Abstract
The completion of the Human Genome Project was a landmark achievement, but as clinical genetic testing becomes more mainstream, the extent of remarkable genetic variation is increasingly being appreciated. Newer DNA sequencing technology can now complete the sequencing of an entire human genome several times over in a matter of days, but this will undoubtedly add new challenges to the difficulty of distinguishing true pathogenic variants from benign variants in diagnostic genetics and in the research setting. The recent discovery of the role of titin gene (TTN) mutations in dilated cardiomyopathy (DCM) will make genetic testing in this disease more efficient. Furthermore, better understanding of genotype-phenotype associations will assist clinicians in identifying early stages of disease and providing more appropriate treatments. This high level of complexity requires an expert genetic team to offer counseling and to manage, deliver, and follow-up over time the results of genetic testing, which is particularly important for screening of family members potentially at risk. In DCM, genetic testing may be useful for the identification of non-carriers and asymptomatic carriers, as well as for prevention strategies, sport recommendations, and defibrillator implantation. It can also guide reproductive decision-making including utilization of pre-implantation genetic diagnostic strategies.
Introduction
Dilated cardiomyopathy (DCM) is the most common form of heart muscle disease and a leading cause of cardiac transplantation (Taylor et al., 2007). For many years, the cause of the disease has been considered unknown, possibly autoimmune, or due to a viral myocarditis, in some cases running in families and considered to be of genetic origin only in rare instances. However, the role of genetic factors has gained more importance over the last two decades, with the progressive discovery of a higher frequency of familial cases than previously anticipated (up to 30–50%), a large number genes (over 40) and gene mutations associated with the disease phenotype, and a clear evidence that even the so called “sporadic” cases may harbor a genetic defect (Hershberger et al., 2009). In particular in the last few years, impressive technological advances, large multicenter studies, and novel insights into the phenotype have significantly expanded our understanding of the origin of this common disorder and the opportunity for molecular diagnosis (Mestroni and Taylor, 2011).
Genes and DCM: a New Role for Titin Gene in Dilated Cardiomyopathy
Initially, when the first genetic causes of cardiomyopathies were identified, it was a common understanding that while hypertrophic cardiomyopathy (HCM) was a disease caused by sarcomere dysfunction, DCM was a disease localized to the cytoskeleton (Towbin, 1998). Instead, it is clear now that the complexity of DCM is well beyond this simplistic hypothesis: genetic research over the past decade has shown that mutations in virtually every structure and pathway of the cardiac myocyte may lead to a dilated and poorly contracting ventricle. Currently, over 40 genes have been associated with DCM; most contributing to a low frequency of the prevalence of DCM, with a few notable exceptions accounting for 5–25% of cases (Sinagra et al., 2012). However, until recently, the translation of this knowledge into the clinical practice was significantly limited by the lack of DNA sequencing technology capable of interrogating dozens of genes in an efficient and affordable manner. According to current guidelines, to be considered for clinical genetic testing, disease genes should be associated with a change in management either in the patient or in his relatives at risk, and a test sensitivity of at least 40% (Ackerman et al., 2011). However, in DCM the expected test sensitivity was approximately only 20% (Lakdawala et al., 2012a), and the technical limitations of the traditional sequencing methods in screening such a large number of low frequency genes made virtually impossible a comprehensive genetic testing. Only recently, the discovery of a high frequency gene (titin) and the availability of high throughput technologies in clinical laboratories (next-generation sequencing) have made clinical testing feasible and comprehensive also in DCM.
The identification of the role of the sarcomeric gene titin has been a major advance in the study of DCM (Herman et al., 2012; McNally, 2012). The giant titin (TTN) gene (~350 exons) encodes the largest-known human protein (~35,000 amino acids, 2 µm long). It is highly expressed in the heart, where it functions as a giant spring, provides passive force, and regulates sarcomere contraction and signaling. Because of the giant size of the gene, after the initial reports of Gerull et al. (2002), TTN mutations were known in only a few families with DCM, but the TTN mutation frequency and therefore clinical impact in the overall DCM population were unknown.
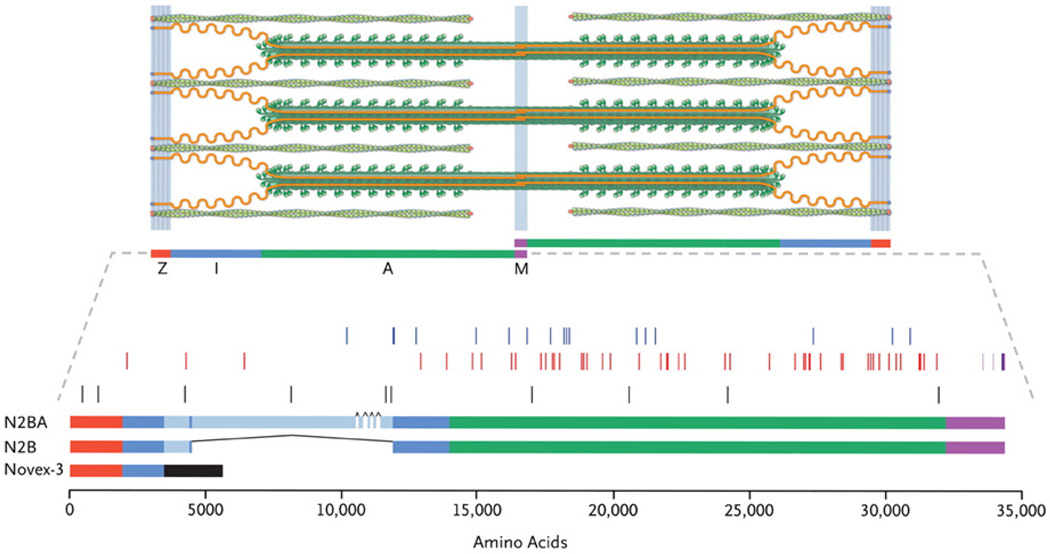
Recently, a multicenter group of investigators from Harvard, University of Colorado, University of Trieste, the Imperial College of London, Mayo Clinic at Rochester, and the University of Washington Seattle answered this question (Herman et al., 2012). The authors studied a large cohort of subjects (792) including 312 subjects with DCM, 231 subjects with HCM, and 249 controls: patients were screened either by next-generation sequencing in the Harvard’s laboratory, or by direct sequencing in the Human Genome Center of Seattle. The results were surprising: of the 72 unique “radical” mutations (Figure 1) found in the TTN gene (25 nonsense, 23 frameshift, 23 splicing, and 1 large CNV), the majority were in the DCM group (over 20%), while in HCM the frequency was similar to the control group (in only 1% and 3%, respectively). In DCM, TTN mutations were over-represented in the titin A-band and absent from the Z-disk and M-band (P≤0.01), suggesting a domain-specific dominant-negative effect, rather than haploinsufficiency: in other words, the mutant protein is incorporated in the sarcomere, but unable to function normally. Furthermore, DCM TTN mutations truncated the protein’s C-terminal kinase domain probably impairing signaling, and some of them were predicted to disrupt recoil. Genotype-phenotype analysis showed a high penetrance (>95%) after 40 years, but no other significant difference except some unusual nuclei on histology and a worse prognosis in male than female carriers (P=4×10−5). Finally, TTN mutations were found both in ~25% of familial DCM and in 18% of sporadic DCM, a difference that was not statistically significant.
Figure 1.
Titin mutations in DCM. Top: the cardiac sarcomere including titin (orange), the thick filaments (green rods with globular heads) and thin filaments (green coiled ovals), Z-disk (red), I-band (blue), A-band (green), and M-band (purple). Middle: DCM TTN splicing and copy-number mutations (blue), nonsense and frameshift mutations (red); truncating mutations in controls and subjects with hypertrophic cardiomyopathy (black), truncating mutations in congenital myopathy (light purple) or limb-girdle muscular dystrophy (dark purple). Bottom: TTN isoforms and sequence variants. From Herman et al., with permission (Herman et al., 2012).
The important role of TTN in the pathogenesis of the dilated phenotype is not surprising: in a recent work, Taylor et al. (2011) found a significant TTN mutation frequency in arrhythmogenic right ventricular cardiomyopathy (ARVC). Furthermore, functional investigations on a novel DCM gene, RBM20, showed that this protein is a key component of the titin pathway by allowing the alternative splicing and post-translational regulation of TTN (Guo et al., 2012). Overall, these discoveries suggest an important role of this gene in the origin of dilated cardiomyopathies.
Advances in the Phenomics of Dilated Cardiomyopathy: from Better Diagnosis to Better Risk Stratification
Recent studies have also brought more insights into the association between the genotype and the phenotype in DCM, information that is eventually critical in the management of genetic testing as we discuss below. McNair et al. (2011) investigated the frequency and type of cardiac sodium channel (SCN5A) mutations in a large DCM cohort. The frequency of rare variants was found in the expected range of 2% and not surprisingly, the carriers of SCN5A putative mutations had a remarkable tendency toward an arrhythmogenic trait, including conduction defects, atrial fibrillation, and ventricular tachycardia, while the prevalence of arrhythmia was significantly lower in non-carrier DCM patients. In the clinical setting, these features may be “red flags” of channelopathy-related DCM and may represent a useful tool in the clinical setting when estimating the pathogenicity of a novel SCN5A rare variant in patients with DCM.
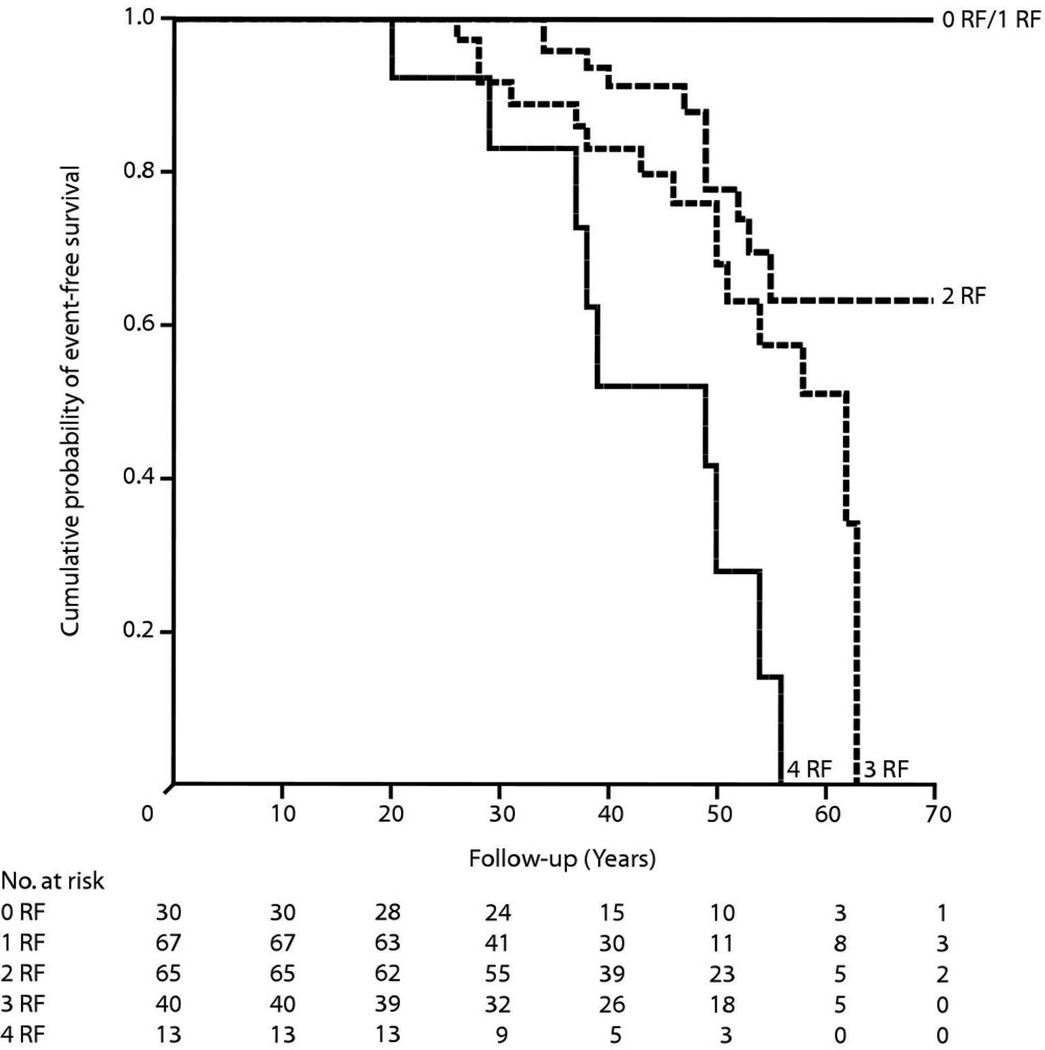
Another recent study has examined the incidence of malignant arrhythmia in a cohort of carriers of lamin A/C (LMNA) gene mutations (van Rijsingen et al., 2012). LMNA encodes the two intermediate filaments lamin A and C, which provide the backbone network supporting the nuclear wall in cells in heart, muscle, and other organs. The phenotype resulting from LMNA mutations can range from an isolated cardiomyopathy to a muscular dystrophy, and frequently these clinical features can produce “overlapping phenotypes,” again a precious information when evaluating the possible pathogenic role of a rare variant in a patient with DCM. LMNA mutations occur in approximately 8% of cases and in these cases a poor prognosis and a risk of sudden death were known (Meune et al., 2006; Taylor et al., 2003). In their study, van Rijsingen et al. (2012) collected a large multicenter European cohort of DCM patient carriers of LMNA mutations and examined their arrhythmic events during a mean follow up of 4 years. The investigators found that life-threatening events occurred in 18% of cases, and that they were associated with 4 independent risk factors for malignant ventricular arrhythmia (MVA): non-sustained ventricular tachycardia, left ventricular ejection fraction less than 45%, a truncation mutation, and the male gender. Finally, they discovered that the four risk factors had a cumulative effect which increased as more risk factors were present, ranging from a normal survival if zero or one risk factor was present, to 50% mortality in two years at less than 40 years of age if all four were present, even in the absence of severe left ventricular dysfunction (Figure 2).
Figure 2.
Survival in patients with LMNA mutations. Kaplan-Meier event-free survival from the date of first visit stratified by 4 independent risk factors (RF): non-sustained ventricular tachycardia, left ventricular ejection fraction <45% at the first visit to the cardiologist, being male, and truncation mutations. The event was defined as occurrence of malignant ventricular arrhythmias (appropriate implantable cardioverter-defibrillator treatment, cardiopulmonary resuscitation, or sudden cardiac death). From van Rijsingen et al., with permission (van Rijsingen et al., 2012).
Finally, another recent study has focused on the phenotype of early “subclinical” stages of disease (LVEF ≥55%) (Lakdawala et al., 2012b). Lakdawala et al. studied a cohort of 62 subjects belonging to families with DCM caused by sarcomeric gene mutations: of those, 12 were subclinical carriers, 21 had overt signs of disease, and 29 were non-carriers. Detailed echocardiographic investigations showed that subclinical carriers have subtle but significant changes in systolic myocardial tissue velocity, longitudinal, circumferential and radial strain, and longitudinal and radial strain rate, but no changes in diastolic parameters compared to the non-carriers. Interestingly, these changes had an opposite pattern compared to hypertrophic cardiomyopathy subclinical carriers, where the initial signs of disease are those of diastolic dysfunction. These findings suggest that an accurate state-of-the-art clinical examination can contribute to the comprehensive assessment of carriers of rare variants suspected to be causal of the disease.
New High-throughput Technologies and Genome-wide Approaches
A dramatic advance in sequencing technology, called next-generation sequencing, recently brought the efficiency of the assay from sequencing 1 gene in 1 week to 50 genes overnight, dropping the cost and the turnaround time and increasing the sensitivity of genetic testing in genetic disorders such as DCM. Next-generation sequencing in the research setting has been used to sequence the whole exome, which is the portion of genome coding for all proteins of the human body. This approach has already produced important results in the study of DCM, when the causal gene was unknown. Theis et al. (2011) identified, by linkage analysis and exome sequencing, GATAD1 as a gene for a rare form of autosomal recessive DCM. GATAD1 binds to a histone modification site that regulates gene expression. Norton et al. (2011) discovered, by genome-wide copy number variation analysis in an autosomal dominant DCM family, a mutation in another novel disease gene, the heat shock protein cochaperone BCL2-associated athanogene 3 (BAG3). They confirmed the presence of BAG3 gene rare variants in ~2% of their DCM study population.
From Bench to Bedside: Clinical Impact of Genomic Advances
Current guidelines may provide an important help to clinicians in selecting the appropriate genetic testing approach in cardiomyopathies. In 2009, on behalf of the Heart Failure Society of America, a group of U.S. experts published the “Practice Guideline” for the Genetic Evaluation of Cardiomyopathy (Hershberger et al., 2009). In 2010, a group of European experts published a position statement on genetic counseling and testing in cardiomyopathies on behalf of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases (Charron et al., 2010). Finally, in 2011, the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) published an expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies (Ackerman et al., 2011). Therefore, the clinician can currently count on a number of international updated guidelines to provide an appropriate management to patients with genetic heart muscle diseases: here we summarize the most important principles (Ackerman et al., 2011; Charron et al., 2010; Hershberger et al., 2009). Class I recommendation (”is recommended”) for genetic testing include: (a) index cases with cardiomyopathy, and (b) family members for the mutation identified in the proband when therapy/protective measures/lifestyle adaptations may be adopted. Interestingly, Class IIa recommendation is when results of genetic testing may be useful for reproductive counseling or instances in which genetic testing is requested by the patient who wants to know his/her mutation status. Genetic counseling should be part of proper management of genetic cardiomyopathies and is recommended for all patients and relatives with the familial heart diseases and should include discussion of the risks, benefits, and options available for clinical testing and/or genetic testing. Importantly, treatment decisions should not rely solely on the genetic test result but should be based on an individual’s comprehensive clinical evaluation. All guidelines agree that at the current status of knowledge, it is preferable for pre-genetic test counseling, genetic testing, and the interpretation of genetic test results to be performed in centers experienced in the genetic evaluation and in family-based management of the heritable cardiomyopathies (Ackerman et al., 2011; Charron et al., 2010; Hershberger et al., 2009).
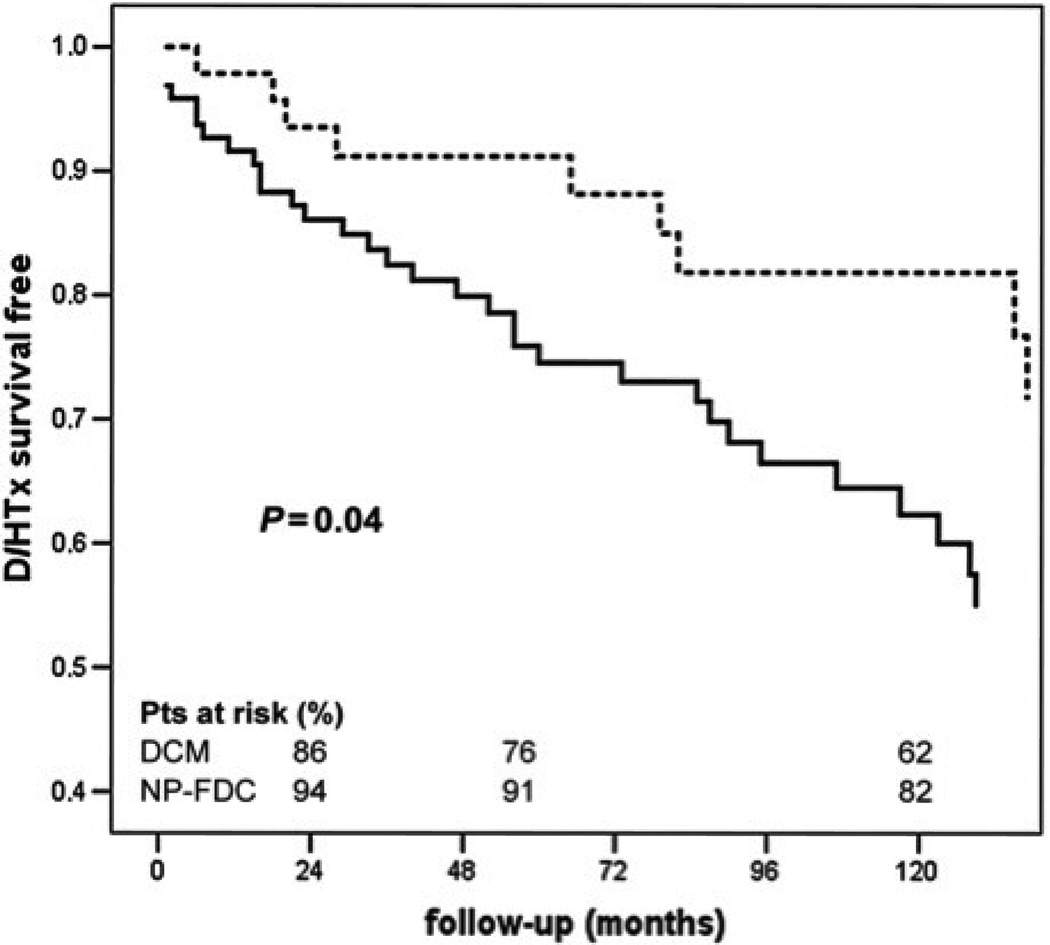
Family screening is an important part of a correct clinical approach to DCM. Besides the opportunity to perform genetic testing, family screening with clinical examination of family members at risk, with the possibility of catching early stages of the disease, may have significant prognostic impact. Moretti et al. (2010) compared the natural history of subjects with non-familial DCM to affected relatives of DCM families identified by family screening. As shown in Figure 3, the survival free from death or heart-transplant in sporadic DCM (solid line) was worse compared to the relatives of familial DCM (dotted line). This study suggests that family screening itself, with or without an identified mutation, can effectively identify DCM patients at an earlier stage of disease and improve survival.
Figure 3.
Survival in patients with sporadic DCM and familial DCM. Analysis of survival free from heart-transplant (HTx) in sporadic (solid line) and non-proband familial DCM (dotted line). D/HTx, death/heart transplant. From Moretti et al., with permission (Moretti et al., 2010).
Novel technologies now offer unprecedented possibilities to test a large number of genes. Newer DNA sequencing technology (whole-exome and whole-genome sequencing) can now complete the sequencing of an entire human genome several times over in a matter of days -- orders of magnitude faster than the nearly thirteen years required for the initial first-pass done by the Human Genome Project consortium. This technology, which is already used in clinical genetic testing, will undoubtedly add new challenges to the difficulty of distinguishing signal from noise. For instance, the pan cardiomyopathy panel of Harvard Partners laboratory can now test 51 cardiomyopathy genes with a turn-around time of only 8–12 weeks and a cost not significantly different from the cost of sequencing a modest number of genes a few years ago (http://www.hpcgg.org/).
Discerning Rare Benign Variations Versus Pathogenic Mutations in DCM
Meder et al. (2011) have recently tested the power and limitations of next generation sequencing as a gene mutation screening tool with a proof-of-concept study. They screened 10 patients with either HCM or DCM by next-generation sequencing for 47 known or putative DCM genes. They found a remarkable number of novel variants (~60,000) including ~21,000 novel deleterious variants such as insertions/deletions. They were able to successfully identify 6 known disease-causing mutations in their study patients. However, they also identified several novel variants (~30) predicted by all consensus criteria to be “disease-causing mutations,” in 9/10 subjects, underlying the difficulty to discriminate between benign and pathogenic variants.
Indeed, the Human Genome Mutation database contains >100,000 mutations in ~4,000 genes reported as pathogenic, mostly missense mutations. However, the unexpected variability of the human genome represents a challenge in the clinical setting. Two large studies have considered this problem: how frequent in the general population are variants that are considered “pathogenic”? And are those variants true pathogenic mutations with low penetrance or just rare benign variations?
Norton et al. (2012) analyzed the presence of mutations previously reported as causative for DCM in the cohort of 2,439 subjects from the NHLBI GO Exome Sequencing Project (ESP). Reported pathogenic mutations in 30 DCM genes were present in 17% of the ESP population. Of these mutations, 42% had previous functional data to support their pathogenic role, suggesting that they were unlikely to be false-positives. Based on these findings, the authors proposed a series of novel criteria for the evaluation of rare variants in Mendelian disorders, including a novel more stringent cut-off for the allele frequency of ≤0.04%, suggesting that higher frequencies are of less certain pathogenicity.
In another study, Tennessen et al. (2012) examined the exome sequencing data of the 2,440 subjects of the NHLBI GO ESP cohort. In this study the authors examined over 15,000 protein-coding genes to understand the contribution of rare variants to the risk of disease. They found that each subject had ~14,000 SNVs, of which 200 were novel, mainly rare nonsynonymous variants, and that these rare variants predicted to disrupt the protein function of over 300 genes for each person. As the authors note, this abundance of variation is consistent with the explosion of human population growth. They concluded that to detect the effect of rare variants very large sample size and population-specific data are needed.
Another interesting perspective is that offered by Bick et al. (2012), who screened for mutations of 8 sarcomeric genes associated with cardiomyopathy by next-generation sequencing in 2 large cohorts, the Framingham and Jackson Studies Cohorts, for a total of 3,600 subjects. They found rare missense sarcomeric variants in 11% of population, and likely pathogenic variations in 0.6%, twice the previous estimates. Of the 22 individuals found to have likely pathogenic variants, only 4 had cardiac hypertrophy, raising the question if the other variants have low penetrance or are single-nucleotide polymorphisms (SNPs). However, when the authors investigated the risk for cardiovascular events in carriers of any rare missense sarcomeric variants, they found that this was significantly increased (HR 2.3), raising the question that some of these variants are indeed pathogenic.
Overall, these recent studies indicate once more that integration of genetic testing with functional assays, robust bioinformatics, large control cohorts, and expert clinical evaluation can better assist clinicians in discerning pathogenic versus benign rare variants.
Concluding Remarks: the Ying-Yang of the Current Advances
Yang
In the past few months, a number of important scientific and technological advances have changed our approach to the genetics of DCM. We now have approximately 50 cardiomyopathy genes, 40 of which associated with DCM genes, and a novel powerful assay for genetic testing, next-generation sequencing. The frequency for each gene from a low yield (<1% to ~8%) with the identification of TTN is now up to 25% therefore the expected mutation detection rate is >40% of cases. This indicates that genetic testing can now be considered a class I recommendation according to current guidelines (Ackerman et al., 2011). Phenomic studies, such as the identification of red flags/overlapping syndromes (muscular dystrophy, conduction disease, arrhythmias, and other cardiomyopathies in the family), biomarkers, and epigenetic factors are useful diagnostic tools (Piran et al., 2012). Novel data for better risk stratification (such as for LMNA) are able to modify patient management and justify once more genetic testing (class I recommendation) and therapeutic intervention (such as AICD).
Ying
However, the new discoveries and tools generate new challenges for geneticists and clinicians. The unexpected remarkable human genetic variation makes genetic testing a dynamic process: testing reports need to be updated and possibly changed based on new data about rare genetic variants. To ensure that a rare variant is appropriately classified [benign, pathogenic, or of uncertain significance (VUS)] it is evident now that over 2,000–3,000 ethnically matched controls may be needed to control for genomic variability. Fortunately, there are some databases now available to begin to meet these needs: the 1000 Genome Project, the NHLBI Exome Sequencing Project, and the dbSNP.
To reliably assess the pathogenic role of a mutation, more stringent criteria are needed: variants should be rare (new cut-off allele frequency in DCM <0.04%), “radical” (in/del, splice, nonsense), and segregate within the family, compared to large cohorts of ethnically matched controls, analyzed with robust bioinformatics approaches, and possibly confirmed by functional assays. In this regard, induced-pluripotent stem cells (iPS) repositories of patients with DCM could play an important role.
Clearly this high level of complexity requires an expert genetic team for counseling, managing, delivering, and following up over time the results of genetic testing (www.genetest.org; Lakdawala et al., 2012a; Maron et al., 2012; Norton et al., 2012). Genetic testing is in fact particularly important for screening of family members potentially at risk and must start from successful genotyping of the proband. It may be useful for the identification of non-carriers (no need of precautions and surveillance), although exceptions may exist (multiple mutations, mutation/VUS, laboratory errors). Genetic testing may identify genotype+ phenotype−, useful for prevention strategies, sport recommendations, and indications for defibrillator implantation and therapies as discussed above. Finally, genetic testing may be requested by couples to guide reproductive decision-making including utilization of pre-implantation genetic diagnostic strategies.
Footnotes
Disclosure
The authors declare that there are no conflicts of interest, grants, or relationship with industry.
Contributor Information
Luisa Mestroni, Cardiovascular Institute, University of Colorado, Aurora, Colorado 80305, USA
Matthew R.G. Taylor, Adult Clinical Genetics, Department of Medicine, University of Colorado, Aurora, Colorado 80305, USA
References
- Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, Camm AJ, Ellinor PT, Gollob M, Hamilton R, Hershberger RE, Judge DP, Le Marec H, McKenna WJ, Schulze-Bahr E, Semsarian C, Towbin JA, Watkins H, Wilde A, Wolpert C, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Heart Rhythm. 2011;8(8):1308–1339. doi: 10.1016/j.hrthm.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Bick AG, Flannick J, Ito K, Cheng S, Vasan RS, Parfenov MG, Herman DS, Depalma SR, Gupta N, Gabriel SB, Funke BH, Rehm HL, Benjamin EJ, Aragam J, Taylor HA, Jr, Fox ER, Newton-Cheh C, Kathiresan S, O’Donnell CJ, Wilson JG, et al. Burden of rare sarcomere gene variants in the Framingham and Jackson Heart Study cohorts. Am J Hum Genet. 2012;91(3):513–519. doi: 10.1016/j.ajhg.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron P, Arad M, Arbustini E, Basso C, Bilinska Z, Elliott P, Helio T, Keren A, McKenna WJ, Monserrat L, Pankuweit S, Perrot A, Rapezzi C, Ristic A, Seggewiss H, Van Langen I, Tavazzi L. Genetic counseling and testing in cardiomyopathies: a position statement of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J. 2010;31(22):2715–2726. doi: 10.1093/eurheartj/ehq271. [DOI] [PubMed] [Google Scholar]
- Gerull B, Gramlich M, Atherton J, McNabb M, Trombitas K, Sasse-Klaassen S, Seidman J, Seidman C, Granzier H, Labeit S, Frenneaux M, Thierfelder L. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat Genet. 2002;30:201–204. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, Dauksaite V, Vakeel P, Klaassen S, Gerull B, Thierfelder L, Regitz-Zagrosek V, Hacker TA, Saupe KW, Dec GW, Ellinor PT, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18(5):766–773. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, Conner L, Depalma SR, McDonough B, Sparks E, Teodorescu DL, Cirino AL, Banner NR, Pennell DJ, Graw S, Merlo M, Di Lenarda A, Sinagra G, Bos JM, Ackerman MJ, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366(7):619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger RE, Lindenfeld J, Mestroni L, Seidman CE, Taylor MR, Towbin JA. Genetic evaluation of cardiomyopathy -- a heart failure society of America practice guideline. J Card Fail. 2009;15(2):83–97. doi: 10.1016/j.cardfail.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Lakdawala NK, Funke BH, Baxter S, Cirino AL, Roberts AE, Judge DP, Johnson N, Mendelsohn NJ, Morel C, Care M, Chung WK, Jones C, Psychogios A, Duffy E, Rehm HL, White E, Seidman JG, Seidman CE, Ho CY. Genetic testing for dilated cardiomyopathy in clinical practice. J Card Fail. 2012a;18(4):296–303. doi: 10.1016/j.cardfail.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakdawala NK, Thune JJ, Colan SD, Cirino AL, Farrohi F, Rivero J, McDonough B, Sparks E, Orav EJ, Seidman JG, Seidman CE, Ho CY. Subtle abnormalities in contractile function are an early manifestation of sarcomere mutations in dilated cardiomyopathy. Circ Cardiovasc Genet. 2012b;5(5):503–510. doi: 10.1161/CIRCGENETICS.112.962761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol. 2012;60(8):705–715. doi: 10.1016/j.jacc.2012.02.068. [DOI] [PubMed] [Google Scholar]
- McNair WP, Sinagra G, Taylor MRG, Di Lenarda A, Ferguson DA, Salcedo EE, Slavov D, Zhu X, Caldwell J, Mestroni L. SCN5A mutations associate with arrhythmic dilated cardiomyopathy and commonly localize to the voltage-sensing mechanism. J Am Coll Cardiol. 2011;57(21):2160–2168. doi: 10.1016/j.jacc.2010.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally EM. Genetics: broken giant linked to heart failure. Nature. 2012;483(7389):281–282. doi: 10.1038/483281a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder B, Haas J, Keller A, Heid C, Just S, Borries A, Boisguerin V, Scharfenberger-Schmeer M, Stahler P, Beier M, Weichenhan D, Strom TM, Pfeufer A, Korn B, Katus HA, Rottbauer W. Targeted next-generation sequencing for the molecular genetic diagnostics of cardiomyopathies. Circ Cardiovasc Genet. 2011;4(2):110–122. doi: 10.1161/CIRCGENETICS.110.958322. [DOI] [PubMed] [Google Scholar]
- Mestroni L, Taylor MR. Pharmacogenomics, personalized medicine, and heart failure. Discov Med. 2011;11(61):551–561. [PubMed] [Google Scholar]
- Meune C, Van Berlo JH, Anselme F, Bonne G, Pinto YM, Duboc D. Primary prevention of sudden death in patients with lamin A/C gene mutations. N Engl J Med. 2006;354(2):209–210. doi: 10.1056/NEJMc052632. [DOI] [PubMed] [Google Scholar]
- Moretti M, Merlo M, Barbati G, Di Lenarda A, Brun F, Pinamonti B, Gregori D, Mestroni L, Sinagra G. Prognostic impact of familial screening in dilated cardiomyopathy. Eur J Heart Fail. 2010;12(9):922–927. doi: 10.1093/eurjhf/hfq093. [DOI] [PubMed] [Google Scholar]
- Norton N, Li D, Rieder MJ, Siegfried JD, Rampersaud E, Zuchner S, Mangos S, Gonzalez-Quintana J, Wang L, McGee S, Reiser J, Martin E, Nickerson DA, Hershberger RE. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am J Hum Genet. 2011;88(3):273–282. doi: 10.1016/j.ajhg.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton N, Robertson PD, Rieder MJ, Zuchner S, Rampersaud E, Martin E, Li D, Nickerson DA, Hershberger RE. Evaluating pathogenicity of rare variants from dilated cardiomyopathy in the exome era. Circ Cardiovasc Genet. 2012;5(2):167–174. doi: 10.1161/CIRCGENETICS.111.961805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piran S, Liu P, Morales A, Hershberger RE. Where genome meets phenome: rationale for integrating genetic and protein biomarkers in the diagnosis and management of dilated cardiomyopathy and heart failure. J Am Coll Cardiol. 2012;60(4):283–289. doi: 10.1016/j.jacc.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Sinagra G, Mestroni L, Camerini F. Genetic Cardiomyopathies. A Clinical Approach. London, UK and New York, New York, USA: Springer, Milan Dordrecht Heidelberg; 2012. [Google Scholar]
- Taylor DO, Edwards LB, Boucek MM, Trulock EP, Aurora P, Christie J, Dobbels F, Rahmel AO, Keck BM, Hertz MI. Registry of the International Society for Heart and Lung Transplantation: twenty-fourth official adult heart transplant report -- 2007. J Heart Lung Transplant. 2007;26(8):769–781. doi: 10.1016/j.healun.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Taylor M, Graw S, Sinagra G, Barnes C, Slavov D, Brun F, Pinamonti B, Salcedo EE, Sauer W, Pyxaras S, Anderson B, Simon B, Bogomolovas J, Labeit S, Granzier H, Mestroni L. Genetic variation in titin in arrhythmogenic right ventricular cardiomyopathy-overlap syndromes. Circulation. 2011;124(8):876–885. doi: 10.1161/CIRCULATIONAHA.110.005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MR, Fain PR, Sinagra G, Robinson ML, Robertson AD, Carniel E, Di Lenarda A, Bohlmeyer TJ, Ferguson DA, Brodsky GL, Boucek MM, Lascor J, Moss AC, Li WL, Stetler GL, Muntoni F, Bristow MR, Mestroni L. Familial Dilated Cardiomyopathy Registry Research Group. Natural history of dilated cardiomyopathy due to lamin A/C gene mutations. J Am Coll Cardiol. 2003;41:771–780. doi: 10.1016/s0735-1097(02)02954-6. [DOI] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, Mcgee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis JL, Sharpe KM, Matsumoto ME, Chai HS, Nair AA, Theis JD, De Andrade M, Wieben ED, Michels VV, Olson TM. Homozygosity mapping and exome sequencing reveal GATAD1 mutation in autosomal recessive dilated cardiomyopathy. Circ Cardiovasc Genet. 2011;4(6):585–594. doi: 10.1161/CIRCGENETICS.111.961052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin JA. The role of cytoskeletal proteins in cardiomyopathies. Curr Opin Cell Biol. 1998;10(1):131–139. doi: 10.1016/s0955-0674(98)80096-3. [DOI] [PubMed] [Google Scholar]
- van Rijsingen IA, Arbustini E, Elliott PM, Mogensen J, Hermans-Van Ast JF, Van Der Kooi AJ, Van Tintelen JP, Van Den Berg MP, Pilotto A, Pasotti M, Jenkins S, Rowland C, Aslam U, Wilde AA, Perrot A, Pankuweit S, Zwinderman AH, Charron P, Pinto YM. Risk factors for malignant ventricular arrhythmias in lamin a/c mutation carriers a European cohort study. J Am Coll Cardiol. 2012;59(5):493–500. doi: 10.1016/j.jacc.2011.08.078. [DOI] [PubMed] [Google Scholar]