Abstract
Background:
Uveal melanoma is the most frequent primary tumour of the eye. It is molecularly clearly distinct from cutaneous melanoma and shows a different pattern of driver mutations. The influence of sunlight ultraviolet (UV) exposure on the aetiology of uveal melanoma is a matter of debate. The recent identification of driver mutations in the promoter of the telomerase reverse transcriptase (TERT) gene with UV-induced cytidine-to-thymidine transitions in cutaneous melanoma prompted us to investigate whether these mutations also occur in uveal melanoma.
Methods:
We analysed 50 cases of uveal melanoma obtained from enucleation surgery for mutations in the genes GNAQ, GNA11, BAP1, SF3B1, EIFAX1 and TERT, measured gene expression using microarrays and analysed gene copy numbers by SNP arrays.
Results:
We detected a TERT mutation in only one case of a 57-year-old white male patient with clinical and histopathological features typical for uveal melanoma. The tumour showed mutations in GNA11 and EIF1AX that are typical for uveal melanoma and absent from cutaneous melanoma. No mutations were detected in GNAQ, BAP1 and SF3B1 that are frequently mutated in uveal melanoma. Both copies of chromosome 3 were retained. Several tumours among which the one carrying the TERT promoter mutation showed elevated TERT expression. Consistent with previous reports, GNAQ is inversely associated with chromosome 3 monosomy and metastasis. BAP1 mutations are significantly associated with chromosome 3 monosomy but not with relapse.
Conclusion:
These data indicate that TERT mutations are rare in uveal melanoma. No conclusion can be drawn on their potential influence on tumour progression.
Keywords: TERT, GNA11, BAP1, UV exposure, sunlight
Uveal melanoma is the most common primary tumour of the eye with an annual incidence of approximately two cases per million in southern European countries to eight cases in northern European countries (Virgili et al, 2007). Incidence increases with latitude in a highly significant manner (Virgili et al, 2007). Whether this association can be attributed to the exposure to sunlight of variable intensity or not, remains a matter of discussion (Gallagher et al, 1985; Pane and Hirst, 2000; Guenel et al, 2001; Singh et al, 2004). Uveal melanoma shows a mutation pattern that is clearly distinct from cutaneous (for recent reviews, see Coupland et al, 2013; Harbour, 2013; Zeschnigk and Lohmann, 2013), mucosal (Furney et al, 2013) and conjunctival melanomas (Griewank et al, 2013c). The mutations typically encountered in cutaneous and conjunctival melanomas, BRAF and NRAS, are rare in uveal melanomas that are characterised by mutations of the G-proteins GNAQ and GNA11 occurring in mutual exclusive manner in ∼85% of the cases (Zeschnigk and Lohmann, 2013). The mutation pattern observed by exome sequencing in cutaneous melanoma is clearly consistent with an aetiological role of sunlight exposure (Hodis et al, 2012). Cytidine-to-thymidine transition frequency has not been addressed by the two exome sequencing studies of uveal melanoma (Harbour et al, 2013; Martin et al, 2013) but the complete lack of overlap among the driver mutations in the two pathologies is consistent with a different aetiology.
Recently, three mutations in the promoter of the telomerase reverse transcriptase (TERT) needed for telomere maintenance in cancer cells, close to the transcriptional start site, have been described for sporadic (Huang et al, 2013) and familiar (Horn et al, 2013) forms of cutaneous melanoma. The familiar cases all derived from one family and showed a mutation different from those detected in sporadic melanomas and melanoma cell lines (Huang et al, 2013). Two mutations were identified in sporadic cases where they occurred in a mutually exclusive manner in over 70% of cases. The same mutations were also present, though less frequently, in other tumours and tumour-derived cell lines. The mutations consisted in cytidine-to-thymidine transitions at a dipyrimidine motif consistent with ultraviolet (UV) light-induced damage. All three mutations created novel binding sites for the transcription factor E-twenty-six (ETS) in the TERT promoter within 100 bp upstream of the transcription start site (TSS) in sporadic cases and closer to the TTS (−57 bp) in the familiar cases. Reporter assays showed the functionality of the new binding sites observed in sporadic cases (Huang et al, 2013). The mutation is highly penetrant in the melanoma family analysed and it is linked to early onset (mean 34 years) and short survival (mean 3.5 years in 8 patients who died from the disease of a total of 11 cases). It is therefore probable that the mutation strongly contributes to the development of aggressively growing cutaneous melanoma.
Griewank et al (2013a) have addressed whether the same mutations are present also in ocular melanomas reporting an incidence of 32% in conjunctival melanomas and the absence from uveal (ciliary body or choroid) melanomas (n=47).
In the present study, we describe the analysis of TERT promoter mutations in a series of 50 uveal melanomas. A mutation was detected in a single case that also carried mutations in GNA11 and EIF1AX, typical for this disease and absent from conjunctival and cutaneous melanomas. The tumour showed disomy of chromosome 3 and had wild-type sequences of GNAQ, BAP1 and SF3B1 which frequently show mutations in uveal melanoma.
Materials and methods
Patients
Tissue samples were obtained from 52 primary uveal melanomas after enucleation surgery upon approval of the institutional bioethics board and informed written consent of the patients. Clinical, pathological and molecular characteristics and follow-up data are reported in Supplementary Table 1.
DNA/RNA extraction, array comparative genome hybridisation, MPLA, gene expression profiling and Sanger sequencing
DNA was extracted from macrodissected tumour material conserved in RNAlater (Ambion, Monza, Italy) or from archival formalin-fixed, paraffin-embedded (FFPE) blocks using QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany). For FFPE samples, two 10-μm-thick sections were cut from each paraffin block and subjected to de-paraffinisation using scalar concentrations (100% to 40%) of Histoclear (Sigma Aldrich, Milan, Italy), followed by re-hydration and overnight digestion with proteinase K (100 μg ml−l, Sigma Aldrich) at 37 °C. DNA concentration and quality were checked in the Nanodrop ND-1000 spectrophotometer and integrity of DNA was tested by agarose gel electrophoresis. Processing of genomic DNA was performed using the GeneChip Mapping 250 K Assay Kit (Affymetrix, Santa Clara, CA, USA) as recently described (Amaro et al, 2013). Briefly, 250 ng of DNA sample were digested with the restriction enzyme NspI. Adapters were ligated using T4 DNA Ligase. Whole genome amplification was performed on a GeneAmp PCR System 9700 (Applied Biosystems–Life Technologies Corporation, Monza, Italy). Ninety micrograms of amplified and normalised PCR product were fragmented and labelled. Hybridisation, washing, staining and scanning of SNP arrays were performed on the Affymetrix fluidic station. Quality of the samples was assessed on agarose gels before the hybridisation step. Affymetrix Genotyping Console (GTC4.1.2) was used to perform genotype call and quality control assessments. Copy number analysis was performed using CNAG3.0 (Yamamoto et al, 2007).
Multiplex ligation-dependent probe amplification (MLPA) was performed according to the manufacturer's instructions (MRC-Holland, Amsterdam, The Netherlands). In short, 5 μl of genomic DNA diluted in TE at a concentration of 20 ng ml−1 were denatured at 98 °C for 5 min, cooled to 25 °C and 3 μl of a 1 : 1 mixture of MLPA buffer and SALSA P027.C1 probe-mix was added. After hybridisation for 16 h at 60 °C, 32 μl ligation mix were added, and the reaction was incubated for 15 min at 54 °C followed by 5 min at 98 °C. Subsequently, 10 μl of the SALSA PCR-mix (FAM label) were added to 40 μl of ligation product and this was amplified by PCR in 35 cycles (30 s, 95 °C; 30 s 60 °C; 60 s 72 °C). PCR products were quantified using the ABI 3130XL (Applied Biosystems-Life Technologies Corporation, Milan, Italy) and coffalyser.net software (MRC-Holland). Raw data were expressed as peak heights, as a measure of peak intensity, for each of the 50 probes (38 test probes and 12 control probes). For each probe, the relative ratio was calculated using population normalisation and using the internal control probe normalisation. The MLPA data were considered reliable if six or more control probes were within the normal range. Loss was identified as MLPA relative ratio <0.70 and gain as MLPA relative ratio >1.3.
For RNA extraction, tumour samples were homogenised in the tissue lyserMixer Mill (Qiagen) in total RNA extraction lysis buffer using RNeasy (Qiagen). RNA quality was assessed in the BioAnalyser (Agilent, St Clara, CA, USA). Gene expression profiling was performed using Affymetrix HGU133plus2 arrays following standard procedures as described (Gangemi et al, 2012). Assignment to prognostic gene expression classes was performed by nearest neighbour classification using the gene expression values for the discriminator genes described (Onken et al, 2004).
PCRs were carried out using 25 ng of genomic DNA in a 25 μl reaction mix including 10 × Platinum PCR Supermix, 1.5 mM MgCl2, 200 μM dNTPs, 1 μM primers and 0.5U Taq Platinum (Invitrogen–Life Technologies Corporation, Monza, Italy) on a Veriti 96 wells Thermal Cycler (Applied Biosystems–Life Technologies Corporation). Thermal cycling conditions were as follows: 94 °C for 4 min, followed by 25 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s and a final extension at 72 °C for 7 min. PCR conditions were slightly modified for EIF1AX gene (2 mM MgCl2 were used for exon 1 and the annealing temperature was 54 °C for exon 1 and 64 °C for exon 2).
Telomerase reverse transcriptase promoter region oligonucleotide primers were synthesised according to the Eukaryotic Promoter Database genomic reference sequence of TERT and designed to amplify a portion of the TERT core promoter (−144 to +43). The amplicon of 187 bp includes the two major recurrent mutations at coordinates chr 5 : 1 295 228 and chr 5: 1 295 250. PCR were performed by using 100–200 ng of genomic DNA in a 50 μl reaction mix with 400 μM dNTPs (Fermentas-Thermo Scientific Life Science, Milan, Italy), 0.75 μM primers, 2 mM MgCl2, 1X Buffer and 2.5 U AmpliTaq Gold 360 DNA Polymerase (Applied Biosystems-Life Technologies Corporation) on a Veriti 96 wells Thermal Cycler (Applied Biosystems, Life Technologies Corporation). Thermal cycling conditions were as follows: 95 °C for 7 min, followed by 42 cycles of 95 °C for 30 s, 56 °C for 30 s, 72 °C for 30 s, and a final extension at 72 °C for 7 min.
All PCR primers (see Supplementary Table 2) except for TERT were designed with a universal sequence at 5′-end (universal forward primer 5′-GTTGTAAAACGACGGCCAGT-3′ and M13 (−48) reverse primer 5′-GTGTGAAATTGTTATCCGCT-3′) to perform single-pass sequencing. Additional primers were synthesised for FFPE samples that did not yield any amplicon using the first set of primers. Mutational screening was carried out by direct sequencing of fragments obtained by PCR using an ABI3130xl and ABI3730 Genetic Analyzer (Applied Biosystems–Life Technologies Corporation). Sequencing data were analysed using SeqScape v2.5 software (Applied Biosystems–Life Technologies Corporation) and MacVector V11 software (MacVector Inc., Cary, NC, USA).
Transcription factor binding analysis was performed using TFSEARCH based on the TRANSFAC database (Heinemeyer et al, 1998).
Results
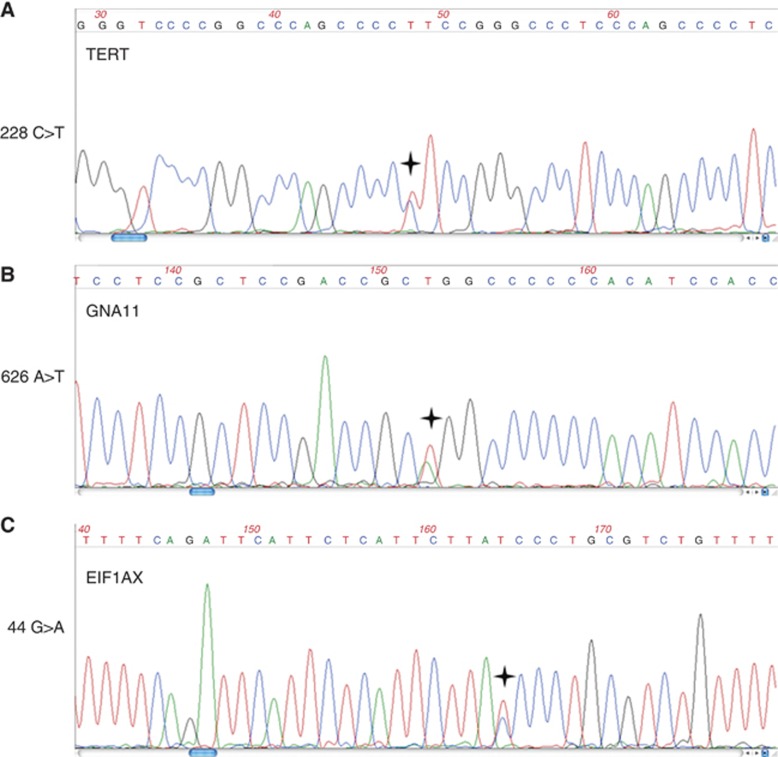
From the available 52 tumours, sufficient amounts of DNA could be obtained from 51 tumours and 50 of which could be successfully sequenced for TERT. One (MU076) of these 50 cases showed a heterozygous mutation in the promoter of TERT, a C to T transition on chromosome 5 in position 1 295 228 (Figure 1A) that increases the likelihood of the sequence to bind ETS transcription factors from 78.4 (wild type) to 86.3 (mutation) as analysed by TFSEARCH and has been shown to drive TERT promoter activity (Huang et al, 2013). The same tumour showed a heterozygous GNA11 mutation in exon 5 (Figure 1B) and wild-type sequences for BAP1 and SF3B1 (data not shown). EIF1AX exon 2 showed a mutated sequence in addition to the wild-type allele (Figure 1C).
Figure 1.
Mutation analysis by Sanger sequencing. The genes TERT (A), GNA11 (B) and EIF1AX (C) were found to be mutated as indicated on the left of each diagram by Sanger sequencing using the primers enlisted in Supplementary Table 2. The asterisk indicates the mutated position. No mutations have been detected in GNAQ, BAP1 and SF3B1 (see text).
The mutation spectrum of genes that are frequently mutated in uveal melanoma is compatible with the frequencies observed in previous reports (Martin et al, 2013; Griewank et al, 2013b). EIF1AX mutations exclusively occurred in disomic, non-metastatic cases. One of the three SF3B1 mutations detected occurred in a metastatic case with uniparental disomy of chromosome 3. GNAQ and GNA11 mutations accounted for ∼84% of the cases. GNAQ was more frequently observed in disomic cases, GNA11 more frequently in monosomic cases. BAP1 was significantly more frequent in monosomic cases, however, due to several monosomic, progression-free cases with long follow-up, BAP1 was not significantly associated with metastasis. GNAQ was inversely associated with metastasis (see Table 1).
Table 1. Mutation frequencies.
| Mutation | Total (%) | Disomic cases (%) | Monosomic cases (%) |
|---|---|---|---|
|
GNAQ (n=45) |
19 (42.2) |
14 (73.7) |
5 (26.3) |
|
GNA11 (n=46) |
15 (32.6) |
6 (40.0) |
9 (60.0) |
|
BAP1 (n=38) |
12 (31.5) |
0 (0.0) |
12 (100.0) |
|
SF3B1 (n=31) |
3 (9.7) |
3 (100.0) |
0 (0.0) |
|
EIF1AX (n=37) |
7 (18.9) |
7 (100.0) |
0 (0.0) |
| TERT (n=50) | 1 (2.0) | 1 (100.0) | 0 (0.0) |
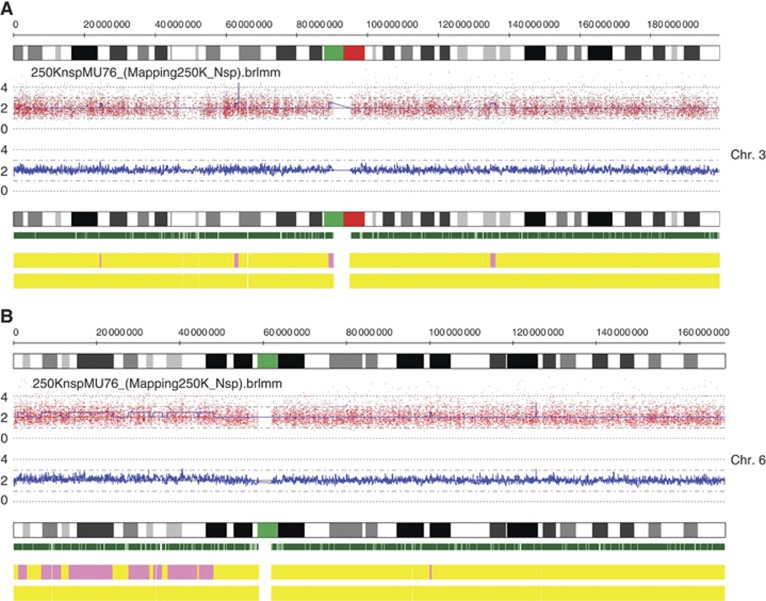
Cytogenetic analyses divide uveal melanomas into two distinct patterns: the first, detected in ∼50% of uveal melanomas, characterised by loss of chromosome 3, often associated with chromosome 8 gain and strongly linked to metastatic disease. The second pattern accounts for 25% of uveal melanomas and is characterised by chromosome 6p gain, often associated with 6p loss but rarely with chromosome 3 loss (Hoglund et al, 2004). Copy number alteration analysis of the uveal melanoma carrying the TERT promoter mutation using Affymetrix 250K SNP arrays revealed disomy of chromosome 3 and copy number gain of the short arm of chromosome 6. No extended regions of homozygosity compatible with uniparental disomy of chromosome 3 could be identified (Figure 2).
Figure 2.
Copy number alteration of chromosomes 3 and 6. DNA extracted from the tumour sample was analysed by arrayCGH on Affymetrix 250 K SNP arrays. Chromosome 3 (A) is present with two copies that, based on SNP analysis, are heterozygous. Short amplified stretches but no deletions are observed. Chromosome 6q (B) shows extended stretches with presumably three copies. On top of each diagram the chromosome bands are represented. The plot below shows the actual intensities; the blue plot corresponds to the copy number call applying a five SNP window. The green vertical bars show the positions of the single probe sets of the array, the upper yellow bar indicates disomy (=yellow) or gain (=pink) and the lower bar indicates probability of loss of heterozygosity (<30%=yellow).
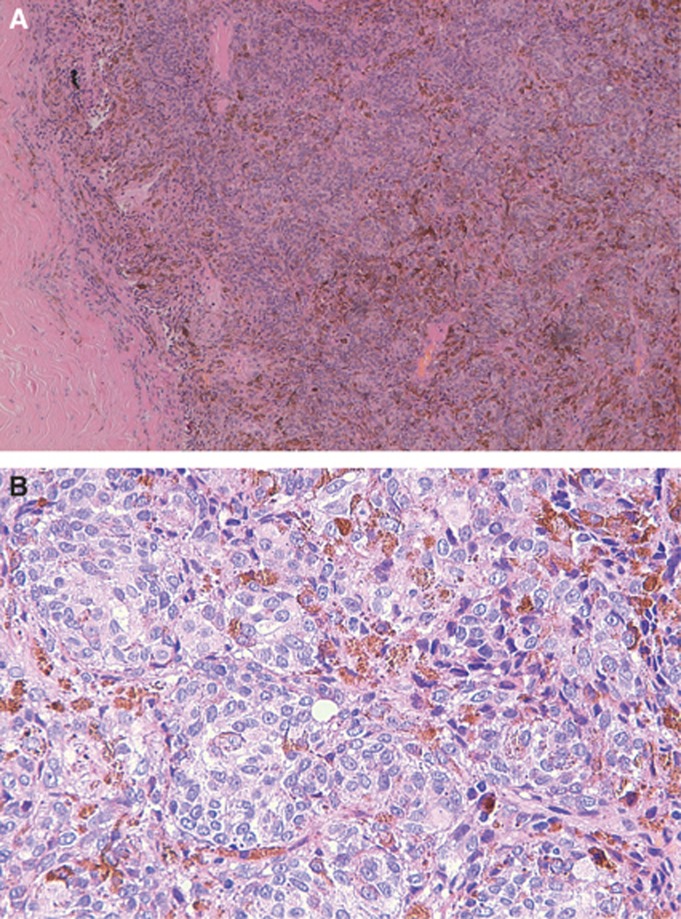
The tumour occurred in the posterior chamber of the right eye of a 57-year-old male patient of European ancestry, had a depth of 11.97 mm and a width of 12.62 mm, showed the characteristic mushroom shape, epitheloid histology and a low mitotic index (Figure 3). The tumour was removed by enucleation of the eye. The patient is free of disease 31 months after diagnosis.
Figure 3.
Clinical and histopathological features of the tumour carrying the TERT promoter mutation. (A) Solid growth pattern of atypical epitheloid melanocytes. Irregular distribution of melanin. (B) Confluent nests of atypical epitheloid melanocytes. Very rare mitoses.
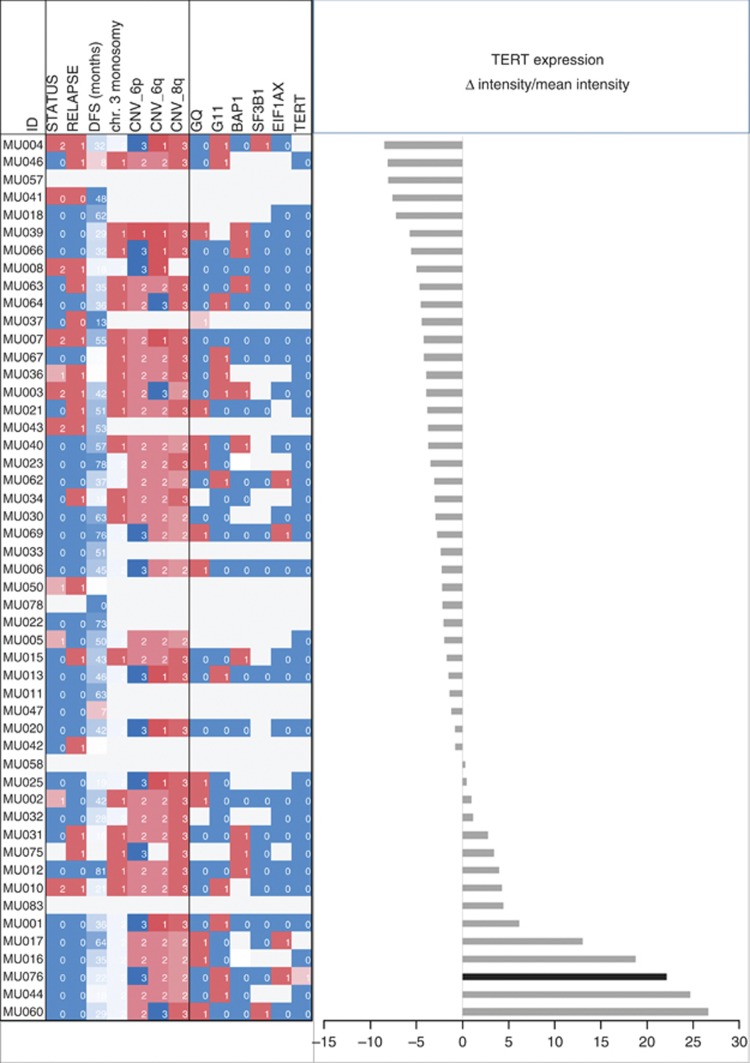
The application of the prognostic gene set described by Onken et al (2004) to this case classifies it as class 1 tumour associated with low risk. The expression of TERT as revealed by microarray gene expression data shows that the tumour carrying the mutation, as well as several other tumours with wild-type TERT promoters, has elevated levels of TERT expression as compared with the mean of TERT expression in the whole cohort (Figure 4). The six tumours with the highest TERT expression levels did not metastasise (mean follow-up 33.3 months, range 16–64 months). Four of these tumours have two chromosomes 3, this information is not available for the other two. However, despite this trend, high TERT expression levels are not significantly associated with disease-free survival (P=0.338) or other clinical, pathological or molecular features of the tumours (Figure 4; supplementary table 1).
Figure 4.
TERT gene expression and tumour characteristics. The differences between microarray intensity values measured for the single cases and the mean of the whole cohort are plotted (right panel). The case carrying the TERT promoter mutation is indicated in black. Clinical, histopathological and molecular characteristics are indicated (left panel): status (0=alive, 1=death of other causes, 2=death of uveal melanoma metastasis), relapse (0=relapse free, 1=relapse), CNVs (1=monosomic or losses, 2=disomic, 3=trisomic or gains), mutations (0=wild type, 1=mutated). For details see Supplementary Table 1.
Discussion
The analysis of the recently identified uveal melanoma driver mutations in the genes GNAQ, GNA11, BAP1, SF3B1 and EIF1AX generally confirms the observed mutation frequencies and the prevalence of SF3B1 and EIF1AX mutations in disomic, non-metastatic cases. Also the mutual exclusive mutations of GNAQ and GNA11 and the association of the latter with a more aggressive tumour phenotype could be confirmed. Mutations of the tumour suppressor gene BAP1 are associated with monosomy of chromosome 3 yet in our cohort, there are several cases without metastases despite monosomy of chromosome 3 and BAP1 mutation. Although this is probably due to chance it underlines the limited prognostic value of mutation analyses.
In addition, we show here a case of uveal melanoma that carries one of the two mutations in the promoter region of TERT that have been described for sporadic cutaneous melanoma that closely resembles cutaneous melanoma with which it shares BRAF and NRAS mutations (Griewank et al, 2013a) (and references therein). In fact, TERT mutations in conjunctival melanomas also occurred in concomitance with BRAF and NRAS mutations (Griewank et al, 2013a). However, Griewank et al (2013a) did not detect TERT promoter mutations in 47 uveal melanomas. Hence, the TERT mutation identified here has an estimated frequency of 1 in 97 (∼1%) in uveal melanoma. The tumour carrying this mutation can clearly be identified as a uveal melanoma as it was located in the posterior chamber of the eye, showed the typical epitheloid morphology and carries a mutation in the GNA11 gene. GNA11 mutations occur in 33.2% of uveal melanomas and in 6.5% of blue nevi but are absent from other nevi (n=105) and extraocular melanomas (n=273) (Van Raamsdonk et al, 2010; see Supplementary Table 2 for mutation frequencies of GNAQ, GNA11, BAP1, SF3B1 and EIF1AX as obtained from the Catologue of somatic mutations in Cancer (COSMIC) database; http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/). We can therefore be certain of the uveal origin of the tumour that carries the TERT mutation.
The mutation in EIF1AX has also been shown to be frequent in uveal melanoma. EIF1AX is located on the X chromosome and, being the patient of male sex (confirmed by analysis of X chromosome number by Devyser QF-PCR and several Y-chromosome markers; data not shown), the appearance of two alleles can most likely be attributed to contaminating non-tumoral cells present in the sample or to the presence of the mutation in only a subclone of the tumour. The latter appears more likely as the presence of non-tumoral components is not evident for GNA11 and TERT mutations. We can exclude contamination with the Y-chromosome paralogue, EIF1AY, as the primers used are identical to those described by Martin et al (2013) and do not amplify this gene. The occurrence of an EIF1AX mutation in a disomic case with a GNA11 mutation but without BAP1 and SF3B1 mutations is consistent with the original report of this mutation (Martin et al, 2013).
Huang et al (2013) have shown that the TERT promoter mutation determines increased promoter activity in a luciferase reporter system. We show here for the first time that the mutation in the promoter is actually associated with increased expression of the gene in the tumour. However, several wild-type tumours also show increased TERT expression indicating that there are other means to increase the expression of the catalytic subunit of human telomerase. Telomere maintenance might therefore be important in at least a small subgroup of uveal melanomas. Increased TERT expression is expected to facilitate immortalisation of somatic cells as an essential step in neoplastic transformation as the replicative potential of normal cells is limited by telomere shrinkage (Bodnar et al, 1998). Telomerase reverse transcriptase promoter mutations have been observed in a variety of tumours and it has been speculated that these mutations could be more frequent in tumours derived from tissues with low self-renewal where tumour development might be hindered by the inherently lower telomerase activity (Killela et al, 2013). Telomere shrinkage can also be overcome in cancer cells by mutations in the genes alpha thalassemia/mental retardation syndrome X-linked (ATRX) or death-domain-associated protein (DAXX) leading to recombination-mediated telomere maintenance (Heaphy et al, 2011). No such mutations have been found by exome sequencing of uveal melanomas (Griewank et al, 2013a).
The case carrying this mutation shows disomy of chromosome 3 and wild-type status of the tumour suppressor gene BAP1 that is frequently mutated in tumours that undergo metastasis (Harbour et al, 2010) and that show a chromosome 3 monosomy-associated gene expression profile (Onken et al, 2004, 2012). The epitheloid morphology of the tumour cells is associated with increased risk of metastasis (Singh et al, 2001). Given the strong prognostic effect of chromosome 3 monosomy, this tumour would be considered as at low risk of metastasis despite epitheloid morphology and tumour dimensions. The patient whose uveal melanoma carried the TERT mutation is free of disease at 31 months after diagnosis but an effect of the mutation in the TERT promoter on the metastasis risk cannot be definitely excluded. No information on the association of TERT promoter mutations with prognosis is available, not even for cutaneous melanoma as this has not been addressed by the original studies (Horn et al, 2013; Huang et al, 2013). The mutation detected in the melanoma family showed high penetrance and occurred in aggressive melanomas with early onset and short survival after diagnosis even in some cases of the last generation where present day management of the disease can be assumed (Horn et al, 2013). Yet this specific mutation is different from those detected in sporadic cases and in the present case of uveal melanoma and the precise location within the promoter and/or the location with respect to other promoter elements could affect the extent of promoter activation. On the other hand, several cell lines derived from metastatic melanomas showed the mutation identified in sporadic cases independently of known driver mutations consistent with driver function for the TERT promoter mutation (Huang et al, 2013). It is therefore possible that the mutation influences the overall risk of metastasis.
If so, this mutation could be associated with the rare cases of uveal melanoma with disomy and BAP1 wild type that develop metastases and that, though rare, determine pitfalls in prognostic testing (Zeschnigk and Lohmann, 2013). GNAQ and GNA11 mutations are not sufficient for the generation of a metastatic phenotype of uveal melanoma (Van Raamsdonk et al, 2009, 2010). In most cases, the metastatic phenotype is acquired only after loss of one copy of chromosome 3 and BAP1 mutation (Coupland et al, 2013; Harbour, 2013; Zeschnigk and Lohmann, 2013). The association of the GNA11 mutation with another driver mutation in TERT might drive a more aggressive phenotype independently of chromosome 3 monosomy and BAP1 mutation. Analysis of TERT should therefore be included in the assessment of uveal melanomas in order to reveal its real frequency and to investigate into its association with metastasis.
Acknowledgments
This work was funded by grants from the Compagnia di San Paolo and from the Regione Liguria to UP. AA is the recipient of a fellowship PO CRO Fondo Sociale Europeo Regione Liguria 2007–2013 Asse IV ‘Capitale Umano'.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Amaro A, Mirisola V, Angelini G, Musso A, Tosetti F, Esposito AI, Perri P, Lanza F, Nasciuti F, Mosci C, Puzone R, Salvi S, Truini M, Poggi A, Pfeffer U. Evidence of epidermal growth factor receptor expression in uveal melanoma: inhibition of epidermal growth factor-mediated signalling by Gefitinib and Cetuximab triggered antibody-dependent cellular cytotoxicity. Eur J Cancer. 2013;49 (15:3353–3365. doi: 10.1016/j.ejca.2013.06.011. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279 (5349:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Coupland SE, Lake SL, Zeschnigk M, Damato BE. Molecular pathology of uveal melanoma. Eye. 2013;27 (2:230–242. doi: 10.1038/eye.2012.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furney SJ, Turajlic S, Stamp G, Nohadani M, Carlisle A, Thomas JM, Hayes A, Strauss D, Gore M, van den Oord J, Larkin J, Marais R. Genome sequencing of mucosal melanomas reveals that they are driven by distinct mechanisms from cutaneous melanoma. J Pathol. 2013;230 (3:261–269. doi: 10.1002/path.4204. [DOI] [PubMed] [Google Scholar]
- Gallagher RP, Elwood JM, Rootman J, Spinelli JJ, Hill GB, Threlfall WJ, Birdsell JM. Risk factors for ocular melanoma: Western Canada Melanoma Study. J Natl Cancer Inst. 1985;74 (4:775–778. [PubMed] [Google Scholar]
- Gangemi R, Mirisola V, Barisione G, Fabbi M, Brizzolara A, Lanza F, Mosci C, Salvi S, Gualco M, Truini M, Angelini G, Boccardo S, Cilli M, Airoldi I, Queirolo P, Jager MJ, Daga A, Pfeffer U, Ferrini S. Mda-9/syntenin is expressed in uveal melanoma and correlates with metastatic progression. PLoS One. 2012;7 (1:e29989. doi: 10.1371/journal.pone.0029989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griewank KG, Murali R, Schilling B, Scholz S, Sucker A, Song M, Susskind D, Grabellus F, Zimmer L, Hillen U, Steuhl KP, Schadendorf D, Westekemper H, Zeschnigk M. TERT promoter mutations in ocular melanoma distinguish between conjunctival and uveal tumours. Br J Cancer. 2013;109 (2:497–501. doi: 10.1038/bjc.2013.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griewank KG, van de Nes J, Schilling B, Moll I, Sucker A, Kakavand H, Haydu LE, Asher M, Zimmer L, Hillen U, Thompson JF, Scolyer RA, Schadendorf D, Murali R.2013Genetic and clinico-pathologic analysis of metastatic uveal melanoma Mod Pathole-pub ahead of print 26 July 2013; doi: 10.1038/modpathol.2013.138 [DOI] [PubMed]
- Griewank KG, Westekemper H, Murali R, Mach M, Schilling B, Wiesner T, Schimming T, Livingstone E, Sucker A, Grabellus F, Metz C, Suesskind D, Hillen U, Speicher MR, Woodman SE, Steuhl KP, Schadendorf D. Conjunctival melanomas harbor BRAF and NRAS mutations and copy number changes similar to cutaneous and mucosal melanomas. Clin Cancer Res. 2013;19:3143–3152. doi: 10.1158/1078-0432.CCR-13-0163. [DOI] [PubMed] [Google Scholar]
- Guenel P, Laforest L, Cyr D, Fevotte J, Sabroe S, Dufour C, Lutz JM, Lynge E. Occupational risk factors, ultraviolet radiation, and ocular melanoma: a case-control study in France. Cancer Causes Control. 2001;12 (5:451–459. doi: 10.1023/a:1011271420974. [DOI] [PubMed] [Google Scholar]
- Harbour JW. Genomic, prognostic, and cell-signaling advances in uveal melanoma. Am Soc Clin Oncol Educ Book. 2013;2013:388–391. doi: 10.1200/EdBook_AM.2013.33.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, Council ML, Matatall KA, Helms C, Bowcock AM. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330 (6009:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013;45 (2:133–135. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, Offerhaus GJ, McLendon R, Rasheed BA, He Y, Yan H, Bigner DD, Oba-Shinjo SM, Marie SK, Riggins GJ, Kinzler KW, Vogelstein B, Hruban RH, Maitra A, Papadopoulos N, Meeker AK. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333 (6041:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26 (1:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L. A landscape of driver mutations in melanoma. Cell. 2012;150 (2:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoglund M, Gisselsson D, Hansen GB, White VA, Sall T, Mitelman F, Horsman D. Dissecting karyotypic patterns in malignant melanomas: temporal clustering of losses and gains in melanoma karyotypic evolution. Int J Cancer. 2004;108 (1:57–65. doi: 10.1002/ijc.11558. [DOI] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, Schadendorf D, Kumar R. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339 (6122:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339 (6122:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, Grollman AP, He TC, He Y, Hruban RH, Jallo GI, Mandahl N, Meeker AK, Mertens F, Netto GJ, Rasheed BA, Riggins GJ, Rosenquist TA, Schiffman M, Shih IeM, Theodorescu D, Torbenson MS, Velculescu VE, Wang TL, Wentzensen N, Wood LD, Zhang M, McLendon RE, Bigner DD, Kinzler KW, Vogelstein B, Papadopoulos N, Yan H. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA. 2013;110 (15:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Masshofer L, Temming P, Rahmann S, Metz C, Bornfeld N, van de Nes J, Klein-Hitpass L, Hinnebusch AG, Horsthemke B, Lohmann DR, Zeschnigk M. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45 (8:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onken MD, Worley LA, Char DH, Augsburger JJ, Correa ZM, Nudleman E, Aaberg TM, Jr, Altaweel MM, Bardenstein DS, Finger PT, Gallie BL, Harocopos GJ, Hovland PG, McGowan HD, Milman T, Mruthyunjaya P, Simpson ER, Smith ME, Wilson DJ, Wirostko WJ, Harbour JW. Collaborative Ocular Oncology Group report number 1: prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology. 2012;119 (8:1596–1603. doi: 10.1016/j.ophtha.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64 (20:7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane AR, Hirst LW. Ultraviolet light exposure as a risk factor for ocular melanoma in Queensland, Australia. Ophthal Epidemiol. 2000;7 (3:159–167. [PubMed] [Google Scholar]
- Singh AD, Rennie IG, Seregard S, Giblin M, McKenzie J. Sunlight exposure and pathogenesis of uveal melanoma. Surv Ophthalmol. 2004;49 (4:419–428. doi: 10.1016/j.survophthal.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Singh AD, Shields CL, Shields JA. Prognostic factors in uveal melanoma. Melanoma Res. 2001;11 (3:255–263. doi: 10.1097/00008390-200106000-00007. [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O'Brien JM, Simpson EM, Barsh GS, Bastian BC. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457 (7229:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, Obenauf AC, Wackernagel W, Green G, Bouvier N, Sozen MM, Baimukanova G, Roy R, Heguy A, Dolgalev I, Khanin R, Busam K, Speicher MR, O'Brien J, Bastian BC. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363 (23:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgili G, Gatta G, Ciccolallo L, Capocaccia R, Biggeri A, Crocetti E, Lutz JM, Paci E. Incidence of uveal melanoma in Europe. Ophthalmology. 2007;114 (12:2309–2315. doi: 10.1016/j.ophtha.2007.01.032. [DOI] [PubMed] [Google Scholar]
- Yamamoto G, Nannya Y, Kato M, Sanada M, Levine RL, Kawamata N, Hangaishi A, Kurokawa M, Chiba S, Gilliland DG, Koeffler HP, Ogawa S. Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotide-polymorphism genotyping microarrays. Am J Hum Genet. 2007;81 (1:114–126. doi: 10.1086/518809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeschnigk M, Lohmann DR.2013Prognostic testing in uveal melanomaIn: Pfeffer U (ed). Cancer Genomics: Molecular Classification, Prognosis And Response Prediction Chapter 3Springer Science and Business Media: Dordrecht, The Netherlands; 79–96. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.