of the tumour cells was not sufficient for appropriate measurements. Amplified tumours accounted for 29 out of 45 (64%), and the mean amplification level was 5.6 (0th: 2.0; 25th: 3.1; 50th: 4.7; 75th: 6.2; and 100th: 17). Ninety-one per cent of (21 out of 23) TNBC had an increased number of copies of the EGFR gene, which was significantly (P<0.010) more than 29% that was observed for TNBC-like (8 out of 22) (Table 1).
Amplification through SISH was observed in 29 out of 47 (62%) tumours, whereas 18 out of 47 were negative. TNBC (21 out of 24=87%) were significantly more SISH-positive than TNBC-like tumours (8 out of 23=29%) (P<0.0005) (Table 1). A significant correlation was observed between the SISH and qPCR data (P<0.0001). Among the 29 out of 45 EGFR-SISH-positive cases, 23 were also qPCR amplified and six were non-amplified. Among the 17 out of 45 EGFR-SISH-negative, 16 were also qPCR non-amplified, resulting in 75.6% sensitivity and 94% specificity for qPCR when compared with ISH. All qPCR-positive samples were IHC- and SISH-positive.
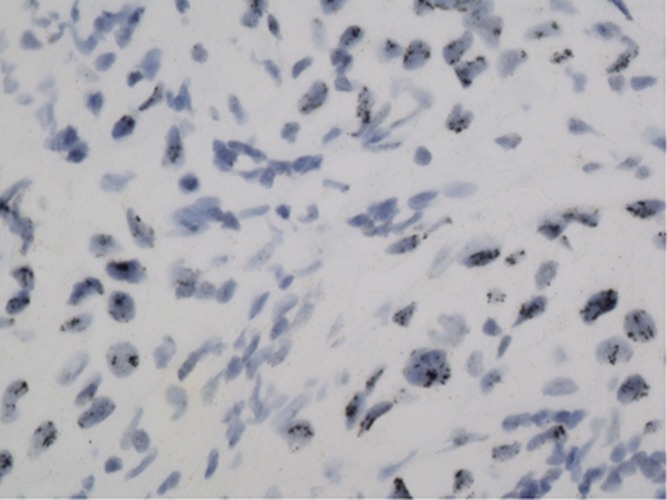
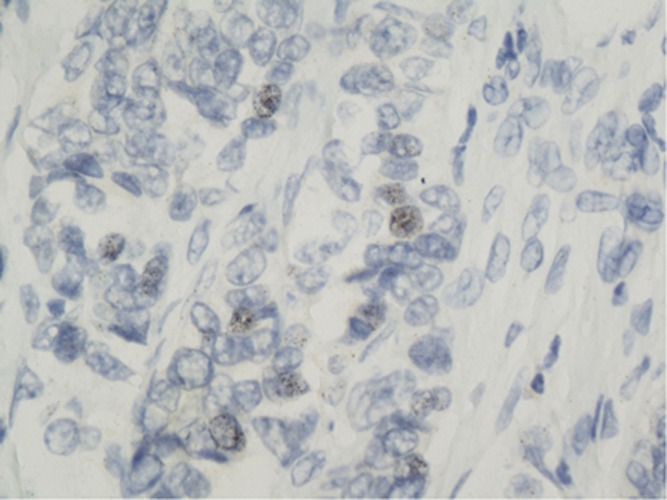
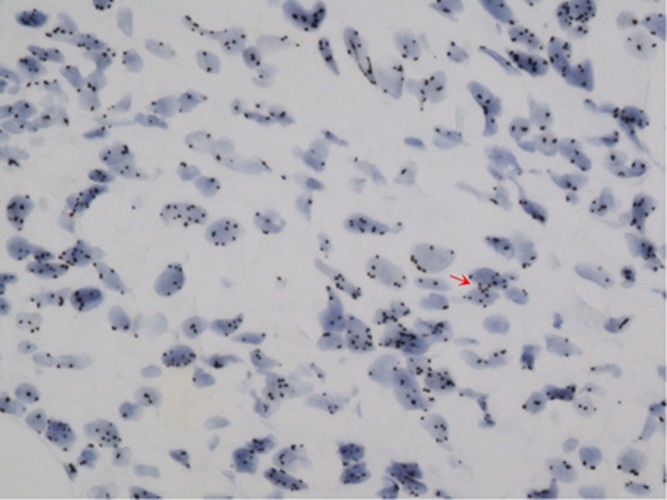
In large sections, SISH was positive in nine cases that had strong reactions of black dot clustering within the cell nucleus (Figure 2), and in 13 cases several dots were loosely distributed throughout the nucleus (Figures 3 and 4). In 4 out of 24 (18%) of the cases, black dots were distributed as short lines or curves in the cell cytoplasm near the nuclear membrane (Figure 5), which produced a ‘double minute' pattern, as described in brain cancer gliomas (Vogt et al, 2004; Gibaud et al, 2010). The DNA molecules (DMs) were intrachromosomal.
Figure 2.
Positive in situ hybridisation with black dot clustering within the cell nucleus.
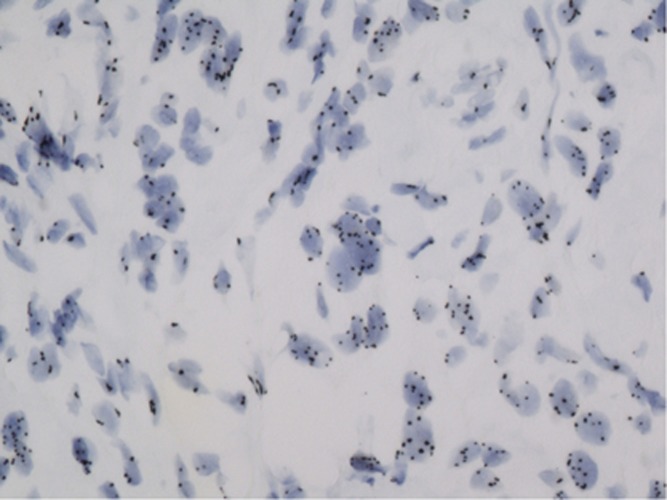
Figure 3.
Positive in situ hybridisation appearing as separated tiny dots distributed in the nucleus of many cells.
Figure 4.
Positive in situ hybridisation within some cells in a triple negative breast carcinoma contrasting with the clustering distribution observed in Figure 2.
Figure 5.
In situ hybridisation showing the double minutes chromosome (arrow) appearing as arciform pattern of positive dots within the cytoplasm close to nuclear membrane.
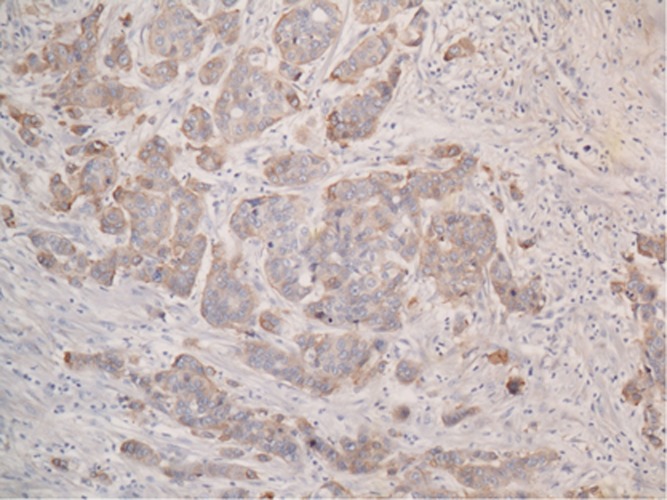
IHC versus SISH: EGFR immunoexpression in the entire series of TNBC and TNBC-like large-tissue sections was observed in 67% (32 out of 47) of the tumours that were evaluated, and the samples had a significantly (P<0.0001) greater (23 out of 24) EGFR-positive subset in TNBC when compared with TNBC-like that accounted for 9 out of 23 tumours (Table 1). Positive staining (Figure 6) was distributed throughout the cell cytoplasm with enhancements in the cell membrane, but were not found within the nuclei.
Figure 6.
EGFR immunohistochemical positive expression in current large-tissue sections of triple negative breast carcinoma.
Among the 29 out of 47 large-sections, EGFR-SISH-positive cases, 28 were also IHC-positive and one was IHC-negative. Furthermore, among the 18 out of 47 negative cases, 14 were also IHC-negative, which resulted in 78% specificity and 97% sensitivity of the IHC compared with SISH. In addition, among the 114 TNBC, 85% (97 out of 114) were EGFR-IHC-positive, and 92% (105 out of 114) were EGFR-SISH-positive by TMA. High correlations between both tests were observed, with 6 out of 114 being IHC-negative and SISH-positive, and 7 out of 114 being IHC-positive and SISH-negative.
Mutations in large sections
EGFR mutations
None of the 47 large tumour sections in the series exhibited an EGFR mutation within exons 18–21. Likewise, the IHC tests for the detection of exon 19 deletions and exon 21 missense mutations were negative in all the 47 cases. In addition, no positive IHC reaction using antibodies for the detection of exon 19 deletion and exon 21 mutation was observed in the 915 tumours that were included in the TMAs.
EGFR downstream pathway mutations
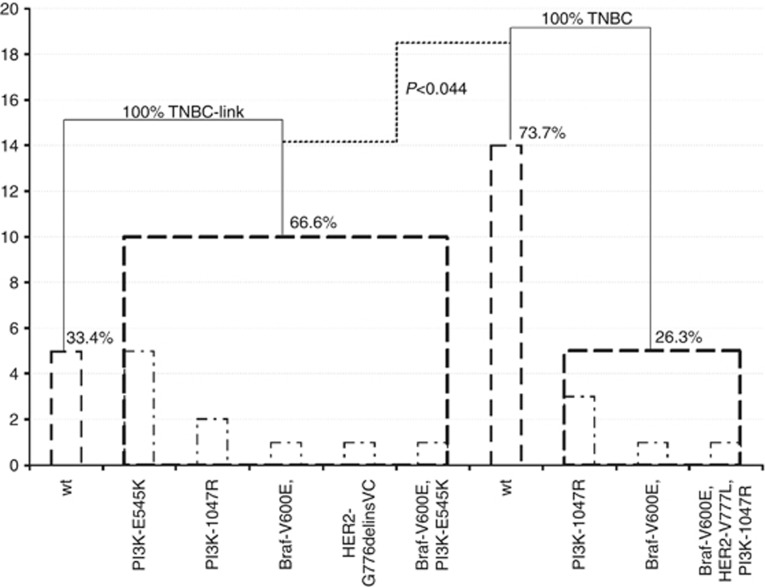
Among the 34 patients who were tested for mutations in the EGFR downstream pathway, 44% (15 out of 34) were mutated and no Kras mutation was observed. However, PI3K mutations were observed in 35% (12 out of 34) of the tumours and an equivalent proportion of mutants were found in exons 9 and 20 regardless of the gene (6 out of 6). Braf mutations were observed in 13.8% (4 out of 29) of cases and HER-2 mutations were detected in 5.9% cases (2 out of 34). One patient had concomitant mutations in BRAF and PI3K, and another one had mutations for BRAF, PI3K and HER-2. No significant differences were observed for gene or mutation type between TNBC-like or TNBC (Figure 7).
Figure 7.
PI3K mutations were observed in 35.3% (12/34) tumours. Overall exons 9 and 20, 10 PI3K mutations are exclusive of all other mutations (5/exon 9 and 5/exon 20), seven are found in TNBC-like with E545K mutation. H1047R mutations are distributed between both the tumour profiles. Braf mutations were observed in 13.8% (4/29) cases, two are exclusive of all other mutations and are distributed between both tumour profiles. HER-2 mutations were detected in 5.9% cases (2/34), one is exclusive of all other mutations and is found in TNBC-like. One classified TNBC-like patient has concomitant mutations for Braf and PI3K and another one for Braf, PI3K and HER-2. In total 66.6% (10/15) of TNBC-like are significantly (P<0.044) mutated compared with 26.3% (5/19) of the TNBC. No EGFR and Kras mutation was detected. wt: Wild type; Association between qualitative variables was assessed with Fisher's exact test.
ALK translocation
None of the 47 TNBC large sections or any of the 114 TNBC by TMA were ALK-positive using the IHC anti-translocated ALK, which was in contrast to the positive control, NSCLC.
Discussion
TNBC accounts for 10–20% of all BCs (Thike et al, 2010; De Ruijter et al, 2011; Teng et al, 2011). Accordingly, in our TMA breast cancer set that was tested for EGFR-IHC overexpression and EGFR-SISH amplification, TNBC accounted for 114 cases (12.5%). Among these 114 TNBC, 105 out of 114 (92%) were found to be EGFR-amplified by SISH and 21 out of 24 (87%) of the large sections were found to be amplified. Our results significantly differed from the literature data in which EGFR amplification was found in 30–52% (Nielsen et al, 2004; Corkery et al, 2009; Gumuskaya et al, 2010; Lamy et al, 2011) of TNBC and in 56–84% of basal-cell breast carcinomas (Da Silva et al, 2007).
The discrepancies likely result from variations in the criteria that were used for defining TNBC. In most studies, a different cut-off for ER receptor positivity is used. However, in our study, we used strict diagnosis criteria to define TNBC, and only TN tumours that were completely negative for ER and PR (i.e., with no positive cells) were considered TNBC and were included. Furthermore, the fact that only 29% (n=8 out of 23) of the TN-like tumours in our large sections set were EGFR-amplified (versus 92% of properly selected TNBC with 0% of ER-positive cells), does support variations in EGFR amplification rates according to the cut-off of ER positivity. Finally, as recommended in the guidelines for ISH that were used for interpreting SISH or CISH for HER-2-negative tumours, only tumours with a ratio between the number of spots (copies) and the number of chromosome seven of less than two, were considered as non-amplified, discarding possible false-positive cases due to polysomia in tumours that were only evaluated by IHC (Gumuskaya et al, 2010).
Using such strict criteria, we obtained an excellent correlation between the positive IHC staining and SISH in TMA and large sections. As expected, this result suggests that, as was currently performed for HER-2, both methods could be combined for EGFR status evaluation in clinical practice. Likewise, the sensitivity and cost efficiency of IHC could serve as the initial tumour screening, and the specificity of SISH could serve to exactly evaluate the degree of amplification and discard polysomia.
Interestingly, we also obtained good correlation in large sections between SISH and qPCR that definitively validated our results and the high frequency of EGFR amplification and overexpression in TNBC. The results also indicated that ISH could be considered a cost-effective procedure for EGFR amplification evaluation, as was recently assessed for HER-2. Bhargava et al, reported similar data in a series of 175 TNBC (Bhargava et al, 2005). As already acknowledged, discrepant data for HER-2 ISH, which was reported in the literature concerning rates of EGFR-amplified carcinomas was likely a result of the lack of standardisation and variations of ISH methods, such as the use of various kits, procedures and cut-offs (Reis-Filho et al, 2006, Ryden et al, 2010). However, our results deserve more studies involving additional series.
In clinical practice, ISH, as used for HER-2 status evaluation, is endowed with significant advantages over qPCR because it can be more rapidly assessed within diagnostic time concomitantly to hormone receptor evaluation in consecutive sections of the tissue; thus, it can be perfectly controlled by the pathologist, who can exclude inappropriate tissue areas such as necrosis, insufficient density of tumour cells and fixation defects. In this regard, it is also noteworthy that all of the tests were performed using paraffin sections of archived blocks that were stored in the libraries of pathology laboratories at room temperature after diagnosis, and are much easier to control and handle than frozen samples.
The clonal and mutational spectrum of primary TNBC has recently been reported (Shah et al, 2012). In our study on TNBC, we did not observe, as others have (Lynch et al, 2004, Bhargava et al, 2005), the EGFR mutations in NSCLC in large sections or in TMA using anti-43B2 or anti-6B6 monoclonal antibodies. However, a recent report (Teng et al, 2011) has shown that 11.4% (70 out of 653) of exon 19 deletion or missense exon 21 mutations affected Asian populations more than Caucasian populations in a series of 653 TNBC (Jacot et al, 2011; Teng et al, 2011). Most literature reports, such as ours, suggest that Caucasian patients with TNBC who lack EGFR mutations but have an EGFR amplification accompanied by EGFR overexpression, could potentially benefit from anti-EGFR treatment (such as cetuximab), as reported in some preliminary clinical trials (O'Shaughnessy et al, 2011; Carey et al, 2012). Nevertheless, further studies are required, particularly those including an accurate selection of patients with true TNBC and EGFR amplification that properly predict the potential response of this disease to EGFR-targeted therapy.
Anti-EGFR therapy of patients with EGFR-amplified glioblastomas has been reported to not significantly improve patient survival (Vogt et al, 2004, Gibaud et al, 2010). In addition, EGFR-amplified glioblastomas, in contrast to TNBC, do not often concomitantly overexpress the EGFR protein, which could explain the lack of an anti-EGFR response when using antibody therapies. Furthermore, those drugs fail to reach their target in the brain (Vogt et al, 2010).
Mutations in downstream EGFR signalling pathways have been reported to be responsible for the resistance of TNBC to EGFR therapies (Martin et al, 2012). We show that 44% of TNBC were mutated for PI3K, Braf, or Her-2. PI3K mutations have been reported in 25% of breast carcinomas (Martin et al, 2012), and we found that 35% of PI3K mutations could also explain the failure of patient response to EGFR-targeted therapy. The effects of EGFR activation of the MAP kinase and PI3Kinase (AKT-mTOR) signalling pathways can be reduced by inhibitors of these molecules, particularly rapamycin, which is an inhibitor of mTOR. Several ongoing trials are investigating patient's responses to mTOR inhibitors (CCI-779, RAD001, AP 235732) (Macaskill et al, 2011). In addition, promising preclinical studies concerning breast cancer cell lines and animal xenografts have recently been reported (Brachmann et al, 2009).
In our study of 4 out of 24 large sections (18%) and 16 out of 114 TMA spots (14%), we observed a particular pattern of EGFR copy distribution in SISH, as was previously reported in some human tumours such as gliomas or lung cancer, and the amplified sequences were generally localised to DMs (Vogt et al, 2004; Gibaud et al, 2010). Up to 50% of glioblastomas exhibit EGFR amplification with small, acentric, circular, extrachromosomal DNA molecules, which are referred to as DMs and contain from a few hundred kilobases to megabases, and are autonomously replicating DNA fragments. DMs may confer a proliferative advantage to cells, such as by carrying amplified oncogenes or drug-resistance genes (Mondello et al, 2010). This accumulation of extrachromosomal DMs has also been observed in some haematological malignancies (Vogt et al, 2004; Gibaud et al, 2010). We report for the first time, such DMs of EGFR-amplified DNA in breast carcinomas and, more specifically, in TNBC.
The EML4-ALK fusion gene has been identified as an oncogene in 11.3% of non-small cell lung cancers (NSCLC) and 24% of colorectal carcinomas. ALK translocation has also been described in some breast carcinomas (2.4%) (Lin et al, 2009). This suggests that ALK kinase inhibitors (such as crizotinib) may represent effective treatments for patients whose tumours contain the EML4-ALK fusion. Unfortunately, our results showed no TNBC-positive staining with the recently commercially available anti-ALK antibody that was used to screen patients with NLCLC for ALK inhibitor therapies. Potential false-negative results due to IHC bias were controlled (1) by a positive immunoreaction of NSCLC tumour samples that were fixed, paraffin-embedded, and immunostained according to the same conditions and procedures as that of our breast cancer samples, and (2) by the correlation of ISH results with visualisation of EML4-ALK translocation through FISH in IHC-positive NSCLC.
In conclusion, our study shows that in TNBC, EGFR is frequently amplified without mutation but is overexpressed, suggesting that antibody anti-EGFR therapy could be a potential therapy for this aggressive breast cancer subset, provided that the tumours are properly characterised and that clinical trials further prove its benefit for appropriately selected patients. Although preliminary reports of preclinical studies have shown that anti-EGFR treatment in amplified tumours does not significantly reduce the progression of breast carcinomas, more extensive studies are required that will accurately evaluate EGFR amplification by ISH and EGFR overexpression by IHC in TNBC. As shown in our study, ISH is a sensitive and specific procedure for the selection of patients for anti-EGFR treatment and could be used as a cost-effective screening method that would be suitable in clinical practice, as it is for HER-2. In addition, we showed that PI3K and Braf mutations might be associated with EGFR amplification and overexpression, suggesting that combined therapies (vemurafenib) could significantly reduce tumour progression.
The authors declare no conflict of interest.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Bhargava R, Gerald WL, Li AR, Pan Q, Lal P, Ladanyi M, Chen B. EGFR gene amplification in breast cancer: correlation with epidermal growth factor receptor mRNA and protein expression and HER-2 status and absence of EGFR-activating mutations. Mod Pathol. 2005;18:1027–1033. doi: 10.1038/modpathol.3800438. [DOI] [PubMed] [Google Scholar]
- Brachmann SM, Hofmann I, Schnell C, Fritsch C, Wee S, Lane H, Wang S, Garcia-Echeverria C, Maira SM. Specific apoptosis induction by the dual PI3K/mTor inhibitor NVP-BEZ235 in HER2 amplified and PIK3CA mutant breast cancer cells. PNAS. 2009;106:22299–22304. doi: 10.1073/pnas.0905152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience. Nat Rev Clin Oncol. 2010;7:683–692. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, Ma CX, Liu MC, Storniolo AM, Rimawi MF, Forero-Torres A, Wolff AC, Hobday TJ, Ivanova A, Chiu WK, Ferraro M, Burows E, Bernard PS, Hoadley KA, Perou CM, Winer EP. TBCRC 001: Randomized phase ii study of cetuximab in combination with carboplatin in stage iv triple-negative breast cancer. J Clin Oncol. 2012;30:2615–2623. doi: 10.1200/JCO.2010.34.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpin C, Giusiano S, Secq V, Carpentier S, Andrac L, Lavaut MN, Allasia C, Bonnier P, Garcia S. Quantitative immunocytochemical profile to predict early outcome of disease in triple-negative breast carcinomas. Int J Oncol. 2009;34:983–993. doi: 10.3892/ijo_00000224. [DOI] [PubMed] [Google Scholar]
- Charpin C, Secq V, Giusiano S, Carpentier S, Andrac L, Lavaut MN, Allasia C, Bonnier P, Garcia S. A signature predictive of disease outcome in breast carcinomas, identified by quantitative immunocytochemical assays. Int J Cancer. 2009;124:2124–2134. doi: 10.1002/ijc.24177. [DOI] [PubMed] [Google Scholar]
- Corkery B, Crown J, Clynes M, O'Donovan N. Epidermal growth factor receptor as a potential therapeutic target in triple-negative breast cancer. Ann Oncol. 2009;20:862–867. doi: 10.1093/annonc/mdn710. [DOI] [PubMed] [Google Scholar]
- Da Silva L, Clarke C, Lakhani SR. Demystifying basal-like breast carcinomas. J Clin Path. 2007;60:1328–1332. doi: 10.1136/jcp.2006.041731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ruijter TC, Veeck J, De Hoon J, Van Engeland M, Tjan-Heijnen VC. Characteristics of triple-negative breast cancer. J Cancer Res Clin Oncol. 2011;137:183–192. doi: 10.1007/s00432-010-0957-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Guerrab A, Zegrour R, Nemlin CC, Vigier F, Cayre A, Penault-Llorca F, Rossignol F, Bignon YJ. Differential impact of EGFR-targeted therapies on hypoxia responses: implications for treatment sensitivity in triple-negative metastatic breast cancer. PLoS One. 2011;6:e25080. doi: 10.1371/journal.pone.0025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Generali D, Leek R, Fox SB, Moore JW, Taylor C, Chambers P, Harris AL. EGFR mutations in exons 18-21 in sporadic breast cancer. Ann Oncol. 2007;18:203–205. doi: 10.1093/annonc/mdl322. [DOI] [PubMed] [Google Scholar]
- Gibaud A, Vogt N, Hadj-Hamou NS, Meyniel JP, Hupe P, Debatisse M, Malfoy B. Extrachromosomal amplification mechanisms in a glioma with amplified sequences from multiple chromosome loci. Hum Mol Gene. 2010;19:1276–1285. doi: 10.1093/hmg/ddq004. [DOI] [PubMed] [Google Scholar]
- Gumuskaya B, Alper M, Hucumenoglu S, Altundag K, Uner A, Guler G. EGFR expression and gene copy number in triple-negative breast carcinoma. Canc Genet Cytogenet. 2010;203:222–229. doi: 10.1016/j.cancergencyto.2010.07.118. [DOI] [PubMed] [Google Scholar]
- Guzel O, Guner El. ISO 15189 accreditation; Requirements for quality and competence of medical laboratories, experience of a laboratory I. Clin Biochem. 2009;42:274–278. doi: 10.1016/j.clinbiochem.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Jacot W, Lopez-Crapez E, Thezenas S, Senal R, Fina F, Bibeau F, Romieu G, Lamy P-J. Lack of EGFR-activating mutations in European patients with triple-negative breast cancer could emphasise geographic and ethnic variations in breast cancer mutation profiles. Breast Cancer Res. 2011;13:R133. doi: 10.1186/bcr3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamy PJ, Fina F, Bascoul-Mollevi C, Laberenne AC, Martin PM, Ouafik LH, Jacot W. Quantification and clinical relevance of gene amplification at chromosome 17q12-q21 in human epidermal growth factor receptor 2-amplified breast cancers. Breast Cancer Research. 2011;13:R15. doi: 10.1186/bcr2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamy PJ, Nanni I, Fina F, Bibeau F, Romain S, Dussert C, Penault-Llorca F, Grenier J, Ouafik LH, Martin PM. Reliability and discriminant validity of HER2 gene quantification and chromosome 17 aneusomy analysis by real-time PCR in primary breast cancer. Int J Biol Markers. 2006;21:20–29. doi: 10.1177/172460080602100104. [DOI] [PubMed] [Google Scholar]
- Lin E, Li L, Guan Y, Soriano R, Rivers CS, Mohan S, Pandita A, Tang J, Modrusan Z. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol Cancer Res. 2009;7:1466–1476. doi: 10.1158/1541-7786.MCR-08-0522. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Macaskill EJ, Bartlett JM, Sabine VS, Faratian D, Renshaw L, White S, Campbell FM, Young O, Williams L, Thomas JS, Barber MD, Dixon JM. The mammalian target of rapamycin inhibitor everolimus (RAD001) in early breast cancer: results of a pre-operative study. Breast Cancer Res Treat. 2011;128:725–734. doi: 10.1007/s10549-010-0967-z. [DOI] [PubMed] [Google Scholar]
- Martin V, Botta F, Zanellato E, Molinari F, Crippa S, Mazzucchelli L, Frattini M. Molecular characterization of EGFR and EGFR-downstream pathways in triple negative breast carcinomas with basal like features. Histol Histopathol. 2012;27:785–792. doi: 10.14670/HH-27.785. [DOI] [PubMed] [Google Scholar]
- Mondello C, Smirnova A, Giulotto E. Gene amplification, radiation sensitivity and DNA double-strand breaks. Mutation Research. 2010;704:29–37. doi: 10.1016/j.mrrev.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Ishikawa Y, Furuya M, Sano T, Ohno Y, Horiguchi J, Oyama T. Protein expression, gene amplification, and mutational analysis of EGFR in triple-negative breast cancer. Breast Cancer. 2012;21 (1:66–74. doi: 10.1007/s12282-012-0354-1. [DOI] [PubMed] [Google Scholar]
- Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, Akslen LA, Ragaz J, Gown AM, Gilks CB, Van de Rijn M, Perou CM. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy J, Osborne C, Pippen JE, Yoffe M, Patt D, Rocha C, Koo IC, Sherman BM, Bradley C. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med. 2011;364:205–214. doi: 10.1056/NEJMoa1011418. [DOI] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Pinheiro C, Lambros MB, Milanezi F, Carvalho S, Savage K, Simpson PT, Jones C, Swift S, Mackay A, Reis RM, Hornick JL, Pereira EM, Baltazar F, Fletcher CD, Ashworth A, Lakhani SR, Schmitt FC. EGFR amplification and lack of activating mutations in metaplastic breast carcinomas. J Pathol. 2006;209:445–453. doi: 10.1002/path.2004. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathol. 2008;52:108–118. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- Rios J, Puhalla S. PARP inhibitors in breast cancer: BRCA and beyond. Oncology. 2011;25:1014–1025. [PubMed] [Google Scholar]
- Ryden L, Jirstrom K, Haglund M, Stal O, Ferno M. Epidermal growth factor receptor and vascular endothelial growth factor receptor 2 are specific biomarkers in triple-negative breast cancer. Results from a controlled randomized trial with long-term follow-up. Breast Cancer Res Treat. 2010;120:491–498. doi: 10.1007/s10549-010-0758-6. [DOI] [PubMed] [Google Scholar]
- Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977;265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Olumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, Bashashati A, Prentice LM, Khattra J, Burleigh A, Yap D, Bernard V, McPherson A, Shumansky K, Grisan A, Giuliany R, Heravi-Moussavi A, Rosner J, Lai D, Birol I, Varhol R, Tam A, Dhalla N, Zeng T, Ma K, Chan SK, Griffith M, Moradian A, Grace Cheng SW, Morin GB, Watson P, Gelmon K, Chia S, Chin SF, Curtis C, Rueda OM, Pharoah PD, Damaraju S, Mackey J, Hoon K, Harkins T, Tadigotla V, Sigaroudinia M, Gascard P, Tlsty T, Costello JF, Meyer IM, Eaves CJ, Wasserman WW, Jones S, Huntsman D, Hirst M, Caldas C, Marra MA, Aparicio S. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siena S, Sartore-Bianchi A, Di Nicolantonio F, Balfour J, Bardelli A. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst. 2009;101:1308–1324. doi: 10.1093/jnci/djp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng YH, Tan WJ, Thike AA, Cheok PY, Tse GM, Wong NS, Yip GW, Bay BH, Tan PH. Mutations in the epidermal growth factor receptor (EGFR) gene in triple negative breast cancer: possible implications for targeted therapy. Breast Cancer Res. 2011;13:R35. doi: 10.1186/bcr2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thike AA, Cheok PY, Jara-Lazaro AR, Tan B, Tan P, Tan PH. Triple-negative breast cancer: clinicopathological characteristics and relationship with basal-like breast cancer. Mod Pathol. 2010;23:123–133. doi: 10.1038/modpathol.2009.145. [DOI] [PubMed] [Google Scholar]
- Vogt N, Gibaud A, Almeida A, Ourliac-Garnier I, Debatisse M, Malfoy B. Relationships linking amplification level to gene over-expression in gliomas. PLoS One. 2010;5:e14249. doi: 10.1371/journal.pone.0014249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt N, Lefevre SH, Apiou F, Dutrillaux AM, Cor A, Leuraud P, Poupon MF, Dutrillaux B, Debatisse M, Malfoy B. Molecular structure of double-minute chromosomes bearing amplified copies of the epidermal growth factor receptor gene in gliomas. PNAS. 2004;101:11368–11373. doi: 10.1073/pnas.0402979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanikkaya-Demirel G. ISO 15189 accreditation: Requirements for quality and competence of medical laboratories, experience of a laboratory II. Clin Biochem. 2009;42:279–283. doi: 10.1016/j.clinbiochem.2008.09.099. [DOI] [PubMed] [Google Scholar]