Abstract
Background:
In preclinical gastric cancer (GC) models, FGFR2 amplification was associated with increased tumour cell proliferation and survival, and drugs targeting this pathway are now in clinical trials.
Methods:
FGFR2 FISH was performed on 961 GCs from the United Kingdom, China and Korea, and the relationship with clinicopathological data and overlap with HER2 amplification were analysed.
Results:
The prevalence of FGFR2 amplification was similar between the three cohorts (UK 7.4%, China 4.6% and Korea 4.2%), and intratumoral heterogeneity was observed in 24% of FGFR2 amplified cases. FGFR2 amplification was associated with lymph node metastases (P<0.0001). FGFR2 amplification and polysomy were associated with poor overall survival (OS) in the Korean (OS: 1.83 vs 6.17 years, P=0.0073) and UK (OS: 0.45 vs 1.9 years, P<0.0001) cohorts, and FGFR2 amplification was an independent marker of poor survival in the UK cohort (P=0.0002). Co-amplification of FGFR2 and HER2 was rare, and when high-level amplifications did co-occur these were detected in distinct areas of the tumour.
Conclusion:
A similar incidence of FGFR2 amplification was found in Asian and UK GCs and was associated with lymphatic invasion and poor prognosis. This study also shows that HER2 and FGFR2 amplifications are mostly exclusive.
Keywords: FGFR amplification, gastric cancer, HER2 amplification, prognosis
Despite a steady decline in incidence, gastric cancer (GC) is the second most common cause of cancer-related deaths worldwide (Jemal et al, 2011; GLOBOCAN statistics 2009). Most GC patients in the East are diagnosed with early-stage disease (Naylor et al, 2006; Jemal et al, 2011; 2008), whereas GC patients in the West present with locally advanced (inoperable), metastatic or recurrent disease and are treated by cytotoxic combination chemotherapy (Cunningham and Oliveira, 2008; Kang and Kauh, 2011; Bang, 2012). Median overall survival (OS) of patients treated with palliative chemotherapy is 10–12 months (Cunningham and Oliveira, 2008). Targeted therapy has been investigated in this patient group, and the combination of trastuzumab with chemotherapy demonstrated a modest OS benefit in patients with HER2-positive advanced GC (Bang et al, 2010). However, no survival benefit was seen when bevacizumab, an antibody against VEGF (Van Cutsem et al, 2012a), everolimus, a drug targeting mTOR (Van Cutsem et al, 2012b), or the EGFR antibodies panitumumab or cetuximab (Lordick et al, 2013; Waddell et al, 2012) were trialed in non-selected GC patients. Because of the poor prognosis of GC patients, there is a need to identify new potential targets and develop diagnostic tests to identify patients most likely to benefit from targeted therapies.
Fibroblast growth factor receptors (FGFR1–4) are transmembrane tyrosine kinase receptors (Eswarakumar et al, 2005; Turner and Grose, 2010; Brooks et al, 2012; Waddell et al, 2012). FGF binding to the monomeric receptor triggers dimerisation and transphosphorylation of tyrosine residues in the kinase domain (Eswarakumar et al, 2005; Katoh and Katoh, 2006; Turner and Grose, 2010; Brooks et al, 2012). This pathway regulates a variety of cellular functions including cell proliferation, migration and differentiation, which are fundamental to embryonic development, angiogenesis and wound healing (Eswarakumar et al, 2005; Fukumoto, 2008; Turner and Grose, 2010; Brooks et al, 2012).
Dysregulation of the FGFR signalling pathway due to receptor overexpression, gene amplification, mutation or aberrant transcriptional regulation is associated with cancer development and progression in multiple myeloma and cancers of the breast, bladder, lung, endometrium and prostate (Jang et al, 2001; Davies et al, 2005; Grose and Dickson, 2005; Stephens et al, 2005; Katoh, 2010; Turner and Grose, 2010; Brooks et al, 2012).
In preclinical models of GC, FGFR2 amplification was associated with increased tumour cell proliferation and survival, and conferred sensitivity to drugs targeting this pathway, such as the FGFR selective small molecule inhibitors AZD4547 and BGJ398, and anti-FGFR2 antibodies (Bai et al, 2010; Zhao et al, 2010; Gavine et al, 2012; Guagnano et al, 2012; Zhang et al, 2012; Xie et al, 2013). Studies have reported FGFR2 amplification in up to 10% of Asian GC patients (Deng et al, 2012; Jung et al, 2012; Matsumoto et al, 2012), and FGFR2 amplification was recently described in Western GC cohorts (Deng et al, 2012; Dulak et al, 2012; Nadauld et al, 2012). Each of these studies employed a different platform to assess gene amplification, including RT-PCR, fluorescence in situ hybridisation (FISH) and SNP arrays.
This study used FISH to compare the frequency of FGFR2 amplification in large series of GCs from UK, Chinese and Korean patients, the overlap of FGFR2 and HER2 amplification, and the association of FGFR2 amplification with clinicopathological variables and OS.
Material and methods
Patient cohorts
The UK, Chinese and Korean GC cohorts consisted of 408, 197 and 356 patients, respectively, with sporadic gastric adenocarcinoma who underwent surgical resection at Leeds General Infirmary, the United Kingdom (1970––2004), Shanghai Renji Hospital, China (2007–2010) and Seoul National University Hospital, South Korea (1996), respectively (Table 1). Clinical outcome was determined from date of surgery until last seen or mortality status obtained in 2009, 2011 and 2003, for the UK, Chinese and Korean cohorts, respectively. At the end of the study period, 73% and 33% of UK and Chinese patients had died. Median (range) follow-up time was 1.7 years (0–20.5 years), 2.4 years (1 month–4.6 years) and 5.5 years (2 months–8 years) for UK, Chinese and Korean cohorts, respectively.
Table 1. Comparison of the clinicopathological characteristics between UK, Chinese and Korean gastric cancer cohorts.
| |
UK cohort (n=408) |
Chinese cohort (n=197) |
Korean cohort (n=356) |
|||
|---|---|---|---|---|---|---|
| Characteristic | n | % | n | % | n | % |
|
Age (years) | ||||||
| Median | 70 | 62 | 59 | |||
| Range |
13–96 |
|
18–87 |
|
28–82 |
|
|
Gender | ||||||
| Male | 255 | 62.5 | 133 | 67.5 | 247 | 69.4 |
| Female |
153 |
37.5 |
64 |
32.5 |
109 |
30.6 |
|
Grade of differentiation | ||||||
| G1 | 45 | 11.0 | 3 | 1.5 | 20 | 5.6 |
| G2 | 103 | 25.2 | 40 | 20.3 | 173 | 48.6 |
| G3 | 250 | 61.3 | 133 | 67.5 | 161 | 45.2 |
| G4 | 1 | 0.2 | 21 | 10.7 | 2 | 0.6 |
| Unknown |
9 |
2.2 |
0 |
0.0 |
0 |
0.0 |
|
Laurén subtype | ||||||
| Intestinal | 244 | 59.8 | 66 | 33.5 | 170 | 47.8 |
| Diffuse | 96 | 23.5 | 87 | 44.2 | 172 | 48.3 |
| Mixed | 64 | 15.7 | 44 | 22.3 | 14 | 3.9 |
| Unknown |
4 |
1.0 |
0 |
0.0 |
0 |
0.0 |
|
Stage | ||||||
| I | 115 | 28.2 | 15 | 7.6 | 121 | 34.0 |
| II | 81 | 19.9 | 45 | 22.8 | 83 | 23.3 |
| III | 151 | 37.0 | 100 | 50.8 | 89 | 25.0 |
| IV | 60 | 14.7 | 37 | 18.8 | 63 | 17.7 |
| unknown |
1 |
0.2 |
0 |
0.0 |
0 |
0.0 |
|
Depth of invasion (pT) | ||||||
| T1 | 56 | 13.7 | 7 | 3.6 | 67 | 18.8 |
| T2 | 140 | 34.3 | 20 | 10.2 | 167 | 46.9 |
| T3 | 201 | 49.3 | 157 | 79.7 | 114 | 32.0 |
| T4 |
11 |
2.7 |
13 |
6.6 |
8 |
2.2 |
|
Lymph node status (pN) | ||||||
| N0 | 136 | 33.3 | 51 | 25.9 | 121 | 34.0 |
| N1 | 163 | 40.0 | 88 | 44.7 | 127 | 35.7 |
| N2 | 68 | 16.7 | 34 | 17.3 | 58 | 16.3 |
| N3 |
40 |
9.8 |
24 |
12.2 |
50 |
14.0 |
|
Distant metastasis (pM) | ||||||
| M0 | 391 | 95.8 | 182 | 92.4 | 334 | 93.8 |
| M1 |
17 |
4.2 |
15 |
7.6 |
22 |
6.2 |
|
Adjuvant chemotherapy | ||||||
| No | 408 | 100.0 | 36 | 18.3 | 146 | 41.0 |
| Yes | 0 | 0.0 | 129 | 65.5 | 188 | 52.8 |
| Unknown | 0 | 0.0 | 32 | 16.2 | 22 | 6.2 |
Tissue microarray construction
Haematoxylin/eosin-stained sections of resected specimens were reviewed, and blocks with the highest tumour cell density selected for tissue microarray (TMA) construction. TMAs were constructed by random sampling of 3–6, 0.6 mm cores from each tumour and three cores from matched normal mucosa (UK cohort), one 1-mm core from each tumour (Korean cohort), two to four 0.6-mm cores from each tumour and two from matched normal mucosa (Chinese cohort). Four (Korean/Chinese) or 5 μm (UK) sections were cut from each TMA for gene copy-number analysis. Full sections were cut from 26 UK FGFR2-amplified GC specimens to assess amplification heterogeneity within individual tumours. TMA and full sections were quality controlled by an experienced histopathologist.
FGFR2 FISH
The FGFR2 FISH probe was generated in house by AstraZeneca by directly labelling BAC RP11-62L18 (Invitrogen, Grand Island, New York, USA) DNA with Spectrum Red (ENZO, Exeter, UK, 02N34-050) using a nick translation-based method (Abbott Park, IL, USA, 07J00-001) according to the manufacturer's instructions. Pericentromeric Spectrum Green labelled chromosome 10 probe (CEP10, Vysis, 32-132010) was used as an internal control. FISH was performed as described previously (Xie et al, 2013). Sections were deparaffinized and pretreated using the SpotLight Tissue Kit (Invitrogen, 00–8401) according to the manufacturer's instructions. Sections and FGFR2/CEP10 probes were co-denaturated at 80 °C for 5 min and hybridised at 37 °C for 48 h. Excess probe was removed with post-hybridisation wash buffer (0.3% NP40/1 × SSC) at 75.5 °C for 5 min, then 2 × SSC at room temperature for 2 min. Sections were counterstained with 0.3 μg ml−1 DAPI (Vector, H-1200) and coverslipped. FGFR2 and CEP10 signals were scored under a fluorescence microscope (Olympus, Center Valley, PA, USA, BX61). Scoring was adopted from (Varella-Garcia, 2006). Fifty nuclei were evaluated/case. FGFR2 gene copy-number and FGFR2/CEP10 ratio was classified as follows: FGFR2 amplification (score 6): FGFR2/CEP10 ratio ⩾2 or FGFR2 gene clusters in ⩾10% tumour cells; high polysomy (score 5): FGFR2/CEP10 ratio <2 and ⩾4 copies of FGFR2 in ⩾40% tumour cells; low polysomy (score 4): FGFR2/CEP10 ratio <2 and ⩾4 copies of FGFR2 in 10–39% tumour cells; high trisomy (score 3): FGFR2/CEP10 ratio <2 and 3 copies of FGFR2 in ≥40% tumour cells and <10% tumour cells having ⩾4 copies of FGFR2; low trisomy (score 2): FGFR2/CEP10 ratio <2 and 3 copies of FGFR2 in 10–39% tumour cells and <10% tumour cells having ⩾4 copies of FGFR2; disomy (score 1): two copies of FGFR2 in 90% of tumour cells. Scoring was performed independently by two observers at AstraZeneca.
Assessment of FGFR2 amplification heterogeneity
Intratumoral FGFR2 amplification heterogeneity was assessed in TMA and full sections from 26 UK cases with FGFR2 amplification, and was defined as the presence of areas with different FISH scores within the same tumour in full sections and presence of different FISH scores in cores from the same tumour in TMA sections. Scoring was performed independently by two observers.
HER2 FISH
HER2/CEP17 probe (Vysis, 30-161060) was used according to the manufacturer's instructions. Fifty tumour nuclei were scored/case. Tumours with an average HER2 gene copy number >6 or a HER2/CEP17 ratio ⩾2 were defined as HER2 amplified. Scoring was performed independently by two observers.
Combined FGFR2 and HER2 FISH
To detect FGFR2 and HER2 copy number simultaneously, a four-colour FISH probe was generated. The above FGFR2/CEP10 probes were combined with a HER2 probe generated by labelling BAC RP11-94L15 DNA (Invitrogen) with Spectrum Gold (ENZO, ENZ-42843) and a CEP17 Spectrum Aqua probe (Vysis, 32-111017) as internal control using experimental conditions described for FGFR2 FISH. This analysis was performed only in cases identified as FGFR2 and HER2 amplified in the TMA. Scoring was performed independently by two observers.
Statistical analysis
Data from each cohort were analysed individually. The following variables were used for statistical analysis: tumour histology type (Laurén classification, (Lauren, 1965)) tumour grade of differentiation (WHO classification, (Hamilton and Aaltonen, 2000)) depth of invasion (pT), lymph node status (pN), distant metastasis status (pM), resection margin status (R) and stage (TNM classification sixth edition) (Sobin and Wittekind, 2002).
Chi-square tests were used to compare clinicopathological characteristics between cohorts. For association of FGFR2 amplification with clinicopathological characteristics, logistic regression models were fitted in both univariate and multivariate analysis, and P-values were computed by log-likelihood Chi-square test. In multivariate models, age, gender and factors that showed significant association in univariate analysis (pN and grade of differentiation) were included.
For OS, patients were categorised by FGFR2 status into amplified (FISH score 6) and non-amplified (FISH score 1–5). Data were analysed using Kaplan–Meier (Kaplan and Meier, 1958) and log-rank statistics. Univariate and multivariate Cox proportional hazard models were fitted to evaluate FGFR2 status including variables for age, gender, grade of differentiation and stage (Sobin and Wittekind, 2002). Statistical tests were two-sided, P<0.05 was considered significant. Analyses were carried out using R (version 2.10.1).
All studies were performed with the approval of Local Research Ethics committees and were conducted in accordance with the Declaration of Helsinki.
Results
Comparison of patient characteristics and clinicopathological variables between GC cohorts are detailed in Table 1. UK patients were significantly older at the time of diagnosis compared with Asian cohorts (UK/Chinese P<0.0001, UK/Korean P=0.0003). The frequency of intestinal type GC was significantly higher in the UK cohort compared with Asian cohorts (UK/Chinese P<0.0001, UK/Korean P<0.0001). The frequency of well, moderately and poorly differentiated GC was different between all cohorts (UK/Chinese P<0.0001, UK/Korean P<0.0001, Chinese/Korean P<0.0001). There was a significant difference in disease stage distribution between cohorts, with stage III disease more common in Chinese patients (UK/Chinese P<0.0001, Chinese/Korean P<0.0001). All UK patients were treated by surgery alone, while 66% of Chinese and 53% of Korean patients received adjuvant chemotherapy. No patients received neoadjuvant chemotherapy or radiotherapy.
FGFR2 copy number
Results were obtained from a total of 961 cases (Table 2). FGFR2 amplification frequency was 7.4%, 4.6% and 4.2% in the UK, Chinese and Korean cohorts, respectively, and did not differ significantly (UK/Chinese P=0.258, UK/Korean P=0.092, Chinese/Korean P=0.983, UK/Chinese/Korean P=0.586). FGFR2 polysomy was observed in 35.1%, 44.2% and 21.3% of UK, Chinese and Korean cohorts, respectively, and was significantly lower in the Korean cohort (UK/Korean P<0.0001, Korean/Chinese P<0.0001, UK/Chinese/Korean P<0.0001). Correspondingly, there was a significantly higher frequency of FGFR2 disomy in the Korean cohort (UK/Korean P<0.0001, Korean/Chinese P<0.001, UK/Chinese/Korean P<0.001).
Table 2. FGFR2 FISH analysis.
| |
UK cohort (n=408) |
Chinese cohort (n=197) |
Korean cohort (n=356) |
|||
|---|---|---|---|---|---|---|
| FGFR2 FISH score | n | % | n | % | n | % |
| Disomy |
142 |
34.8 |
39 |
19.8 |
190 |
53.4 |
| Low trisomy |
65 |
15.9 |
51 |
25.9 |
63 |
17.7 |
| High trisomy |
28 |
6.9 |
11 |
5.6 |
12 |
3.4 |
| Low polysomy |
87 |
21.3 |
51 |
25.9 |
66 |
18.5 |
| High polysomy |
56 |
13.7 |
36 |
18.3 |
10 |
2.8 |
| Gene amplification | 30 | 7.4 | 9 | 4.6 | 15 | 4.2 |
| |
P-valuesa
(univariate) |
P-valuesb
(multivariate) |
||
|---|---|---|---|---|
| FGFR2 FISH score | UK vs Chinese | UK vs Korean | Chinese vs Korean | UK vs Chinese vs Korean |
| 1–5 vs 6 |
0.2584 |
0.0921 |
0.9827 |
0.5855 |
| 1–3 vs 4–5 | 0.0664 | 8.96 × 10−8 | 1.90 × 10−8 | 3.73 × 10−9 |
Abbreviations: FGFR2=fibroblast growth factor receptor 2; FISH=fluorescent in situ hybridisation.
P-values contrasted for a given pair of cohorts are computed from χ2-test by collapsing score 1–5 subjects in 1–5 vs 6 comparison and score 1–3 and score 4–5 subjects, respectively, in 1–3 vs 4–5 comparison.
Multivariate P-values are from a log-likelihood ratio test after adjusting for age, gender, stage, grade and Lauren subtype.
Association of FGFR2 copy number with clinicopathological parameters
The relationship between FGFR2 amplification and clinicopathological parameters was analogous between the cohorts; hence, a combined analysis of all three cohorts is presented. Univariate and multivariate analyses of all 961 patients showed that FGFR2 amplification was significantly more common in patients with higher pN category (P<0.0001). For the analysis of grade of tumour differentiation, the small number of patients with grade 4 tumours (n=24) were grouped together with grade 3 tumours. Statistical analysis showed that the prevalence of FGFR2 amplification was significantly lower in grade 2 (moderately differentiated) tumours compared with grade 1 (well differentiated) or grade 3 and 4 (poorly differentiated/undifferentiated) tumours (P<0.01). There was no association of FGFR2 amplification with age, gender, histological subtype (Table 3) or tumour location (P=0.716, data not shown).
Table 3. Comparisons of clinicopathological characteristics by FGFR2 amplification status.
| FGFR2 non-amplified (FISH 1–5) | FGFR2 amplified (FISH 6) | |||||
|---|---|---|---|---|---|---|
|
Characteristics |
n |
% |
n |
% |
P-value (uni) |
P-value (multi)a |
|
Age | ||||||
| <median age | 444 | 95 | 25 | 5 | ||
| ⩾median age |
462 |
94 |
29 |
6 |
0.1873 |
|
|
Gender | ||||||
| Male | 602 | 95 | 33 | 5 | ||
| Female |
305 |
94 |
21 |
6 |
0.7480 |
|
|
Grade | ||||||
| 1 | 64 | 94 | 4 | 6 | ||
| 2 | 309 | 98 | 7 | 2 | ||
| 3 | 501 | 92 | 43 | 8 | ||
| 4 |
24 |
100 |
0 |
0 |
0.0176 |
0.0073 |
|
Laurén subtype | ||||||
| Intestinal | 459 | 96 | 21 | 4 | ||
| Diffuse | 329 | 93 | 26 | 7 | ||
| Mixed |
115 |
94 |
7 |
6 |
0.1793 |
0.2248 |
|
Stage | ||||||
| 1, 2 | 442 | 96 | 18 | 4 | ||
| 3, 4 |
464 |
93 |
36 |
7 |
0.1021 |
0.1442 |
|
T | ||||||
| 1 | 126 | 97 | 4 | 3 | ||
| 2 | 306 | 94 | 21 | 6 | ||
| 3 | 444 | 94 | 28 | 6 | ||
| 4 |
31 |
97 |
1 |
3 |
0.5671 |
0.4805 |
|
N | ||||||
| 0 | 304 | 99 | 4 | 1 | ||
| 1 | 356 | 94 | 22 | 6 | ||
| 2 | 146 | 91 | 14 | 9 | ||
| 3 |
100 |
88 |
14 |
12 |
1.22 × 10−5 |
<0.0001 |
|
M | ||||||
| 0 | 856 | 94 | 51 | 6 | ||
| 1 | 51 | 94 | 3 | 6 | 0.4456 | 0.4281 |
P-value is calculated from logistic regression adjusting for age and gender.
Association of FGFR2 copy number with overall survival
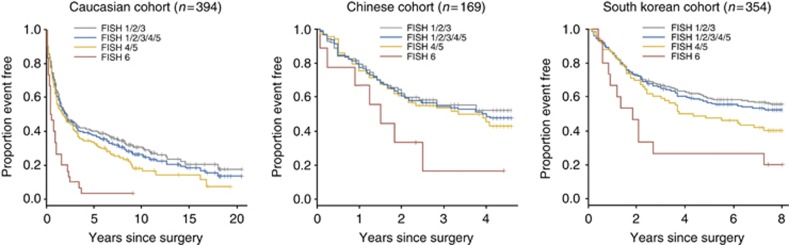
Type of treatment and other patient characteristics differed significantly between the three cohorts. Therefore, the relationship between FGFR2 FISH status and overall survival was analysed separately. Median OS was significantly shorter in patients with FGFR2 amplified GC compared with patients with FGFR2 non-amplified GC in UK (P<0.0001) and Korean (P=0.0073) cohorts by univariate analysis (Table 4). A similar trend was observed for the Chinese cohort but did not achieve significance (P=0.0646; Figure 1). Multivariate survival analysis from the Cox proportional hazard model adjusting for age, gender and grade of tumour differentiation confirmed FGFR2 amplification status as an independent prognostic marker in the UK cohort (P =0.0002; Table 4).
Table 4. Analysis of prognostic value of FGFR2 amplification (FISH 6) or polysomy (FISH 4–5) status for overall survival (Cox proportional hazard model).
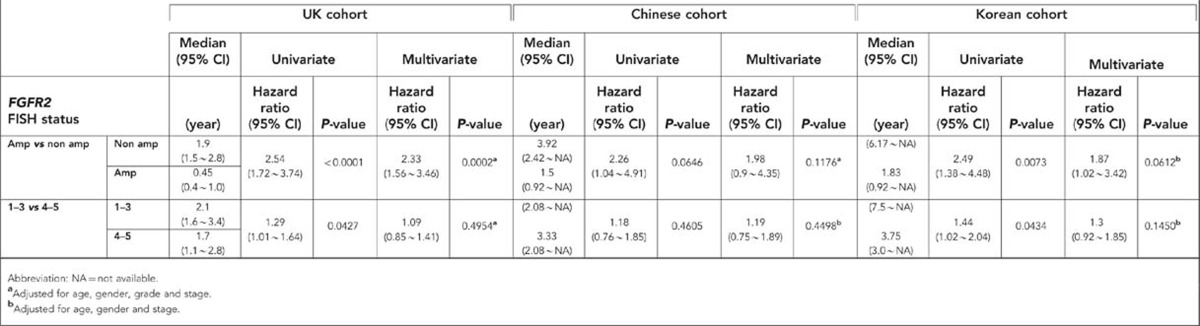
Figure 1.
Kaplan–Meier OS analysis using FGFR2 FISH score in three separate patient cohorts. Median OS and 95% CI for pairwise comparisons are provided.
To assess whether there was an effect of adjuvant chemotherapy treatment on the prognostic value of FGFR2 amplification, a subset analysis was performed in patients treated by surgery only vs patients treated by adjuvant chemotherapy after surgery. Results from Korean and Chinese cohorts were pooled for this analysis. FGFR2 amplification was similarly predictive for shorter OS in patients treated with adjuvant chemotherapy (P=0.002) and the surgery only group (P=0.03).
UK and Korean patients with FGFR2 polysomy GC (score 4–5) had a significantly shorter OS when compared with others with FGFR2 non-amplified disease (score 1–3) in univariate analysis (UK cohort P=0.0427; Korean cohort P=0.0434) that was not significant in multivariate analysis adjusting for age, gender, grade of differentiation and stage (Table 4). No relationship was seen in the Chinese cohort.
FGFR2 amplification heterogeneity
Seven of 29 UK FGFR2 amplified GCs (24.1%) displayed intratumoral heterogeneity within TMAs. Tissue sections were unavailable for 3/29 GCs. Analysis of full sections confirmed FGFR2 amplification in 23/26 cases and intratumoral heterogeneity in six cases, which also showed heterogeneity in the TMA study. Three cases found to be FGFR2 amplified in the TMA study showed no evidence of FGFR2 amplification in full sections, most likely related to FGFR2 amplification heterogeneity.
FGFR2 and HER2 amplification are predominantly exclusive
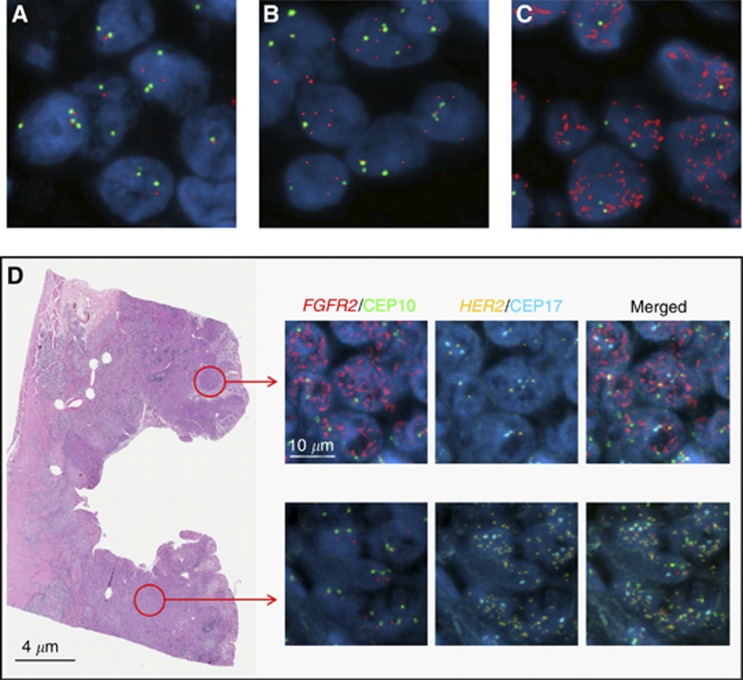
HER2 FISH results were available from 204 and 338 of Chinese and Korean GCs; amplification was present in 14.7% and 8% of GCs, respectively. Co-amplification of HER2 and FGFR2 was present in one Chinese GC but not detected in any GCs from the Korean cohort. In the UK cohort, the frequency of HER2 and FGFR2 co-amplification was investigated in 26 full sections of GCs found to be FGFR2 amplified in the TMA study. Two of the 26 UK GC cases showed FGFR2 and HER2 co-amplification. In order to assess whether co-amplification was present in the same cell, FGFR2/HER2 four-colour FISH was performed. High-level amplifications of HER2 and FGFR2 (FGFR2 average copy number >100 and HER2/CEP17 ratio >10) were found in both UK GCs, and HER2 amplification was seen in tumour cells that were FGFR2 non-amplified and vice versa (Figure 2D). In contrast, HER2 and FGFR2 amplifications occurred in the same tumour cell in the single Chinese GC, but amplifications were of low level and only just satisfied the minimal amplification criteria (FGFR2 average copy number 5.52 and HER2/CEP17 ratio 2.26) (Supplementary Figure 1).
Figure 2.
Dual-colour FISH shows FGFR2 copy-number normal (A), copy-number increase (B) and amplification (C). Red and green signals highlight FGFR2 gene and centromere 10 probes, respectively. Four-colour FISH reveals distinct tumour regions with either FGFR2 or HER2 amplification (D). Gold and aqua probes highlight HER2 and centromere 17.
Discussion
FGFR amplification has been reported in various cancers, including FGFR1 amplification in ER+breast cancer and squamous cell lung cancer, and FGFR2 amplification in triple negative breast cancer and GCs (Andre et al, 2009; Turner et al, 2010a, 2010b; Weiss et al, 2010; Deng et al, 2012; Dulak et al, 2012; Jung et al, 2012; Matsumoto et al, 2012; Nadauld et al, 2012). Hence, there is significant interest in FGFR2 as a therapeutic target for FGFR2-amplified GCs, and clinical trials of FGFR inhibitors are ongoing.
There is variability in the reported incidence of FGFR2 overexpression in GC; FGFR2 protein overexpression was reported to be 30%–40% by immunohistochemistry, (Hattori et al, 1996) while the incidence of FGFR2 amplification varies from 3 to 10% (Mor et al, 1993; Hara et al, 1998; Deng et al, 2012; Dulak et al, 2012; Jung et al, 2012; Matsumoto et al, 2012). Gene copy-number evaluation in solid tumour cells by FISH is widely accepted as a ‘gold-standard approach' for clinical application, and the current study is the largest to date assessing FGFR2 amplification by FISH in patients with resectable GC from three different countries, Korea, China and UK. The prevalence of FGFR2 amplification was investigated in a total of 961 GCs and was 7.4%, 4.6% and 4.2% in UK, Chinese and Korean GCs, respectively, with no significant difference of incidence between cohorts. Our results are similar to those reported previously for Korean (Jung et al, 2012) and Japanese cohorts (Matsumoto et al, 2012). In addition to FGFR2 amplification, we found a significant incidence of FGFR2 polysomy in GC, which was significantly higher in UK (35.1%) and Chinese (44.2%) GC cohorts than in the Korean cohort (21.3%). Further work is required to demonstrate whether FGFR2 polysomy is related to tumour growth, survival and sensitivity to therapeutic intervention.
The observation that FGFR2 amplification was significantly associated with lymph node disease suggests that this molecular aberration may contribute to the development of metastasis. An association between FGFR2 amplification and lymphatic invasion was recently reported (Jung et al, 2012).
It has been reported that FGFR2 amplification is more frequently found in diffuse type GC compared with intestinal type GC (Nakatani et al, 1990). In contrast, our study did not find an association of FGFR2 amplification with histological subtype according to the Laurén classification, which is in agreement with findings recently reported for another Korean GC cohort (Jung et al, 2012). This contrasts with HER2 gene amplification, which is associated with the intestinal subtype of gastric cancer (Bang, 2012).
The current study suggests that FGFR2 amplification is a molecular factor related to poor prognosis in patients with resectable GC, irrespective of ethnic origin and irrespective of the underlying significant differences in clinicopathological parameters, survival and treatment between cohorts from Asia and the United Kingdom. The potential usefulness of FGFR2 amplification as a predictive factor for response to FGFR2 targeting therapies remains to be evaluated in patients with GC.
Amplification of HER2 has been identified in 6–35% of patients with GC (Bang, 2012); however, the majority of recent studies have reported incidences of 6–13% (Okines et al, 2012; Terashima et al, 2012; Aizawa et al, 2014; Narita et al, 2013; Warneke et al, 2013). HER2 amplification has led to the successful development and approval of trastuzumab in patients with GC (Bang et al, 2010). In the present study, we assessed the overlap between FGFR2 and HER2 amplification. Only 3/50 FGFR2-amplified samples (3/961 of total population samples) were also HER2 amplified, confirming that HER2 and FGFR2 amplifications are usually mutually exclusive (Deng et al, 2012). Interestingly, low-level HER2 and FGFR2 amplifications were detected in the same tumour cells in one Chinese GC, whereas in two UK GCs tumour cells with high-level HER2 amplification were located in a different area to tumour cells with high-level FGFR2 amplification. The findings in these latter two GCs suggest that FGFR2- and HER2- amplified tumour cells may have developed from different tumour cell clones with differing genetic characteristics. In these patients, combined FGFR2 and HER2 inhibitor therapies might be required for durable and potent antitumour responses.
We found evidence for intratumoral heterogeneity of FGFR2 amplification in about 25% of GCs, indicating that there is a potential for missing areas of amplification especially when analysis is performed on small biopsies, rather than sections from resection specimens. Similar intratumoral heterogeneity has been reported for HER2 amplification in GC, leading to a recommendation that multiple biopsies should be assessed to determine HER2 status of a tumour.
In conclusion, this is the largest study of FGFR2 FISH in GC and the first study to compare the incidence of FGFR2 amplification in UK and Asian cohorts, demonstrating a similar incidence across cohorts. Furthermore, our data show that FGFR2 amplification is associated with lymph node metastasis and related to poor OS. Overall, this study suggests that FGFR2 may represent an attractive therapeutic target in a subgroup of GCs, irrespective of ethnicity, and FISH methodology could be used for patient selection.
Acknowledgments
This study was sponsored by AstraZeneca. Editorial assistance was provided by Claire Routley from Mudskipper Business Ltd, funded by AstraZeneca.
YJB and SAI have received honoraria and research funding from, and acted within advisory roles for, AstraZeneca. WHK and HIG have received research funding from AstraZeneca. XS, PZ, PRG, SM, CW and EK are employees of and own stocks in AstraZeneca. The remaining authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Part of this work has been presented at the Annual Meeting of the Pathological Society of Great Britain and Ireland in Sheffield, July 2012 and at the ASCO Annual Meeting in Chicago, June 2012.
Supplementary Material
References
- Aizawa M, Nagatsuma AK, Kitada K, Kuwata T, Fujii S, Kinoshita T, Ochiai A. Evaluation of HER2-based biology in 1,006 cases of gastric cancer in a Japanese population. Gastric Cancer. 2014;17 (1:34–42. doi: 10.1007/s10120-013-0239-9. [DOI] [PubMed] [Google Scholar]
- Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, Richon C, Yan K, Wang B, Vassal G, Delaloge S, Hortobagyi GN, Symmans WF, Lazar V, Pusztai L. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res. 2009;15:441–451. doi: 10.1158/1078-0432.CCR-08-1791. [DOI] [PubMed] [Google Scholar]
- Bai A, Meetze K, Vo NY, Kollipara S, Mazsa EK, Winston WM, Weiler S, Poling LL, Chen T, Ismail NS, Jiang J, Lerner L, Gyuris J, Weng Z. GP369, an FGFR2-IIIb-specific antibody, exhibits potent antitumor activity against human cancers driven by activated FGFR2 signaling. Cancer Res. 2010;70:7630–7639. doi: 10.1158/0008-5472.CAN-10-1489. [DOI] [PubMed] [Google Scholar]
- Bang YJ. Advances in the management of HER2-positive advanced gastric and gastroesophageal junction cancer. J Clin Gastroenterol. 2012;46:637–648. doi: 10.1097/MCG.0b013e3182557307. [DOI] [PubMed] [Google Scholar]
- Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, Aprile G, Kulikov E, Hill J, Lehle M, Ruschoff J, Kang YK. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- Brooks AN, Kilgour E, Smith PD. Molecular pathways: fibroblast growth factor signaling: a new therapeutic opportunity in cancer. Clin Cancer Res. 2012;18:1855–1862. doi: 10.1158/1078-0432.CCR-11-0699. [DOI] [PubMed] [Google Scholar]
- Cunningham D, Oliveira J. Gastric cancer: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol. 2008;19 (Suppl 2:ii23–ii24. doi: 10.1093/annonc/mdn075. [DOI] [PubMed] [Google Scholar]
- Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, Teague J, Butler A, Edkins S, Stevens C, Parker A, O'Meara S, Avis T, Barthorpe S, Brackenbury L, Buck G, Clements J, Cole J, Dicks E, Edwards K, Forbes S, Gorton M, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jones D, Kosmidou V, Laman R, Lugg R, Menzies A, Perry J, Petty R, Raine K, Shepherd R, Small A, Solomon H, Stephens Y, Tofts C, Varian J, Webb A, West S, Widaa S, Yates A, Brasseur F, Cooper CS, Flanagan AM, Green A, Knowles M, Leung SY, Looijenga LH, Malkowicz B, Pierotti MA, Teh BT, Yuen ST, Lakhani SR, Easton DF, Weber BL, Goldstraw P, Nicholson AG, Wooster R, Stratton MR, Futreal PA. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- Deng N, Goh LK, Wang H, Das K, Tao J, Tan IB, Zhang S, Lee M, Wu J, Lim KH, Lei Z, Goh G, Lim QY, Tan AL, Sin Poh DY, Riahi S, Bell S, Shi MM, Linnartz R, Zhu F, Yeoh KG, Toh HC, Yong WP, Cheong HC, Rha SY, Boussioutas A, Grabsch H, Rozen S, Tan P. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut. 2012;61:673–684. doi: 10.1136/gutjnl-2011-301839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulak AM, Schumacher SE, van LJ, Imamura Y, Fox C, Shim B, Ramos AH, Saksena G, Baca SC, Baselga J, Tabernero J, Barretina J, Enzinger PC, Corso G, Roviello F, Lin L, Bandla S, Luketich JD, Pennathur A, Meyerson M, Ogino S, Shivdasani RA, Beer DG, Godfrey TE, Beroukhim R, Bass AJ. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012;72:4383–4393. doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Fukumoto S. Actions and mode of actions of FGF19 subfamily members. Endocr J. 2008;55:23–31. doi: 10.1507/endocrj.kr07e-002. [DOI] [PubMed] [Google Scholar]
- Gavine PR, Mooney L, Kilgour E, Thomas AP, Al-Kadhimi K, Beck S, Rooney C, Coleman T, Baker D, Mellor MJ, Brooks AN, Klinowska T. AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 2012;72:2045–2056. doi: 10.1158/0008-5472.CAN-11-3034. [DOI] [PubMed] [Google Scholar]
- GLOBOCAN statistics 2008. Available at http://globocan.iarc.fr/ ( 2009Fluorescence In Situ Hybridization (FISH) Application Guide Springer [Google Scholar]
- Grose R, Dickson C. Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth Factor Rev. 2005;16:179–186. doi: 10.1016/j.cytogfr.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Guagnano V, Kauffmann A, Wohrle S, Stamm C, Ito M, Barys L, Pornon A, Yao Y, Li F, Zhang Y, Chen Z, Wilson CJ, Bordas V, Le Douget M, Gaither LA, Borawski J, Monahan JE, Venkatesan K, Brummendorf T, Thomas DM, Garcia-Echeverria C, Hofmann F, Sellers WR, Graus Porta D. FGFR genetic alterations predict for sensitivity to NVP-BGJ398,a selective pan-FGFR inhibitor. Cancer Discov. 2012;2:1118–1133. doi: 10.1158/2159-8290.CD-12-0210. [DOI] [PubMed] [Google Scholar]
- Hamilton SR, Aaltonen LA. World Health Organization Classification of Tumours. Pathology and Genetics. Tumours of the Digestive System. IARC Press: Lyon; 2000. pp. 38–52. [Google Scholar]
- Hara T, Ooi A, Kobayashi M, Mai M, Yanagihara K, Nakanishi I. Amplification of c-myc, K-sam, and c-met in gastric cancers: detection by fluorescence in situ hybridization. Lab Invest. 1998;78:1143–1153. [PubMed] [Google Scholar]
- Hattori Y, Itoh H, Uchino S, Hosokawa K, Ochiai A, Ino Y, Ishii H, Sakamoto H, Yamaguchi N, Yanagihara K, Hirohashi S, Sugimura T, Terada M. Immunohistochemical detection of K-sam protein in stomach cancer. Clin Cancer Res. 1996;2:1373–1381. [PubMed] [Google Scholar]
- Jang JH, Shin KH, Park JG. Mutations in fibroblast growth factor receptor 2 and fibroblast growth factor receptor 3 genes associated with human gastric and colorectal cancers. Cancer Res. 2001;61:3541–3543. [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Jung EJ, Jung EJ, Min SY, Kim MA, Kim WH. Fibroblast growth factor receptor 2 gene amplification status and its clinicopathologic significance in gastric carcinoma. Hum Pathol. 2012;43:1559–1566. doi: 10.1016/j.humpath.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Kang H, Kauh JS. Chemotherapy in the treatment of metastatic gastric cancer: is there a global standard. Curr Treat Options Oncol. 2011;12:96–106. doi: 10.1007/s11864-010-0135-z. [DOI] [PubMed] [Google Scholar]
- Kaplan EL, Meier P. Nonparametric-estimation from incomplete observations. Amer Statistical Assoc. 1958;53:457–481. [Google Scholar]
- Katoh M. Genetic alterations of FGF receptors: an emerging field in clinical cancer diagnostics and therapeutics. Expert Rev Anticancer Ther. 2010;10:1375–1379. doi: 10.1586/era.10.128. [DOI] [PubMed] [Google Scholar]
- Katoh M, Katoh M. FGF signaling network in the gastrointestinal tract (review) Int J Oncol. 2006;29:163–168. [PubMed] [Google Scholar]
- Lauren P. The two histological main types of gastriccarcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- Lordick F, Kang YK, Chung HC, Salman P, Oh SC, Bodoky G, Kurteva G, Volovat C, Moiseyenko VM, Gorbunova V, Park JO, Sawaki A, Celik I, Goette H, Melezinkova H, Moehler M. Cetuximab in combination with capecitabine and cisplatin as first-line treatment in advanced gastric cancer: Randomized controlled phase III EXPAND study. Lancet Oncol. 2013;14:490–499. doi: 10.1016/S1470-2045(13)70102-5. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Arao T, Hamaguchi T, Shimada Y, Kato K, Oda I, Taniguchi H, Koizumi F, Yanagihara K, Sasaki H, Nishio K, Yamada Y. FGFR2 gene amplification and clinicopathological features in gastric cancer. Br J Cancer. 2012;106:727–732. doi: 10.1038/bjc.2011.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor O, Ranzani GN, Ravia Y, Rotman G, Gutman M, Manor A, Amadori D, Houldsworth J, Hollstein M, Schwab M, Shiloh Y. DNA amplification in human gastric carcinomas. Cancer Genet Cytogenet. 1993;65:111–114. doi: 10.1016/0165-4608(93)90217-a. [DOI] [PubMed] [Google Scholar]
- Nadauld L, Regan JF, Miotke L, Pai RK, Longacre TA, Kwok SS, Saxonov S, Ford JM, Ji HP. Quantitative and sensitive detection of cancer genome amplifications from formalin fixed paraffin embedded tumors with droplet digital pcr. Transl Med (Sunnyvale) 2012;2:pii: 1000107. doi: 10.4172/2161-1025.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani H, Sakamoto H, Yoshida T, Yokota J, Tahara E, Sugimura T, Terada M. Isolation of an amplified DNA sequence in stomach cancer. Jpn J Cancer Res. 1990;81:707–710. doi: 10.1111/j.1349-7006.1990.tb02631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Seshimo A, Suzuki M, Murata J, Kameoka S. Status of Tissue Expression and Serum Levels of HER2 in Gastric Cancer Patients in Japan. Hepatogastroenterology. 2013;60:1083–1088. doi: 10.5754/hge121022. [DOI] [PubMed] [Google Scholar]
- Naylor GM, Gotoda T, Dixon M, Shimoda T, Gatta L, Owen R, Tompkins D, Axon A. Why does Japan have a high incidence of gastric cancer? Comparison of gastritis between UK and Japanese patients. Gut. 2006;55:1545–1552. doi: 10.1136/gut.2005.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okines AF, Thompson LC, Cunningham D, Wotherspoon A, Reis-Filho JS, Langley RE, Waddell TS, Noor D, Eltahir Z, Wong R, Stenning S. Effect of HER2 on prognosis and benefit from peri-operative chemotherapy in early oesophago-gastric adenocarcinoma in the MAGIC trial. Ann Oncol. 2012;24:1253–1261. doi: 10.1093/annonc/mds622. [DOI] [PubMed] [Google Scholar]
- Sobin LH, Wittekind C.2002TNM Classification of Malignant Tumours6th edn.Wiley-Liss: New York [Google Scholar]
- Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, Bignell G, Teague J, Smith R, Stevens C, O'Meara S, Parker A, Tarpey P, Avis T, Barthorpe A, Brackenbury L, Buck G, Butler A, Clements J, Cole J, Dicks E, Edwards K, Forbes S, Gorton M, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jones D, Kosmidou V, Laman R, Lugg R, Menzies A, Perry J, Petty R, Raine K, Shepherd R, Small A, Solomon H, Stephens Y, Tofts C, Varian J, Webb A, West S, Widaa S, Yates A, Brasseur F, Cooper CS, Flanagan AM, Green A, Knowles M, Leung SY, Looijenga LH, Malkowicz B, Pierotti MA, Teh B, Yuen ST, Nicholson AG, Lakhani S, Easton DF, Weber BL, Stratton MR, Futreal PA, Wooster R. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- Terashima M, Kitada K, Ochiai A, Ichikawa W, Kurahashi I, Sakuramoto S, Katai H, Sano T, Imamura H, Sasako M. Impact of expression of human epidermal growth factor receptors EGFR and ERBB2 on survival in stage II/III gastric cancer. Clin Cancer Res. 2012;18:5992–6000. doi: 10.1158/1078-0432.CCR-12-1318. [DOI] [PubMed] [Google Scholar]
- Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, Geyer FC, van KM, Kreike B, Mackay A, Ashworth A, van d V, Reis-Filho JS. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013–2023. doi: 10.1038/onc.2009.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, Natrajan R, Marchio C, Iorns E, Mackay A, Gillett C, Grigoriadis A, Tutt A, Reis-Filho JS, Ashworth A. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70:2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cutsem E, De HS, Kang YK, Ohtsu A, Tebbutt NC, Ming XJ, Peng YW, Langer B, Delmar P, Scherer SJ, Shah MA. Bevacizumab in combination with chemotherapy as first-line therapy in advanced gastric cancer: a biomarker evaluation from the AVAGAST randomized phase III trial. J Clin Oncol. 2012;30:2119–2127. doi: 10.1200/JCO.2011.39.9824. [DOI] [PubMed] [Google Scholar]
- Van Cutsem E, Yeh K-H, Bang YJ, Shen L, Ajani JA, Bai Y-X, Chung HC, Pan H-M, Chin K, Muro K, Kim YH, Smith H, Costantini C, Musalli S, Rizvi S, Sahmoud T, Ohtsu A. Phase III trial of everolimus (EVE) in previously treated patients with advanced gastric cancer (AGC): GRANITE-1. J Clin Oncol. 2012;30 (15S:abst LBA3. doi: 10.1200/JCO.2012.48.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varella-Garcia M. Stratification of non-small cell lung cancer patients for therapy with epidermal growth factor receptor inhibitors: the EGFR fluorescence in situ hybridization assay. Diagn Pathol. 2006;1:19. doi: 10.1186/1746-1596-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell T, Chau I, Barbachano Y, Gonzalez de Castro D, Wotherspoon A, Saffery C, Middleton G, Wadsley J, Ferry D, Mansoor W, Crosby T, Coxon F, Smith D, Waters J, Iveson T, Falk S, Slater S, Okines A, Cunningham D.2012A randomized multicenter trial of epirubicin, oxaliplatin, and capecitabine (EOC) plus panitumumab in advanced esophagogastric cancer (REAL3) J Clin Oncol 30(15Sabst LBA4000Ref Type: Abstract. [Google Scholar]
- Warneke VS, Behrens HM, Boger C, Becker T, Lordick F, Ebert MP, Rocken C. Her2/neu testing in gastric cancer: evaluating the risk of sampling errors. Ann Oncol. 2013;24:725–733. doi: 10.1093/annonc/mds528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, Ullrich RT, Menon R, Maier S, Soltermann A, Moch H, Wagener P, Fischer F, Heynck S, Koker M, Schottle J, Leenders F, Gabler F, Dabow I, Querings S, Heukamp LC, Balke-Want H, Ansen S, Rauh D, Baessmann I, Altmuller J, Wainer Z, Conron M, Wright G, Russell P, Solomon B, Brambilla E, Brambilla C, Lorimier P, Sollberg S, Brustugun OT, Engel-Riedel W, Ludwig C, Petersen I, Sanger J, Clement J, Groen H, Timens W, Sietsma H, Thunnissen E, Smit E, Heideman D, Cappuzzo F, Ligorio C, Damiani S, Hallek M, Beroukhim R, Pao W, Klebl B, Baumann M, Buettner R, Ernestus K, Stoelben E, Wolf J, Nurnberg P, Perner S, Thomas RK. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Su X, Zhang L, Yin X, Tang L, Zhang X, Xu Y, Gao Z, Liu K, Zhou M, Gao B, Shen D, Zhang LH, Ji JF, Gavine PR, Zhang J, Kilgour E, Zhang X, Ji Q. FGFR2 gene amplification in gastric cancer predicts sensitivity to the selective FGFR inhibitor AZD4547. Clin Cancer Res. 2013;19:2572–2583. doi: 10.1158/1078-0432.CCR-12-3898. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang L, Su X, Li M, Xie L, Malchers F, Fan S, Yin X, Xu Y, Liu K, Dong Z, Zhu G, Qian Z, Tang L, Zhan P, Ji Q, Kilgour E, Smith PD, Brooks AN, Thomas RK, Gavine PR. Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin Cancer Res. 2012;18:6658–6667. doi: 10.1158/1078-0432.CCR-12-2694. [DOI] [PubMed] [Google Scholar]
- Zhao WM, Wang L, Park H, Chhim S, Tanphanich M, Yashiro M, Kim KJ. Monoclonal antibodies to fibroblast growth factor receptor 2 effectively inhibit growth of gastric tumor xenografts. Clin Cancer Res. 2010;16:5750–5758. doi: 10.1158/1078-0432.CCR-10-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.