SUMMARY
The CARMA1/Bcl10/MALT1 (CBM) signalosome mediates antigen receptor-induced NF-κB signaling to regulate multiple lymphocyte functions. While CARMA1 and Bcl10 contain caspase recruitment domains (CARDs), MALT1 is a paracaspase with structural similarity to caspases. Here we show that the reconstituted CBM signalosome is a helical filamentous assembly in which substoichiometric CARMA1 nucleates Bcl10 filaments. Bcl10 filament formation is a highly cooperative process whose threshold is sensitized by oligomerized CARMA1 upon receptor activation. In cells, both cotransfected CARMA1/Bcl10 complex and the endogenous CBM signalosome are filamentous morphologically. Combining crystallography, nuclear magnetic resonance, and electron microscopy, we reveal the structure of the Bcl10 CARD filament and the mode of interaction between CARMA1 and Bcl10. Structure-guided mutagenesis confirmed the observed interfaces in Bcl10 filament assembly and MALT1 activation in vitro and NF-κB activation in cells. These data support a paradigm of nucleation-induced signal transduction with threshold response due to cooperativity and signal amplification by polymerization.
INTRODUCTION
NF-κB family transcription factors are master regulators of immune, inflammatory, and apoptotic responses and play crucial roles in many aspects of mammalian biology including innate immunity, adaptive immunity, and embryonic development. In adaptive immunity, the ternary complex of CARMA1 (also known as CARD11), Bcl10, and MALT1 (CBM) acts as the signalosome to mediate NF-κB activation in T and B cells (Jiang and Lin, 2012). Analogously, the CARMA1-related proteins CARMA2 (also known as CARD14), CARMA3 (also known as CARD10 or Bimp1), and CARD9 form NF-κB-activating signalosomes with Bcl10 and MALT1 (Jiang and Lin, 2012). While CARMA3 is a critical component in G protein-coupled receptor and receptor tyrosine kinase-induced NF-κB activation pathways, CARD9 was shown to mediate innate immune responses in the RIG-I and the C type lectin receptor pathways (Jiang and Lin, 2012; Strasser et al., 2012). Furthermore, CARMA protein family-induced Bcl10 and MALT1 activation has become a hub for NF-κB activation by multiple additional pathways including IL-2, BAFF, and saturated fat-induced insulin resistance (Jiang and Lin, 2012).
The CARMA1/Bcl10/MALT1 (CBM) complex is a prototype of these NF-κB activation signalosomes. Upon interaction of B cell receptor (BCR) with antigens and engagement of T cell receptor (TCR) with MHC-bound antigen peptides, CARMA1 is recruited to the immunological synapse together with protein kinase C β (PKCβ) in B cells or PKCθ in T cells. CARMA1 contains an N-terminal CARD, coiled-coil domains, and a C-terminal MAGUK domain likely responsible for its association with the cytoplasmic membrane (Figure 1A). PKCβ or PKCθ phosphorylates CARMA1 at the interdomain linker between the coiled coil and the MAGUK domains, leading to release of CARMA1 autoinhibition (Jiang and Lin, 2012). Activated CARMA1 recruits Bcl10 via a CARD/CARD interaction with CARD belonging to the death domain fold superfamily important for many protein/protein interactions in immune signaling (Ferrao and Wu, 2012). The C-terminal Ser/Thr-rich domain of Bcl10 associates with the N-terminal Ig domains of MALT1. MALT1 contains a C-terminal paracaspase domain, which shares similarity with caspases but cleaves specifically after arginine residues (Wiesmann et al., 2012; Yu et al., 2011). The CBM signalosome further recruits signaling components such as TRAF6 for nondegradative polyubiquitination to activate the IκB kinase (IKK), resulting in IκB phosphorylation and degradation, and release of NF-κB proteins for nuclear translocation (Jiang and Lin, 2012). Formation of the CBM signalosome also enables the paracaspase activity of MALT1, which cleaves negative regulators to further enhance NF-κB activity (Coornaert et al., 2008; Hailfinger et al., 2011; Rebeaud et al., 2008; Staal et al., 2011). Nondegradative polyubiquitination that leads to association of Bcl10 with autophagosomes and feedback degradative polyubiquitination of Bcl10 may both result in its gradual degradation and homeostatic control of the pathway (Moreno-García et al., 2013; Paul et al., 2012).
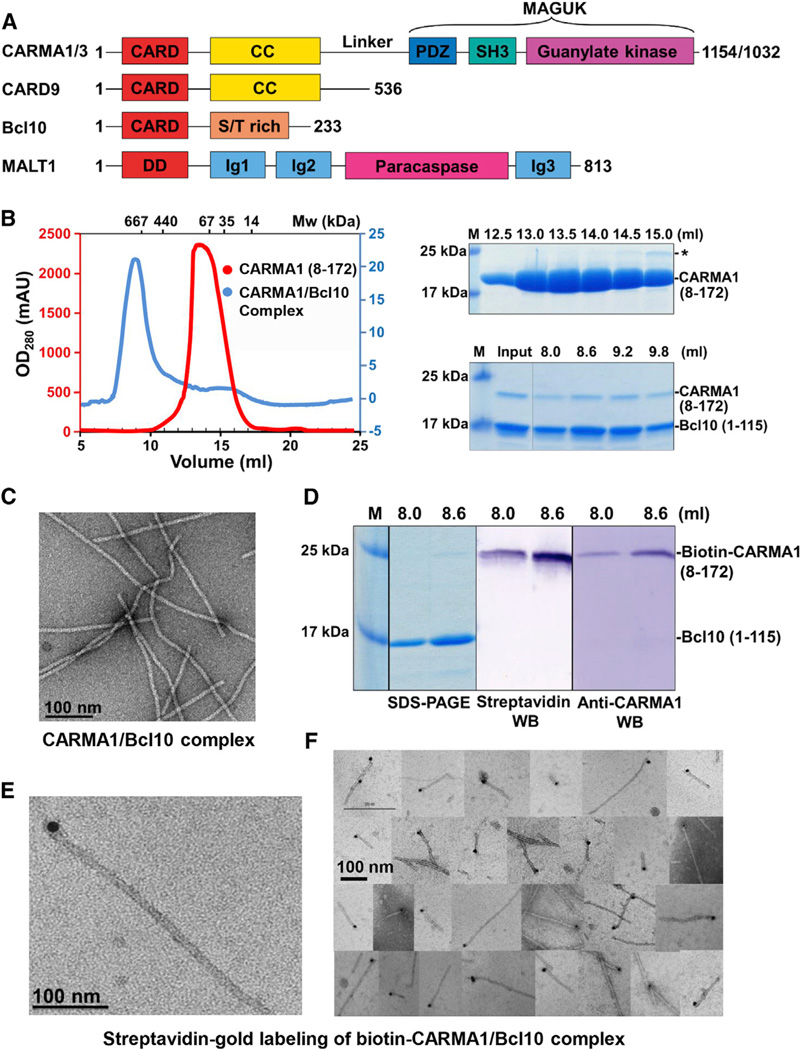
Figure 1. The CARMA1/Bcl10 Complex Is Filamentous with End Localization of CARMA1.
(A) Domain organizations. Abbreviations are as follows: CARD, caspase recruitment domain; CC, coiled-coil domain; PDZ, PSD95, DLG, and ZO1 homology domain; SH3, Src homology 3 domain; MAGUK, membrane-associated guanylate kinase domain; S/T rich, Ser/Thr rich; DD, death domain; and Ig, immunoglobulin-like domain. The number of residues in each protein is labeled.
(B) Gel filtration profiles of CARMA1 and the CARMA1/Bcl10 complex (left) and SDS-PAGE gels of peak fractions (right). Because Bcl10 CARD does not contain Trp residues, the OD280 reading of the complex peak is low (blue, right of the graph).
(C) An EM image of the CARMA1 (8–172)/Bcl10 complex.
(D) SDS-PAGE and western blots (WB) of biotinylated CARMA1/Bcl10 complex.
(E) One EM image of streptavidin gold-labeled biotinylated CARMA1/Bcl10 complex.
(F) A collage of EM images of streptavidin gold-labeled biotinylated CARMA1/Bcl10 complex.
See also Figure S1.
The genes encoding CARMA1, Bcl10, and MALT1 are bona fide oncogenes as missense mutations and chromosomal translocations of these genes are associated with non-Hodgkin’s lymphomas, such as MALT lymphoma and diffuse large B cell lymphoma (DLBCL). Missense mutations of CARMA1 are most frequently found at the coiled-coil region of CARMA1, which correlate with constitutive NF-κB activation (Lenz et al., 2008). Chromosomal translocations of both Bcl10 and MALT1 have been found to either position Bcl10 and MALT1 genes to the Ig locus for overexpression, or create a fusion protein comprising the BIR domains of cIAP2 and the paracaspase domain of MALT1, leading to constitutive NF-κB activation (Rosebeck et al., 2011). The paracaspase activity of MALT1 is essential for lymphomagenesis of DLBCL (Hailfinger et al., 2009), and peptide and small molecule inhibitors against MALT1 showed effectiveness against the disease (Ferch et al., 2009; Fontan et al., 2012; Nagel et al., 2012).
Previous studies have revealed that endogenous Bcl10 displays a diffuse cytosolic distribution, whereas overexpression of Bcl10 induces formation of filament-like structures shown by fluorescence microscopy (Guiet and Vito, 2000; Yan et al., 1999). TCR signaling also leads to formation of punctate structures critical for NF-κB activation; they have been named POLKADOTS and contain components of the CBM signalosome (Rossman et al., 2006). These data support the formation of supramolecular structures in CBM signaling, but the exact molecular architecture and the mechanism of assembly are unknown. In addition, full-length MALT1 is in an inactive, monomeric form, but active MALT1 paracaspase is dimeric (Wiesmann et al., 2012; Yu et al., 2011). Bcl10 was thought to be involved in MALT1 activation, but the molecular mechanism remains unknown.
Here we present a comprehensive study to reveal the molecular architecture of the CBM complex formed upon receptor triggering. We showed surprisingly using electron microscopy (EM) that both the CARMA1/Bcl10 and the CBM complexes form filaments and that CARMA1 resides on one end of the Bcl10 filament to nucleate the helical CARD filamentous structure. Polymerization assays revealed a cooperative Bcl10 filament assembly whose threshold is greatly sensitized by activated, oligomerized CARMA1. Bcl10 filaments significantly enhanced the paracaspase activity of the bound MALT1, likely by providing the platform for its dimerization and activation. We determined the crystal structure of CARMA1 CARD, the nuclear magnetic resonance (NMR) structure of a monomeric mutant of Bcl10 CARD, and the 3D helical reconstructed EM structure of the Bcl10 filament. Fitting Bcl10 subunits into the EM density resulted in a modeled structure of a CARD filament in the DD fold superfamily. Modeling of the interaction between CARMA1 and Bcl10 CARDs suggests that coiled-coil oligomerized CARMA1 may form preassembled short filaments for direct nucleation of Bcl10. Nucleation of Bcl10 by CARMA1 reveals a new hierarchical cooperative paradigm in signal transduction and amplification that lead to threshold responses in signaling.
RESULTS
The CARMA1/Bcl10 Complex Is a Filamentous Assembly with CARMA1 as the Nucleator of Bcl10 Polymerization
We began our studies by reconstituting the interaction between human CARMA1 and Bcl10. Because CARD9 does not have the C-terminal MAGUK domain yet is still able to recruit Bcl10 (Bertin et al., 2000), we generated CARMA1 constructs containing the N-terminal CARD and variable lengths of the CC domain and selected a well-expressed construct (8–172) (Figure 1B). Coexpression and purification of the CARMA1 construct with Bcl10 CARD (1–115) showed that the copurified sample eluted at the void position of a Superdex 200 gel filtration column (Figure 1B), suggesting formation of a large "aggregate." Although there was much less CARMA1 than Bcl10 in the complex, the elution position shift of CARMA1 and comigration of CARMA1 with Bcl10 were evident (Figure 1B). Similarly, CARMA1 CARD alone (8–117) also formed a complex with Bcl10 (see Figure S1A online). We used EM to visualize the negatively stained CARMA1/Bcl10 complex, which revealed filaments with uniform diameters of 9.9 ± 1.1 nm (Figure 1C, Figure S1A). These filaments showed helical diffraction (see below).
To elucidate the substoichiometric molar presence of CARMA1 in the CARMA1/Bcl10 complex (Figure 1B), we generated a construct of CARMA1 that is fused with a peptide substrate for E. coli biotin holoenzyme synthetase to allow enzymatic biotinylation of CARMA1 during expression. Coexpression of the CARMA1 construct with Bcl10 CARD generated a complex with specific biotinylation of CARMA1 as shown by both streptavidin and anti-CARMA1 western blots (Figure 1D). The biotinylated CARMA1/Bcl10 complex still formed filaments of similar width (Figure S1B). Remarkably, labeling biotinylated CARMA1 by 10 nm streptavidin gold particles showed that CARMA1 is localized only at one end of the filaments (Figures 1E and 1F). In contrast, as a negative control, the nonbiotinylated CARMA1/Bcl10 complex did not show any labeling (Figure S1C).
The finding of the end location of CARMA1 suggested its role as the nucleator of the CARMA1/Bcl10 filament, leading to directional elongation of Bcl10 polymerization in a manner analogous to nucleated polymerization of the cytoskeleton. For example, in microtubule polymerization, the γ-tubulin ring complex in microtubule-organizing centers provides the template for addition of α/β tubulin dimers to generate directional microtubule growth (Kollman et al., 2011). To quantitatively assess Bcl10 polymerization, we set up a fluorescence polarization (FP) assay using Alexa 488-labeled Bcl10. Both unlabeled and Alexa 488-labeled MBP-Bcl10 contained a prominent monomeric peak (Figures S1D and S1E). Polymerization of monomeric Bcl10 was initiated by addition of the TEV protease to remove MBP from the fusion protein, and the increase in FP was monitored as a function of time (Figure 2A). In the absence of CARMA1, Bcl10 polymerization exhibited a significant time lag of ~70 min. In the presence of increasing amounts of substoichiometric CARMA1 (1/200 to 1/5 molar ratios), Bcl10 polymerization was dramatically enhanced. At the 1/5 molar ratio of CARMA1, the time lag nearly disappeared, with an approximate 3-fold increase in the change of FP at the end of the measurement period of 200 min in comparison with Bcl10 polymerization in the absence of CARMA1. Removal of MBP appeared to have completed by 10 min, and CARMA1 did not cause a change in the rate of the cleavage (Figure S1F). The CARMA1/Bcl10 filaments generated from the polymerization assay (Figure S1G) showed a similar morphology as those of the coexpressed and copurified complex (Figure 1C) and of the Bcl10 sample upon removal of MBP (Figure S1G).
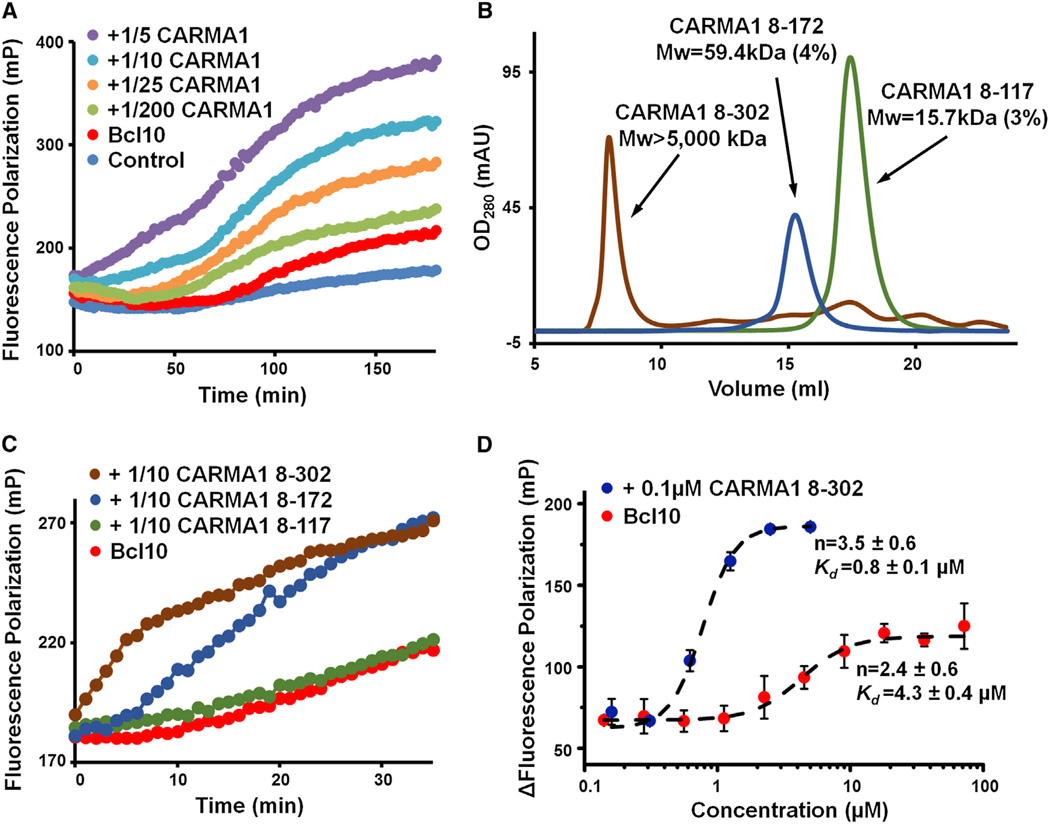
Figure 2. CARMA1 Nucleates Bcl10 Filaments Cooperatively.
(A) Fluorescence polarization assay of Alexa 488-labeled MBP-Bcl10 (C29A/C57A double mutant) in the presence and absence of substoichiometric amounts of CARMA1 (8–172) upon MBP removal by the TEV protease.
(B) Elution profiles of three CARMA1 constructs from a Superdex 200 10/300GLgelfiltration column. Multiangle light scattering (MALS) measurements are shown for two of the constructs, with numbers in parentheses representing fitting errors. The molecular mass of CARMA1 (8–302) is estimated from the fractionation limit of Superose 6 (Figure S2B).
(C) Same as (A), but with three different CARMA1 constructs.
(D) Concentration dependence of Bcl10 polymerization in the presence and absence of CARMA1. N, Hill coefficient. Kd, microscopic dissociation constant. Data are represented as mean ± SD from three independent measurements.
See also Figure S2.
Oligomerization, Threshold, and Cooperativity in CARMA1-Nucleated Bcl10 Polymerization
CARMA1 contains a long coiled-coil region that contains four predicted coiled-coil segments ending at around residues 175, 300, 350, and 440, respectively (Figure S2A). The CARMA1 construct (8–172) comprising the N-terminal CARD, and the first coiled-coil segment showed a measured molecular mass of 59.4 kDa (4% error) using multiangle light scattering (MALS) (Figure 2B). Because the construct has a monomer molecular weight of 21.7 kDa, the data indicate that it exists in solution as trimers. In contrast, CARMA1 CARD (8–117, monomer MW 15.2 kDa) is essentially monomeric, with a measured molecular mass of 15.7 kDa (3% error) (Figure 2B). To determine the functions of the additional CC segments, we generated CARMA1 constructs 8–302, 8–342, and 8–449. CARMA1 (8–302) produced soluble proteins that eluted at the void positions of both Superdex 200 and Superose 6 gel filtration columns (Figure 2B, Figure S2B), indicating a higher oligomerization state than CARMA1 (8–172). The 8–342 and 8–449 constructs produced only insoluble proteins under the same expression and purification conditions, suggesting an even more enhanced tendency to form higher-order aggregates. Because the fractionation limit of a Superose 6 column is 5 MDa, the average CARMA1 (8–302) oligomer would consists of >100 molecules. Comparison of CARMA1 (8–302) and CARMA1 (8–172) showed that the former had enhanced ability in nucleating Bcl10 filaments (Figure 2C), suggesting the requirement for CARMA1 oligomerization in this process. In contrast, monomeric CARMA1 (8–117) did not significantly enhance Bcl10 polymerization.
To address the biophysical properties of Bcl10 polymerization, we measured the concentration dependence in the increase of FP. Both in the presence and the absence of CARMA1 (8–302), Bcl10 polymerization showed a sigmoidal increase as a function of Bcl10 concentration (Figure 2D). The fitted Hill coefficients (n) are 3.5 ± 0.6, and 2.4 ± 0.6, respectively, in the presence and absence of CARMA1 (Figure 2D), demonstrating that Bcl10 filament formation is highly cooperative. Additionally, in the presence of a substoichiometric amount of CARAM1, the Bcl10 concentration reaching half saturation (Kd) was determined to be 0.8 ± 0.1 µM, much lower than 4.3 ± 0.4 µM, the Kd in the absence of CARMA1. These data suggest that nucleation by CARMA1 greatly decreases the threshold for Bcl10 polymerization, which would be important under physiological conditions when Bcl10 concentration is relatively low.
The CBM Signalosome Is a Filamentous Assembly with Enhanced MALT1 Activity Both In Vitro and in Cells
To reconstitute the CBM ternary complex, we mixed E. coli expressed MBP-Bcl10 with E. coli expressed CARMA1 (8–172) and insect cell-expressed His-tagged full-length MALT1, and added the TEV protease to cleave off the MBP tag from Bcl10 and the His tag from MALT1. Removal of MBP releases inhibition on Bcl10 polymerization and allows assembly of the CBM complex. The ternary complex could be purified from the void fractions of a Superdex 200 gel filtration column because neither CARMA1 nor MALT1 migrates at the void position (Figures 3A and 1B, S3A). In these fractions, MALT1 and Bcl10 appeared to be approximately 1:1 in molar ratio, while CARMA1 was still a minor species in the complex. EM analysis showed that like the binary CARMA1/Bcl10 complex, the ternary CBM signalosome is also filamentous (Figure 3A). Measurement of the diameter of these filaments in comparison with that of the binary complex filaments showed that the formeris27.5 ±2.5 nm while the latter is 9.9 ± 1.1 nm. The larger diameter of the ternary complex suggests that MALT1 is bound to the periphery of the binary complex and adds bulk to the filaments. Because CARMA1 resides at the end of the filaments, only Bcl10 and MALT1 contribute to the diameters of the complexes. The calculated cross section area ratio of the ternary over the binary complex (27.52/9.92 = 7.7), which represents the ratio in the mass per unit length, is consistent with the ratio (7.5) of the molecular weights of Bcl10 (1–115, 15.7 kDa) and the Bcl10/MALT1 complex (full-lengths, 26.2 kDa and 91.1 kDa, respectively) in the respective filaments.
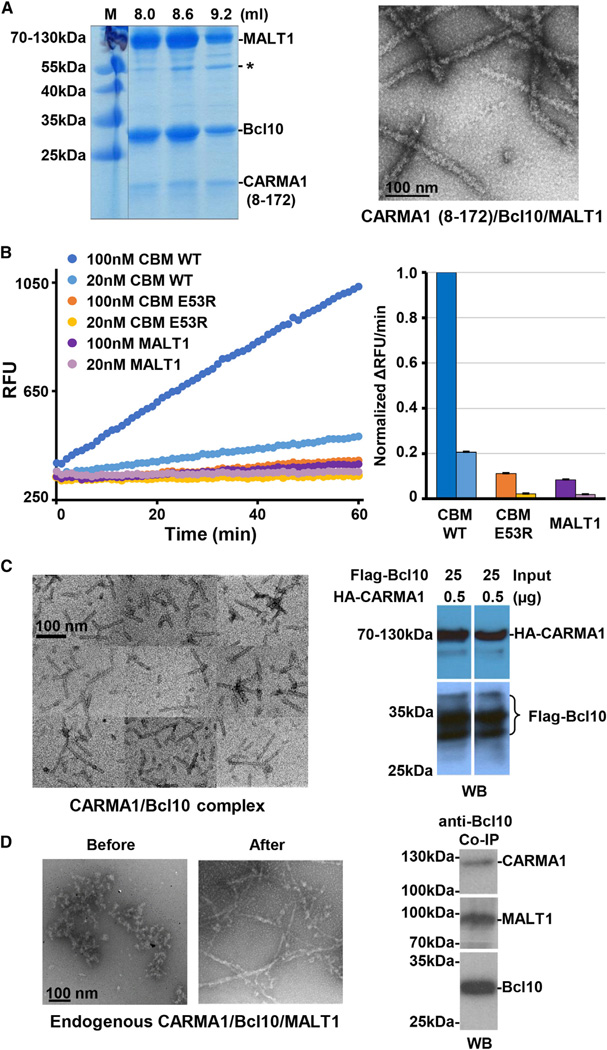
Figure 3. CBM Complexes In Vitro and in Cells.
(A) SDS-PAGE gel of reconstituted CARMA1/Bcl10/MALT1 complex fractions from gel filtration chromatography (left) and an EM image of the complex (right). Asterisk indicates a contaminant.
(B) Paracaspase activities of full-length MALT1, the WT CBM complex, and the CARMA1/E53R Bcl10/MALT1 complex (CBM E53R) using Ac-LRSR-AMC as the substrate. RFU, relative fluorescence unit. Data are represented as mean ± SD from three independent measurements.
(C) An EM image of the immunoprecipitated coexpressed HA-CARMA1 and Flag-Bcl10 complex in 293T cells (left) and western blots of anti-Flag immunoprecipitation of coexpressed HA-CARMA1 and Flag-Bcl10 (right).
(D) EM images of the immunoprecipitated endogenous CBM complex before and after limited proteolysis (left) and western blots of immunoprecipitation using biotinylated anti-Bcl10 antibody, showing the pull-down of CARMA1 and MALT1 from HBL-1 cell lysates.
See also Figure S3.
Assaying MALT1 catalytic activity using the fluorogenic Ac-LRSR-AMC substrate showed that MALT1 alone exhibited low activity and that incorporation into the complex with CARMA1 and Bcl10 greatly enhanced MALT1 activity as shown by the more than 10-fold increase in rates of substrate conversion (Figure 3B). The Bcl10 filaments likely provide the platform for bringing MALT1 into proximity for their activation through dimerization. Consistent with this hypothesis, purified full-length MALT1 and its paracaspase domain are both predominantly monomeric (Figures S3A and S3B). However, like caspases, the paracaspase domain of MALT1 is only active in its dimeric state as shown by biochemical and structural studies of the paracaspase domain of MALT1 (Hachmann et al., 2012). Control experiments performed with a previously reported monomeric Bcl10 mutant (E53R) (Yan et al., 1999) showed no significant effect on MALT1 activity (Figure 3B), supporting that formation of Bcl10 filaments is crucial for MALT1 activation.
To determine whether these filaments also form in mammalian cells, we first transfected full-length HA-tagged CARMA1 and full-length Flag-tagged Bcl10 in 293T cells and purified the complex using anti-Flag immunoprecipitation. Anti-HA and anti-Flag western blots confirmed complex formation between CARMA1 and Bcl10 (Figure 3C). The immunoprecipitated CARMA1 and Bcl10 complex is also filamentous, with diameter similar to that of the recombinant complex (Figure 3C). In contrast, pull-down with an isotype control antibody or single transfection of CARMA1 alone did not reveal filamentous structures (Figures S3C and S3D). Because some lymphoma cell lines exhibit constitutive CBM signaling, we purified the endogenous CBM complex from the lymphoma cell line HBL-1, which harbors an activating missense mutation in the BCR coreceptor CD79 chain B. Pull-down of the complex using anti-Bcl10 showed that CARMA1, Bcl10, and MALT1 were all in the same immunoprecipitates (Figure 3D).EM imaging of the endogenous complex showed convoluted aggregates that may be consistent with filaments. Because the endogenous pull-down could contain additional signaling components such as TRAF6 and IKK, we used limited proteolysis to unmask the core CBM assembly, which revealed much clearer filamentous structures (Figure 3D, Figure S3E). Silver staining of the immunoprecipitated endogenous complex showed bands for Bcl10 and MALT1 and the corresponding western blots showed bands for all components of the CBM complex (Figure S3F). Upon limited proteolysis, Bcl10 was partially degraded to a molecular weight that is consistent with Bcl10 CARD (Figure S3F), which should still form filaments, suggesting the resistance of Bcl10 filaments to protease treatment. Limited proteolysis of in vitro reconstituted full-length Bcl10 filaments also showed resistance to such treatment (Figure S3G). Furthermore, knocking down of TRAF6, a protein downstream of the CBM complex, did not compromise filament formation (Figures S3H and S3I). Collectively, these data confirm that the endogenous CBM complex has a filamentous morphology.
Extended α3-α4 Loop of CARMA1 and Unique Structure and Dynamics of Bcl10
To acquire structural information on the mode of assembly of the CARMA1/Bcl10 CARD filament, we first determined the crystal structure of human CARMA1 CARD at 1.8 Å resolution using single wavelength anomalous diffraction (Figure 4A, Table 1). Like its closest structural homolog Apaf-1 CARD, CARMA1 exhibits a kink in its first helix in the conserved six-helical bundle fold. However, unlike Apaf-1 and other CARD structures, CARMA1 CARD has an extended loop between α3 and α4 with high B factors (Figure 4A, Figure S4A). This loop is involved in the predicted interaction with Bcl10 CARD (see below). The human CARMA1 CARD structure is highly similar to the recently published crystal structure of mouse CARMA1 CARD (Li et al., 2012).
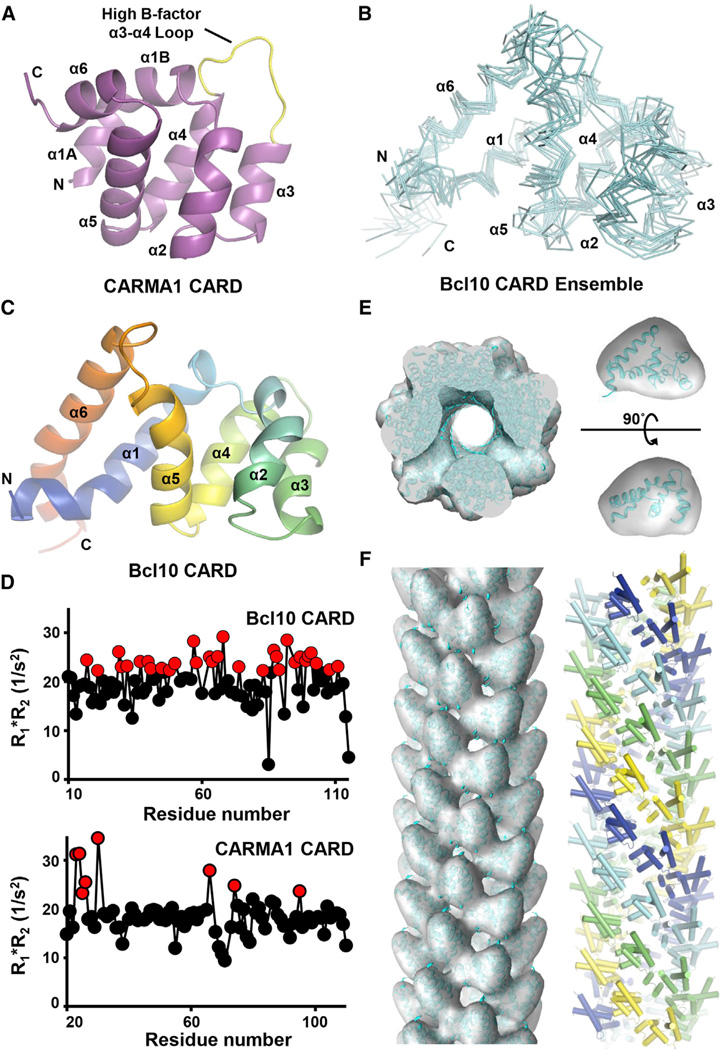
Figure 4. EM Structure of the Bcl10 Filament.
(A) Ribbon diagram of the crystal structure of CARMA1 CARD. Secondary structures and the α3-α4 loop are labeled.
(B) Superimposed Cα traces of minimized Bcl10 CARD NMR structures.
(C) Ribbon diagram of one Bcl10 CARD NMR model, showing the long α1 and α6 helices and the resulting pear shape of the molecule. The color mode is rainbow.
(D) R1*R2 relaxation analysis of Bcl10 and CARMA1 CARDs by NMR. Elevated values indicate the presence of ms-µs chemical exchange. A cutoff value of 21, which is 10% above the maximum calculated R1*R2 product at 600 MHz, is applied.
(E) Fitting of Bcl10 CARD structure (cyan) into the segmented EM density (gray, pear shaped in one orientation) and the entire 3D volume of the Bcl10 filament.
(F) 3D volume of the Bcl10 filament fitted with Bcl10 CARD structure viewed along the helical axis (left) and the structural model of the Bcl10 filament in which the prominent left-handed four-start helical strands are shown in cyan, blue, yellow, and green, respectively (right).
See also Figure S4, Table S1, and Table S2.
Table 1.
Crystallographic Statistics of CARMA1 CARD
| Data Collection | Native | KI Derivative |
|---|---|---|
| Beamline | R-AXIS IV | R-AXIS IV |
| Space group | P4132 | P4132 |
| Cell dimensions | ||
| a, b, c (Å) | 82.0, 82.0, 82.0 | 82.4, 82.4, 82.4 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 25.9–1.8 | 27.5–2.0 |
| Wavelength (Å) | 1.5418 | 1.5418 |
| Rsym (%) | 4.5 (27.5) | 7.3 (37.5) |
| I/σI | 78.9 (8.4) | 87.0 (13.2) |
| Completeness (%) | 99.1 (90.0) | 99.9 (100) |
| Redundancy | 19.3 (18.0) | 38.4 (36.6) |
| Refinement | ||
| Resolution (Å) | 25.9–1.8 | |
| Number of reflections | 9,329 | |
| Rwork/Rfree | 0.199/0.223 | |
| Number of protein atom | 744 | |
| Rmsds | ||
| Bond lengths (Å)/angles(°) | 0.007/1.03 | |
| Ramachandran plot | ||
| Most favored/allowed (%) | 100/100 | |
Values in parentheses are for the highest-resolution shell.
Because Bcl10 CARD forms filaments, we had to engineer a defined Bcl10 species for structural elucidation using crystallography or NMR. We attempted but failed to crystallize the MBP-Bcl10 fusion protein that is monomeric in solution. We then generated a number of Bcl10 mutants according to previous mutagenesis results on Bcl10 function (Yan et al., 1999). The E53R mutant of Bcl10 CARD was completely monomeric, and we determined its structure using NMR (Figure 4B, Table S1). A DALI structural homology search showed that Bcl10 CARD is quite different from other DD superfamily structures with unusually long α1 and α6 and a resultant pear shape (Figure 4C), which facilitated its fitting into the EM density (see below). Instead of having structural homologs with highest Z scores of >20 such as for Apaf-1 CARD, the closet structural homolog of Bcl10 is Apaf-1 with a pairwise Z score of 7.7.
The dynamics of Bcl10 CARD was studied by measuring longitudinal (R1) and transverse (R2) relaxation rates using a series of 1H-15N-HSQC experiments. Close to one-third of the residues in Bcl10 CARD are in chemical exchange, shown by high values of the product of R1 and R2 (R1*R2), suggesting that it may be in a highly dynamic state in the monomeric form. Considering that Bcl10 consists of a CARD domain and a likely unstructured S/T rich region, the full-length Bcl10 may also be highly dynamic by itself. In comparison, HSQC experiments showed that CARMA1 CARD has only a few residues identified to be in chemical exchange, including residues at the beginning of α1 and in the mobile loop between α3 and α4 (Figure 4D). In agreement with the high R1*R2 value, W23 of CARMA1 shows alternative conformations in the electron density map of the crystal structure. Additionally, hydrogen exchange measurements of Bcl10 CARD upon buffer-exchanged into 100% D2O resulted in complete HSQC crosspeak disappearance within the 10 min experimental dead time, indicating lack of protection for the backbone amides (Figure S4B). In contrast, CARMA1 CARD showed significant H-D exchange protection (Figure S4C), further supporting the differences in the dynamics of Bcl10 and CARMA1.
Helical Reconstruction of the Bcl10 Filament and Subunit Fitting
EM images of the negatively stained binary CARMA1/Bcl10 CARD complex showed helical diffraction (Figure S4D). A 3D map at ~20 Å resolution was obtained using a single-particle approach to helical reconstruction (Egelman, 2000). Because CARMA1 only localizes at one end of the filaments, the reconstruction represents a map of the Bcl10-only filament. To generate a model of the filament, we segmented the 3D map into subunits, rigidly fit one Bcl10 CARD domain structure into the segment density using Chimera’s fit-to-segment tool (Z score = 10.0) and applied the helical symmetry (Figures 4E and 4F). The fitting did not assume a priori similarity of assembly to previously determined death domain complex structures (Ferrao and Wu, 2012; Lin et al., 2010; Park et al., 2007). The validity of the fitting may be supported by the poor fitting (Z score = 2.9) of Bcl10 to the 3D map of the opposite hand and the poor fittings (Z scores = 1.7–4.0) of other CARD structures into 3D maps of either hand (Table S2). The long α1 and α6 helices may have been crucial in defining the pear shape of the Bcl10 molecule to obtain decent match to the density. Removal of the extended part of α6 from the Bcl10 CARD structure rendered the fitting poor (Table S2).
Unlike the previously determined death domain complex structures (Ferrao and Wu, 2012), the Bcl10 CARD filament is hollow with an inner diameter of ~25 Å and an outer diameter of ~100 Å (Figure 4E). The structure can be explained by a shallow left-handed single-stranded helical symmetry with ~3.6 subunit/turn and ~5 Å rise/subunit in which the adjacent subunits do not touch. The cross-section of the filament contains approximately seven subunits that correspond to two consecutive turns. Alternatively, the filament can be viewed as possessing a left-handed four-start prominent connected helical arrangement (Figure 4F), each with ~8.3 subunits/turn. Given the large percentage of the Bcl10 CARD surface that is buried in the filament and the dynamic state of the monomeric Bcl10 subunit, it is likely that Bcl10/Bcl10 interactions and stabilization of the subunit structure both contribute to the energetics of Bcl10 filament formation.
Detailed Interactions within the Bcl10 Filament
The fitted Bcl10 model allowed us to analyze the atomic interactions in the Bcl10 filament. The 3D map, as well as the model, indicated that there are two types of major interfaces between the Bcl10 sub-units in the filament (Figures 5A and 5B). One interface connects the adjacent sub-units in the four-start helical strands (Figure 5C). The other interface connects the subunits between the four-start helical strands (Figure 5D). Remarkably, despite the difference in overall helical arrangement, the intrastrand and the interstrand interfaces are similar to the type II and type I interactions, respectively (Figures S5A and S5B), which are conserved in all DD superfamily complexes (Ferrao and Wu, 2012). This similarity also lends support to the structure model of the Bcl10 filament. Both interfaces are rich in charge/charge interactions, including those involving R36 and E50-D51-E53-E54 in the intrastrand interface, and E30, D39, R42-K44-K45 and R62-K63-R65 in the interstrand interface. Supporting the structural model, E53 has been shown to be important for Bcl10 function (Yan et al., 1999) and was the site we mutated to generate monomeric Bcl10 for structure determination by NMR. Residues E50, E53, and E54 have also been mutated based on a Bcl10 homology model and shown to compromise Bcl10-induced NF-κB activation (Li et al., 2012) (Figure 5E). Other previously known mutations that disrupt Bcl10 function include those on L28, L41, I46, L47, and I55 (Yan et al., 1999), which are all buried in the Bcl10 structure and would have caused structural perturbation when mutated (Figure 5E). Almost all residues at the Bcl10/Bcl10 interfaces are conserved among different species (Figure S5C). In addition, we show that Bcl10 polymerization is progressively inhibited by higher salt concentrations using both FP and EM (Figure 5F, Figure S5D), consistent with the observed highly charged interactions.
Figure 5. Detailed Interactions in the Bcl10 Filament.
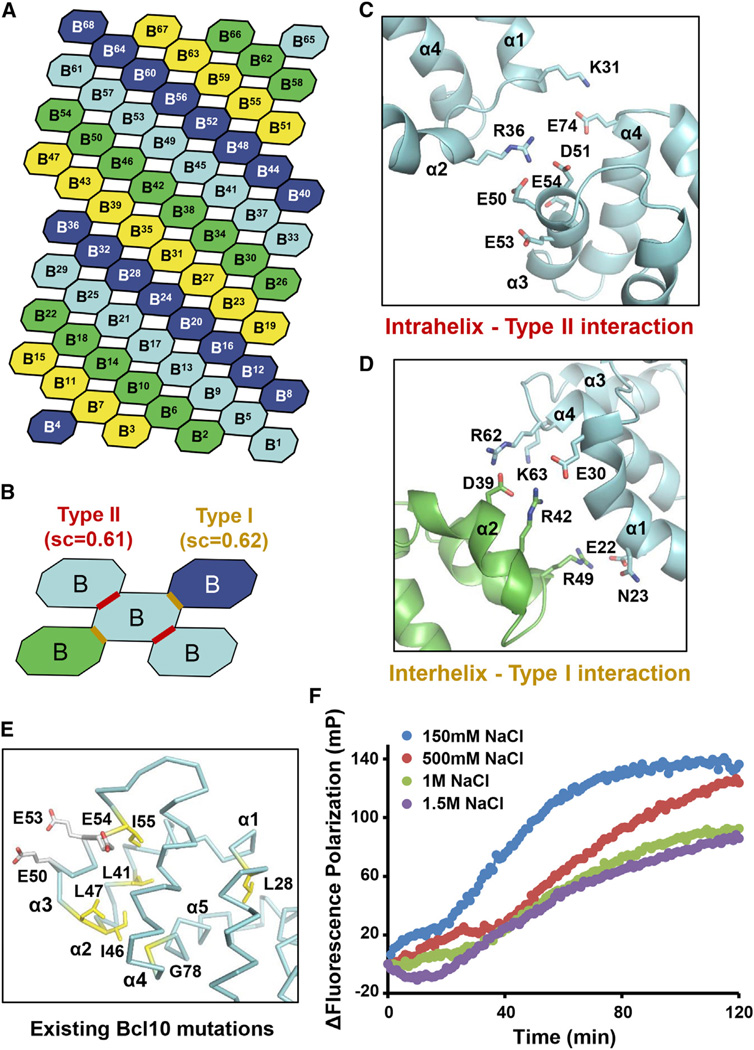
(A) A schematic model showing the helical symmetry in the Bcl10 filament. Bcl10 subunits are shown by octagons and sequentially labeled from B1 to B68 according to the single-start helical symmetry of 3.6 subunit per turn. The four colors represent the prominent connected left handed four-start helices.
(B) Interactions around a Bcl10 subunit (cyan), showing the intrahelix type II and the interhelix type I interactions (marked with red and yellow lines, respectively) and their shape complementarity scores (sc).
(C) Detailed intrahelix type II interactions between two subunits in cyan that are shown in (B).
(D) Detailed interhelix type I interactions between a cyan subunit and a green subunit that are shown in (B).
(E) Mapping of existing mutation sites that compromise Bcl10 function (Li et al., 2012; Yan et al., 1999) onto the structure. Yellow, buried residues; white, surface residues involved in the intrahelix interaction.
(F) Bcl10 polymerization under different NaCl concentrations using fluorescence polarization.
See also Figure S5.
Modeling the Interaction between CARMA1 and Bcl10
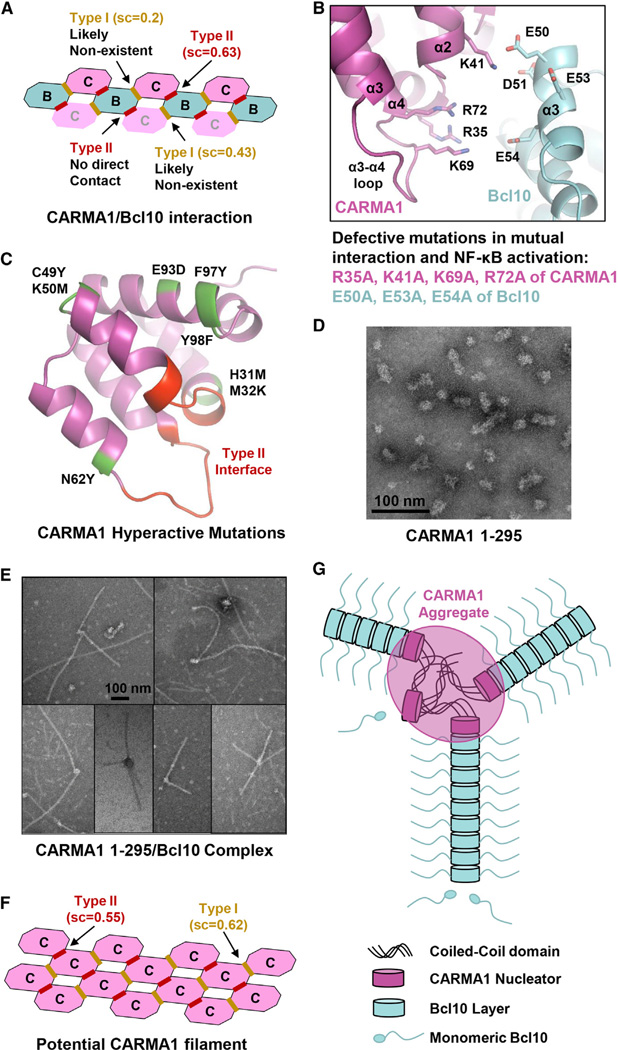
Since CARMA1 only resides at one end of the CARMA1/Bcl10 filament, we could not obtain CARMA1 density from an EM reconstruction. However, previous DD superfamily complex structures showed that the different DD proteins assemble together using the same helical symmetry such as the case for the Myddosome in which MyD88, IRAK4, and IRAK2 form a continuous helix with six MyD88, four IRAK4, and four IRAK2 molecules (Ferrao and Wu, 2012; Lin et al., 2010). To model how CARMA1 may interact with Bcl10 in the helical filament, we added one turn of CARMA1 to either end of a Bcl10 filament model (Figure 6A). Calculation of shape complementarity (sc) suggested that one side (top face) of the filament is more probable due to the high sc score of 0.63, which predicted a type II interface between CARMA1 and Bcl10 (Figure 6B). In comparison, the type I and type II Bcl10/Bcl10 interfaces have sc scores of 0.62 and 0.61, respectively.
Figure 6. Detailed Interactions between CARMA1 and Bcl10.
(A) Potential types of interactions if CARMA1 continues the helical symmetry in either direction of the Bcl10 filament. Shape complementarity (sc) scores are shown for each of the interfaces. The most probable interface is boxed in red.
(B) The modeled type II interactions between CARMA1 and Bcl10, which precisely match previous mutagenesis data (Li et al., 2012).
(C) Mapping of existing CARMA1 hyperactive mutation sites that do not disrupt CARMA1 filament formation or Bcl10 interaction (Chan et al., 2013) onto the structure (green). The sites of type II interaction with Bcl10 are shown in red.
(D) An EM image of CARMA1 (8–302), showing the heterogeneous clusters of 20–40 nm in size.
(E) EM images of CARMA1 (8–302)/Bcl10 filaments showing both single- and multiarmed filaments.
(F) Potential CARMA1 filament built based on the Bcl10 filament with reasonable sc scores.
(G) A schematic diagram of short CARMA1 filament-mediated Bcl10 polymerization.
See also Figure S6.
Remarkably, the CARMA1/Bcl10 interface predicted from the helical symmetry matched completely with that predicted from docking between mouse CARMA1 structure and a homology model of Bcl10 (Li et al., 2012). Positively charged residues at this interface, R35, K41, K69, and R72 of CARMA1, interact with negatively charged residues E50, D51, E53, and E54 of Bcl10. The flexible α3-α4 loop of CARMA1 is at this interface and contributes positively charged residues K69 and R72 (Figure 6B). These residues have been shown to be important for the interaction between CARMA1 and Bcl10, and for NF-κB reporter activity in transfection experiments (Li et al., 2012) (Figure 6B). In addition, mutations identified from a high-throughput quantitative signaling screen that overcome CARMA1 autoinhibition (Chan et al., 2013) are mapped away from the interface (Figure 6C) and therefore can be hyperactive. The predicted interfacial residues in CARMA1 CARD are highly conserved among the different CARMA family members that also include CARMA2, CARMA3, and CARD9 (Figure S6), suggesting that they might all nucleate Bcl10 filaments similarly. The same Bcl10 residues also participate in the type II interaction in filament formation. Collectively, these data demonstrate the validity of the CARMA1/Bcl10 interaction.
The CARMA1/Bcl10 interaction model suggests that three CARMA1 molecules may be a minimal requirement for promotion of Bcl10 polymerization (Figure 6A). The trimeric CARMA1 (8–172) construct (Figure 2B) may therefore serve as a minimal nucleation platform. In contrast, CARMA1 (8–302) is a higher-order oligomer, estimated to contain an average of >100 CARMA1 protomers (Figure 2B). EM of negatively stained CARMA1 (8–302) showed relatively compact but heterogeneous cluster-like structures with average sizes of ~20–40 nm in dimensions, larger than the diameter of Bcl10 filaments (Figure 6D). Interestingly, the CARMA1 (8–302)/Bcl10 complex existed as both single and "multiarmed" filaments (Figure 6E), suggesting that each CARMA1 (8–302) cluster has the potential to nucleate multiple filaments. The CARMA1 "head" from which the filaments stem shows diameters larger than Bcl10, consistent with dimensions of CARMA1 (8–302) (Figure 6D). The number of Bcl10 filaments that each CARMA1 (8–302) cluster nucleates appeared to be random and varied from 1 to a few. The ability to form multiarmed filaments may explain the crisscross morphology of the coexpressed CARMA1/Bcl10 filaments from cells and the convoluted morphology of the endogenous CBM complex under EM (Figures 3C and 3D, Figures S3E and S3I).
Given the large number of CARMA1 molecules in the 8–302 construct and the relatively small number of Bcl10 filaments it nucleates, we wondered whether the coiled-coil oligomerized CARMA1 forms one or several short filaments within each cluster. Formation of Bcl10-like filaments in CARMA1 (8–302), but not in CARMA1 (8–172), could explain the enhanced nucleation capability of the former construct by prealigning the CARMA1 subunits for optimal interaction with Bcl10 subunits. Calculation of shape complementarity shows that type I and type II CARMA1/CARMA1 interactions possess sc scores of 0.62 and 0.55, respectively (Figure 6F), which are representative of feasible protein-protein interaction surfaces, supporting the possibility. Therefore, in this scenario, CARMA1, with its multiple coiled-coil domains, may form short helical segments to nucleate Bcl10 filaments (Figure 6G). Because of the importance of the coiled-coil domains in this process, it is not surprising that activating mutations in DLBCL map to these domains (Lenz et al., 2008), likely through both overcoming autoinhibition and enhancing oligomerization.
Structure-Guided Bcl10 Mutations In Vitro: Disruption and Complementation
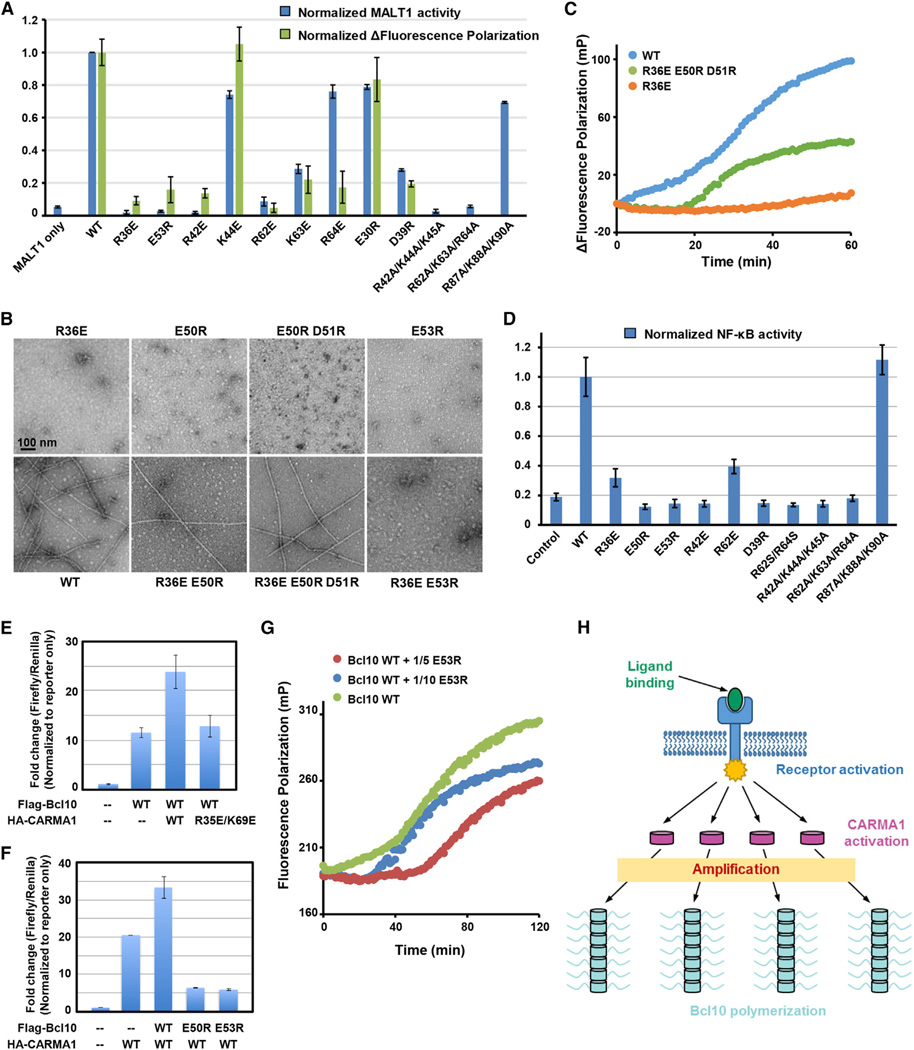
To determine whether the observed Bcl10/Bcl10 interaction affects Bcl10 filament formation in vitro and in cells, we selected charged residues that bury the most surface area at the interface for mutagenesis. Because many residues at the type II interface such as E50, E53, and E54 have already been tested previously (Li et al., 2012; Yan et al., 1999), we mostly focused on the type I interface. These include mutations R42E, K44E, R62E, K63E, R64E, E30R, D39R, and triple combination mutations at the type I interface, a control triple mutation on residues outside of the interface (R87A/R88A/K90A), and R36E and E53R at the type II interface. The mutants were characterized in two independent assays, in vitro Bcl10 polymerization assessed by FP and MALT1 activity in the context of the CBM complex (Figure 7A). Consistent results were obtained from both assays for all but one mutant, showing that most of the interfacial residues are crucial for Bcl10 polymerization as well as CBM complex formation-induced MALT1 activation. R64E is defective in Bcl10 polymerization but close to normal in MALT1 activity, likely due to the longer time used in assembly of the CBM complex for the MALT1 assay.
Figure 7. Structure-Based Bcl10 Mutagenesis In Vitro and in Cells.
(A) Summary of effects of Bcl10 mutants in polymerization (green bars) and in MALT1 activation (blue bars) normalized to the WT Bcl10.
(B) EM images of defective Bcl10 mutants, and combined Bcl10 mutants that rescued filament formation.
(C) FP assays showing recovery of Bcl10 polymerization through mutation complementation.
(D) Effects of the similar set of Bcl10 mutants in (A) in induction of NF-κB activity upon expression in 293T cells normalized to the WT Bcl10.
(E) NF-κB activity upon expression of Flag-Bcl10 alone and coexpression with WT and mutant HA-CARMA1.
(F) NF-κB activity upon expression of HA-CARMA1 alone and coexpression with WT and mutant Flag-Bcl10.
(G) Dominant-negative inhibition of WT Bcl10 polymerization by substoichiometric amounts of the E53R mutant of Bcl10.
(H) A schematic diagram of nucleation-induced signal amplification as a paradigm in signal transduction.
Data in (A), (D), (E), and (F) are represented as mean ± SD from three independent measurements.
See also Figure S7.
To further test the interaction in the filament model, we designed double and triple mutations to determine if such combinations could rescue the defective phenotypes of the single mutations. In the intrahelix interaction, R36 of one Bcl10 is poised against E50 and D51 of the neighboring Bcl10 (Figure 5C), and we wondered if charge reversal could restore the interaction. We also picked E53, which is adjacent to the negative patch of E50 and E51 but does not appear to directly interact with R36 (Figure 5C). As expected, R36E, E50R, E50R/D51R, and E53R of Bcl10 did not show any filaments under EM (Figure 7B). When combined, the R36E/E50R double mutant showed a small portion of filaments, while the R36E/E50R/D51R triple mutant showed comparable amounts of filaments with WT Bcl10 (Figure 7B), demonstrating mutation complementation that rescued filament formation. In contrast, the R36E/E53R double mutant did not show filament formation (Figure 7B). Quantitative assessment of the R36E/E50R/D51R triple mutant showed that it is ~50% as active as WT Bcl10, while the noncombined mutant R36E is completely inactive in polymerization (Figure 7C). These data on the rescue of Bcl10 interaction by complementary charge reversals further confirmed the validity of the Bcl10 filament model.
Bcl10 Filament Formation Is Critical for Signal Amplification in Cells
To investigate how the CBM complex functions in cells, we transfected into 293T cells Bcl10 mutants defective in activities in vitro, and monitored NF-κB activity changes using a luciferase assay. All mutations at the Bcl10/Bcl10 interfaces, both type I and type II residue mutations, caused significant decrease in NF-κB activity (Figure 7D). In contrast, WT and the control triple mutant R87A/R88A/K90A showed high and comparable luciferase activity. Immunoprecipitated Bcl10 mutants that are impaired in NF-κB activation showed no filament structures under EM comparing to WT Bcl10 (Figure S7), suggesting that the defectiveness in NF-κB activation is a consequence of their inability to form higher-order filamentous structures. These results not only validated the structure but also indicated that signal amplification due to Bcl10 polymerization in cells may be an essential step in the CBM signaling pathway.
Dominant-Negative Inhibition of Bcl10 Filament Assembly In Vitro and In Vivo
When transfected into 293T cells, exogenous CARMA1 expression caused significant enhancement in NF-κB activity, likely through activation of endogenous Bcl10, and coexpression with WT Bcl10 further increased this activity (Figure 7E). Coexpression of WT Bcl10 with CARMA1 mutant that cannot recruit Bcl10 results in similar NF-κB activity with WT Bcl10 transfection alone (Figure 7E), suggesting that mutant CARMA1 does not interfere with Bcl10 signaling. In contrast, coexpression of WT CARMA1 with Bcl10 polymerization-defective mutants E53R and E50R intriguingly not only did not augment but also significantly decreased CARMA1-mediated NF-κB activation (Figure 7F), suggesting dominant-negative inhibition of endogenous CBM signaling by Bcl10 mutants. Because Bcl10 mutants could not be recruited by CARMA1 (Li et al., 2012), this inhibition of CBM signaling is most likely due to disruption of the Bcl10/Bcl10 interaction. To test this hypothesis, we showed in vitro that addition of even substoichiometric amounts of the defective Bcl10 mutant E53R significantly inhibited WT Bcl10 polymerization with increased lag time and decreased FP (Figure 7G). Dominant inhibition against WT proteins is a characteristic of mutants of highly oligomeric assemblies, such as for the autoimmune lymphoproliferative disease, in which the death-inducing signaling complex is inhibited by heterozygotic mutations of the death domain-containing receptor Fas (Wang et al., 2010).
DISCUSSION
Paradigm of Nucleation Induced Threshold Behavior in Signal Transduction
The data presented here show that the CBM complex is a supramolecular filamentous complex in which CARMA1 acts as the nucleator to promote Bcl10 polymerization and MALT1 activation. The biophysical nature of such an assembly dictates that this is a cooperative process, as shown by the Hill coefficients of the polymerization reactions in the absence and presence of CARMA1. This nucleation process provides a mechanism for signal amplification in which a small amount of activated CARMA1 can activate a large number of Bcl10 molecules in the cell (Figure 7H). Both the cooperative process and the amplification mechanism would likely lead to a digital, all-or-none, or threshold mode of signal transduction. In this regard, previous studies have shown that TCR-mediated activation of NF-κB is switch-like in nature, in which analog antigen inputs to TCR are converted to digital response in NF-κB activation (Kingeter et al., 2010). With increasing TCR stimulation, the number of responsive cells increases while the magnitude of NF-κB activation is remarkably constant.
The mechanism described here provides a paradigm of nucleation-induced signal transduction that results in a threshold response. We speculate that many pairs of DD fold superfamily interactions, including those of DDs, CARDs, death effector domains (DEDs), and Pyrin domains, may employ similar mechanism in which one interaction partner nucleates a filamentous structure of the other partner. In caspase-8 activation by FADD in death receptor signaling pathways, it has been shown that the complexes contain substoichiometric amount of FADD relative to caspase-8 (Dickens et al., 2012; Schleich et al., 2012), suggesting that FADD DED nucleates caspase-8 DED filaments. Consistent with this, Fas death receptor signaling shows a threshold mechanism for the regulation of apoptosis (Bentele et al., 2004). In immune signaling pathways, additional higher-order assemblies have been identified, which may employ analogous cooperative and amplification processes to induce threshold responses in signal transduction (Wu, 2013).
Similarities to Polymerization of the Cytoskeleton
The mode of directional nucleation in CBM signaling by formation of platforms using related structures may be mechanistically similar to polymerization of the cytoskeleton. In microtubule polymerization, the γ-tubulin ring complex provides the template for addition of α/β tubulin dimers to generate microtubule growth (Kollman et al., 2011). γ-tubulin is similar to α- and β-tubulins in its overall fold. It interacts with γ-tubulin complex proteins (GCPs) that mediate the assembly of a helical structure with symmetry similar to that of the microtubule composed of α- and β-tubulins.
Conformational regulation of nucleation activity appears to be a central control theme in polymerization of the cytoskeleton. In microtubule polymerization, there is an imperfect match between γ-tubulins in the ring complex and the α/β microtubule. The intrinsic flexibility of the GCPs allows conformational adjustments to generate a perfect match between the γ-tubulin geometry and the microtubule geometry to promote nucleation. For CBM signaling, CARMA1 is kept in an autoinhibited conformation by intricate intramolecular interactions in resting states. Upon receptor engagement, this conformation is antagonized by signal-dependent phosphorylation at the CARMA1 linker region (Figure 1A). In DLBCL and other lymphomas, missense mutations of CARMA1 are found in the coiled-coil domain and known to overcome autoinhibition in the absence of phosphorylation, leading to constitutive NF-κB signaling (Lenz et al., 2008). Additionally, the flexibility in the coiled-coil domains and domain junctions of CARMA1 likely permits helical oligomerization of CARMA1 CARD molecules within the CARMA1 clusters to maximally nucleate Bcl10 filaments for signal transduction. Therefore, signal-dependent, controlled, nucleated formation of large assemblies may be a general feature in multiple biological processes.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Human CARMA1 and Bcl10 constructs and full-length MALT1 were expressed in E. coli and insect cells, respectively, and purified by Ni-NTA resin (QIAGEN) and gel filtration chromatography (Superdex 200 10/300 GL, GE Healthcare).
Electron Microscopy
For negative staining, 5µl of purified samples were applied to copper grids, left on the grids for 1 min followed by negative staining with 5% uranyl acetate for 1 min, and air dried. For helical reconstruction, the CARMA1 (8–172)/Bcl10 (1–115) complex samples were negatively stained using uranyl acetate (2% w/v) and imaged in a Tecnai 12 (FEI) TEM at an accelerating voltage of 80 keV. From the EM images, 13,375 overlapping segments (100 pixels long) were collected, with a shift of 8 pixels between adjacent segments. The IHRSR algorithm (Egelman, 2000) was used for the 3D reconstruction.
Crystallization and Structure Determination
The human CARMA1 CARD (residues 11–103) was crystallized in 1.4 M MgSO4 and 100 mM MES at pH 6.5. For phase determination, crystals were soaked with 1 M potassium iodide (KI), and the initial phases were determined by autoSHARP using single wavelength anomalous diffraction.
NMR Spectroscopy and Structure Calculations
All NMR experiments were recorded at 27°C on Varian Unity Inova 600 MHz spectrometer equipped with cryoprobe.
Supplementary Material
ACKNOWLEDGMENTS
We thank Natacha Opalka, Seth Darst, Leona Cohen-Gould, and Maria Ericsson for help with EM imaging, and Liwei Wang, Ermelinda Damko, and Qiubai Li for technical assistance, and recognize support from the National Institutes of Health (R01AI089882 to H.W. and R01EB001567 to E.H.E.), Cancer Research Institute (Q.Q., C.Y., and L.D.), Spanish Ministry of Science and Innovation (L.F.), and Michael D. Ditzian Research Fund-Lymphoma Research Foundation (L.F.).
Footnotes
ACCESSION NUMBERS
The EM map of Bcl10 filament was deposited to EMDataBank under accession code EMD-5729. The crystal structure of CARMA1 CARD and the NMR structure of Bcl10 CARD were deposited in the Protein Data Base with ID codes 4LWD and 2MB9, respectively.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures, two tables, and Supplemental Experimental Procedures and can be found with this article at http://dx.doi.org/10.1016/j.molcel.2013.08.032.
REFERENCES
- Bentele M, Lavrik I, Ulrich M, Stösser S, Heermann DW, Kalthoff H, Krammer PH, Eils R. Mathematical modeling reveals threshold mechanism in CD95-induced apoptosis. J. Cell Biol. 2004;166:839–851. doi: 10.1083/jcb.200404158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin J, Guo Y, Wang L, Srinivasula SM, Jacobson MD, Poyet JL, Merriam S, Du MQ, Dyer MJ, Robison KE, et al. CARD9 is a novel caspase recruitment domain-containing protein that interacts with BCL10/CLAP and activates NF-kappa B. J. Biol. Chem. 2000;275:41082–41086. doi: 10.1074/jbc.C000726200. [DOI] [PubMed] [Google Scholar]
- Chan W, Schaffer TB, Pomerantz JL. A quantitative signaling screen identifies CARD11 mutations in the CARD and LATCH domains that induce Bcl10 ubiquitination and human lymphoma cell survival. Mol. Cell. Biol. 2013;33:429–443. doi: 10.1128/MCB.00850-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coornaert B, Baens M, Heyninck K, Bekaert T, Haegman M, Staal J, Sun L, Chen ZJ, Marynen P, Beyaert R. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat. Immunol. 2008;9:263–271. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- Dickens LS, Boyd RS, Jukes-Jones R, Hughes MA, Robinson GL, Fairall L, Schwabe JW, Cain K, Macfarlane M. A death effector domain chain DISC model reveals a crucial role for caspase-8 chain assembly in mediating apoptotic cell death. Mol. Cell. 2012;47:291–305. doi: 10.1016/j.molcel.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Ferch U, Kloo B, Gewies A, Pfänder V, Düwel M, Peschel C, Krappmann D, Ruland J. Inhibition of MALT1 protease activity is selectively toxic for activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 2009;206:2313–2320. doi: 10.1084/jem.20091167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Curr. Opin. Struct. Biol. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontan L, Yang C, Kabaleeswaran V, Volpon L, Osborne MJ, Beltran E, Garcia M, Cerchietti L, Shaknovich R, Yang SN, et al. MALT1 small molecule inhibitors specifically suppress ABC-DLBCL in vitro and in vivo. Cancer Cell. 2012;22:812–824. doi: 10.1016/j.ccr.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiet C, Vito P. Caspase recruitment domain (CARD)-dependent cytoplasmic filaments mediate bcl10-induced NF-kappaB activation. J. Cell Biol. 2000;148:1131–1140. doi: 10.1083/jcb.148.6.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachmann J, Snipas SJ, van Raam BJ, Cancino EM, Houlihan EJ, Poreba M, Kasperkiewicz P, Drag M, Salvesen GS. Mechanism and specificity of the human paracaspase MALT1. Biochem. J. 2012;443:287–295. doi: 10.1042/BJ20120035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailfinger S, Lenz G, Ngo V, Posvitz-Fejfar A, Rebeaud F, Guzzardi M, Penas EM, Dierlamm J, Chan WC, Staudt LM, Thome M. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA. 2009;106:19946–19951. doi: 10.1073/pnas.0907511106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailfinger S, Nogai H, Pelzer C, Jaworski M, Cabalzar K, Charton JE, Guzzardi M, Décaillet C, Grau M, Dörken B, et al. Malt1-dependent RelB cleavage promotes canonical NF-kappaB activation in lymphocytes and lymphoma cell lines. Proc. Natl. Acad. Sci. USA. 2011;108:14596–14601. doi: 10.1073/pnas.1105020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Lin X. Regulation of NF-κB by the CARD proteins. Immunol. Rev. 2012;246:141–153. doi: 10.1111/j.1600-065X.2012.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingeter LM, Paul S, Maynard SK, Cartwright NG, Schaefer BC. Cutting edge: TCR ligation triggers digital activation of NF-kappaB. J. Immunol. 2010;185:4520–4524. doi: 10.4049/jimmunol.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Merdes A, Mourey L, Agard DA. Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol. 2011;12:709–721. doi: 10.1038/nrm3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, Dave SS, Zhao H, Xu W, Rosenwald A, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- Li S, Yang X, Shao J, Shen Y. Structural insights into the assembly of CARMA1 and BCL10. PLoS ONE. 2012;7:e42775. doi: 10.1371/journal.pone.0042775. http://dx.doi.org/10.1371/journal.pone.0042775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-García ME, Sommer K, Rincon-Arano H, Brault M, Ninomiya-Tsuji J, Matesic LE, Rawlings DJ. Kinase-independent feedback of the TAK1/TAB1 complex on BCL10 turnover and NF-κB activation. Mol. Cell. Biol. 2013;33:1149–1163. doi: 10.1128/MCB.06407-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel D, Spranger S, Vincendeau M, Grau M, Raffegerst S, Kloo B, Hlahla D, Neuenschwander M, Peter von Kries J, Hadian K, et al. Pharmacologic inhibition of MALT1 protease by phenothiazines as a therapeutic approach for the treatment of aggressive ABC-DLBCL. Cancer Cell. 2012;22:825–837. doi: 10.1016/j.ccr.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Park HH, Logette E, Raunser S, Cuenin S, Walz T, Tschopp J, Wu H. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-kappaB. Immunity. 2012;36:947–958. doi: 10.1016/j.immuni.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeaud F, Hailfinger S, Posevitz-Fejfar A, Tapernoux M, Moser R, Rueda D, Gaide O, Guzzardi M, Iancu EM, Rufer N, et al. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat. Immunol. 2008;9:272–281. doi: 10.1038/ni1568. [DOI] [PubMed] [Google Scholar]
- Rosebeck S, Rehman AO, Lucas PC, McAllister-Lucas LM. From MALT lymphoma to the CBM signalosome: three decades of discovery. Cell Cycle. 2011;10:2485–2496. doi: 10.4161/cc.10.15.16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman JS, Stoicheva NG, Langel FD, Patterson GH, Lippincott-Schwartz J, Schaefer BC. POLKADOTS are foci of functional interactions in T-Cell receptor-mediated signaling to NF-kappaB. Mol. Biol. Cell. 2006;17:2166–2176. doi: 10.1091/mbc.E05-10-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleich K, Warnken U, Fricker N, Oztürk S, Richter P, Kammerer K, Schnölzer M, Krammer PH, Lavrik IN. Stoichiometry of the CD95 death-inducing signaling complex: experimental and modeling evidence for a death effector domain chain model. Mol. Cell. 2012;47:306–319. doi: 10.1016/j.molcel.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Staal J, Driege Y, Bekaert T, Demeyer A, Muyllaert D, Van Damme P, Gevaert K, Beyaert R. T-cell receptor-induced JNK activation requires proteolytic inactivation of CYLD by MALT1. EMBO J. 2011;30:1742–1752. doi: 10.1038/emboj.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser D, Neumann K, Bergmann H, Marakalala MJ, Guler R, Rojowska A, Hopfner KP, Brombacher F, Urlaub H, Baier G, et al. Syk kinase-coupled C-type lectin receptors engage protein kinase C-σ to elicit Card9 adaptor-mediated innate immunity. Immunity. 2012;36:32–42. doi: 10.1016/j.immuni.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yang JK, Kabaleeswaran V, Rice AJ, Cruz AC, Park AY, Yin Q, Damko E, Jang SB, Raunser S, et al. The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nat. Struct. Mol. Biol. 2010;17:1324–1329. doi: 10.1038/nsmb.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesmann C, Leder L, Blank J, Bernardi A, Melkko S, Decock A, D’Arcy A, Villard F, Erbel P, Hughes N, et al. Structural determinants of MALT1 protease activity. J. Mol. Biol. 2012;419:4–21. doi: 10.1016/j.jmb.2012.02.018. [DOI] [PubMed] [Google Scholar]
- Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153:287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Lee J, Schilbach S, Goddard A, Dixit V. mE10, a novel caspase recruitment domain-containing proapoptotic molecule. J. Biol. Chem. 1999;274:10287–10292. doi: 10.1074/jbc.274.15.10287. [DOI] [PubMed] [Google Scholar]
- Yu JW, Jeffrey PD, Ha JY, Yang X, Shi Y. Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region. Proc. Natl. Acad. Sci. USA. 2011;108:21004–21009. doi: 10.1073/pnas.1111708108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.