Abstract
The quantitative analysis of blood vessel volumes from magnetic resonance angiograms (MRA) or μCT images is difficult and time-consuming. This fact, when combined with a study that involves multiple scans of multiple subjects, can represent a significant portion of research time. In order to enhance analysis options and to provide an automated and fast analysis method, we developed a software plugin for the ImageJ and Fiji image processing frameworks that enables the quick and reproducible volume quantification of blood vessel segments. The novel plugin named Volume Calculator (VolCal), accepts any binary (thresholded) image and produces a three-dimensional schematic representation of the vasculature that can be directly manipulated by the investigator.
Using MRAs of the mouse hindlimb ischemia model, we demonstrate quick and reproducible blood vessel volume calculations with 95 – 98% accuracy. In clinical settings this software may enhance image interpretation and the speed of data analysis and thus enhance intervention decisions for example in peripheral vascular disease or aneurysms.
In summary, we provide a novel, fast and interactive quantification of blood vessel volumes for single blood vessels or sets of vessel segments with particular focus on collateral formation after an ischemic insult.
Keywords: Magnetic resonance angiogram, blood vessel volume analysis, 3D analysis, mouse hindlimb ischemia
Introduction
Imaging of the vasculature is of great importance in understanding various human diseases ranging from peripheral vascular disease and cerebrovascular disease to cancer. Improved resolution of non-invasive imaging methods such as magnetic resonance imaging (MRI) have increased the use of imaging for clinical diagnosis and treatment monitoring of these diseases. However, the analysis of complex vascular trees and especially vascular volumes remains challenging and time consuming. The volume analysis of blood vessels has traditionally been a manual or semi-automated process and involves the placement of region-of-interest areas in the blood vessels to be analyzed in slices of 2D scans; this process becomes very labor intensive. For example, a simple experimental set-up of 4 mice per treatment group with sham, untreated, and treated group and analysis of a 5 mm vessel segment could require upwards of 600 magnetic resonance angiogram (MRA) slices (12 mice total, with 100 μm resolution in an MRA - 5 mm yields 50 slices – thus, 12 × 50 = 600 slices).
When constrained by time, a slow manual method can lead to missed opportunities for the data collection of more complex blood vessels. Faced with a similar experimental requirement, we devised a novel image analysis technique that shortens the amount of time needed for measuring vasculature volumes while still maintaining accuracy.
The preparation of a vascular image for subsequent quantification typically involves segmentation followed by or in conjunction with analysis. Segmentation delineates objects of interest and analysis serves to identify vascular components: e.g., vessel segments, bifurcations, thromboids, etc. Segmentation software tools are reviewed in [1,2]. Among those available, the MARACAS tool [3] allows for very fine control over the choice of a vessel section for quantification. Most of this analysis is done semi-automatically with the user selecting starting points for boundaries, thus it does not decrease analysis time. An analysis procedure for angiogenesis after an ischemic injury [4] is similar to our approach. However, their method does not allow the analysis of a single blood vessel segment and their measurements are obtained from μCT images of Microfil blood vessels casts, whereas our study is performed in vivo. Finally, Stefancik et al. [5] focused primarily on segmentation for differentiating arteries from veins. Selection of vessels of interest can be performed on a 3D image, however their software interface does not seem to offer the user the ability to choose multiple, ad hoc sets of measurements.
In contrast to these segmentation techniques, our effort was focused on making the selection of vessel segments for quantification both easy and fast. To that end, we extended existing 3D skeletonization and 3D analysis tools. The extension is based on a simple insight: namely, that any voxel displayed in a segmented version of the MRA could be traced back to a voxel in the original image. That is, if a user chose a vessel segment from the image skeleton, the voxels in that segment could be used to locate the same voxels in the original image. In effect, this is the reverse of skeletonization but with control over what is selected in the image. Thus, here we describe a new functional extension - a plugin – that we developed for the Fiji [6] and ImageJ [7] image processing frameworks. It is named Volume Calculator (VolCal) and it was built to fulfill the requirements of various experimental designs, including those addressing the processes of arteriogenesis, angiog-enesis or pharmacological treatment of vascular disease. Our method provides quick, easy and accurate volume measurements of specific sets of blood vessels. The new plugin accepts a binary (thresholded) version of the original scanned image (MR or μCT image) and automatically produces a graphical depiction of the vasculature that can be manipulated by the user in three dimensions. Subsequently, the user can select a specific segment of a vessel, a path of segments or a set of segments with a mouse pointer and have their volumes (in voxels or other volumetric units) automatically calculated and displayed. The accuracy of this method was established using synthetic images of blood vessel trees [8] and resulted in 99.5 – 99.9% recovery of pixel number and volumes. We further determined accuracy in MRAs of the mouse ischemic hindlimb and validated these numbers of in vivo blood vessels using μCT scans of Microfil vascular bed casts of the mouse hindlimb. We found 98% accuracy (based on voxel counts in VolCal compared to voxel counts in commercial software) in obtaining the correct volume. This value is concordant with other established methods of blood vessel volume measurements [3,5], however our method is not labor-intensive, which is especially helpful for longitudinal studies.
Materials and Methodology
Image Analysis
A method that provides support for the direct user-selection of blood vessels for volume measurement is reported here. The method begins with the construction of an in-memory representation of a thresholded image. After analysis, the in-memory image is presented to the user as a 3-dimensional figure. Finally, the user directly interacts with the presentation, selecting blood vessels of interest whereupon VolCal calculates and displays their volumes. An overview of the steps in the method is shown in Fig. (1).
Fig. (1).
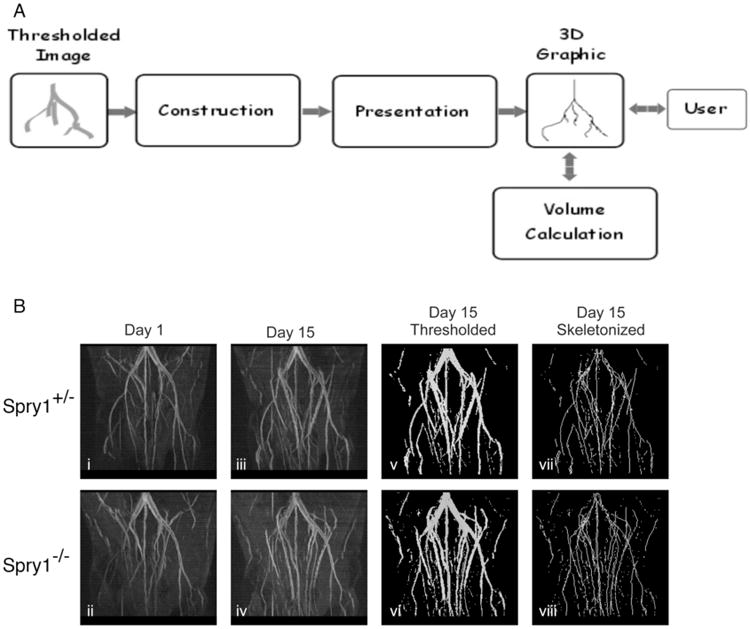
Workflow for image processing using VolCal. A) Graphical overview of the workflow in VolCal. The MR angiogram is thresholded, skeletonized and then analyzed in VolCal. B) Representative images i and ii show MRAs one day post surgery with the ischemic side (left side) clearly showing fewer blood vessels compared to the non-ischemic side (right side). iii and iv are the MRAs after 15 days and show arteriogenesis and collateral formation in the ischemic legs of bothSpry1+/− control and Spry1−/− mice. Images v and vi show the thresholded images after 15 days. Care was taken to use the same threshold for all images of one project. vii and viii show the skeletonized images that were analyzed in VolCal.
A binary (thresholded) image serves as input to the method; this is a requirement of the skeletonization algorithm employed in the construction step. It can be created in several ways, but usually by choosing a threshold value that displays the vessels of interest with a minimum amount of background noise. To reduce any variance in the measurements of the vessel volumes, all MRAs were acquired using the same set of scanning parameters and the threshold value – once a value was determined – was held constant.
Construction
The construction of the in-memory representation of the vasculature is comprised of three steps: skeletonization, tree analysis, and three-dimensional graphical construction.
Skeletonization
In this step the binary image is skeletonized using an existing Fiji/ImageJ plugin: Skeletonize3D [9]. This plugin was designed to operate in three dimensions. It erodes voxels until a vessel is a single voxel wide. The skeletonized image is ultimately what the user sees and manipulates.
Tree Analysis
The skeletonized image becomes input to another Fiji/ImageJ plugin: AnalyzeSkeleton [9]. This plugin analyzes, again in three dimensions, the entire skeleton and determines whether each voxel is a junction (bifurcation, trifurcation, etc.), part of a branch, or the end point of a branch. While this plugin is executing, it is creating an in-memory, schematic representation of the skeletonized image (a forest of trees). In this representation the 3D location of every type of identified element (junction or end point) is noted as well as the elements (branches) to which it is connected. The normal use of this plugin ends here and the user can examine a skeletonized image with junctions displayed in different colors. A table of branch data is also created. These data can be used to measure vessel lengths but not volumes.
Three-dimensional Graphical Construction
After the tree analysis has completed, VolCal uses the in-memory structures, built in the last step, along with the facilities of the Java 3D library [10] to create a graphical (as opposed to a bit-mapped) representation of the vasculature. The advantage of this representation is that each element (edge/segment) of the image exists as a separate object annotated with a unique identification, its 3D position and its connections to other elements. Importantly, because these objects are Java 3D objects, it provides targets (vessel segments) for the user to select with a pointer.
Presentation and User Interaction
To make the 3-dimensional image available to the user for viewing and manipulation, this structure is transferred to the 3D Viewer plugin [11], which is responsible for displaying the image and providing the user with graphical transformations such as panning, zooming and rotation.
VolCal augments the viewer with additional user interface controls that allows for the creation of sets of vessel measurements. Selected vessels are displayed in a color. The total volume of selected vessels for a color is shown in the user control area. VolCal has one predefined color but any number of additional colors can be selected by pushing the New Volume Color button. Any of the available colors may be selected with a push button and subsequent vessel selections will be displayed with the newly chosen color. Next to a color selector is a button that can change that color's vessels back to the original color and at the same time reduce the total number of voxels that have been counted. Finally, selecting an already colored segment with a different selection color will reduce the voxel count of the first color and augment the voxel count for the new color. Fig. (2) show the user interface after the selection of two sets of blood vessels.
Fig. (2). Screenshot of MRA image in VolCal.
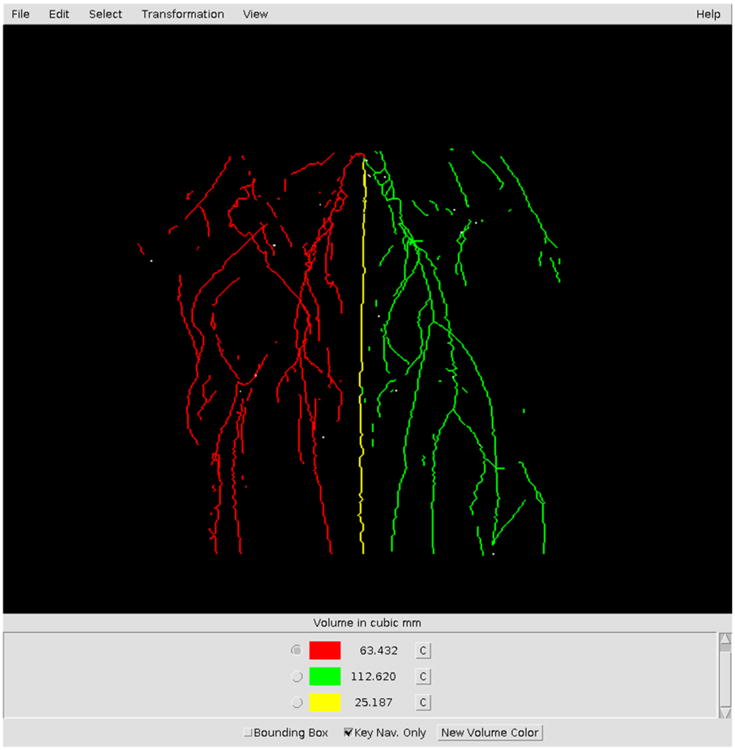
The screenshot shows a skeletonized image in VolCal and how the user can choose blood vessel trees and highlight them in different colors. The user has also the option of picking single blood vessels. In the lower part of the window the total volumes for the vessels of a given color are shown, in this case the ischemic leg is shown in red, the non-ischemic leg in green and the tail in yellow.
Volume Calculation
The user has the option of choosing one or more segments from the three-dimensional representation of the vasculature, a segment being defined as the portion of a vessel between junctions and/or vessel ends. The user initiates volume calculation by clicking twice in the vasculature. If the first selected segment (first click) is not the same as the second selected segment (second click), VolCal will determine if there is a path that can connect these two segments.
The presence of a path is determined by using information maintained by the Java 3D graphics structures. A Scene Graph Path (SGP) is returned with every point selected by the user. The SGP represents the direct path from the selected 3D element “up” a tree to its root. This information was created when the skeletal vasculature was reconstructed by traversing all the vasculature trees identified during the analysis step, from the root down every path. A path between two elements exists only if each path's SGP has at least one shared juncture (vertex).
If there is a path then the volume of all the segments lying in the path is calculated. This is performed by travelling “up” from the two segments until a shared segment is reached. If only one segment has been selected, then the volume of only that segment is calculated. The color of the segment changes (as described above) and VolCal retrieves the segment's three-dimensional coordinates. If there are junctures (at either or both ends of the segment), information about the branches is also obtained.
Next, each three-dimensional location point along the selected line(s), including the end points, is used to navigate to the corresponding voxel in the original binary image. The z dimension in the coordinate indicates the slice number and the x and y values point to a location in the slice plane. From this starting point, all contiguous and adjacent voxels (in the slice plane) are counted. Subsequently, this counting is performed for every slice through which the vessel(s) passes. By summing these slice counts the volume of the selected vessel segments is calculated.
This count of these voxels serves as a starting point for the remainder of the volume calculation. Because we have only the skeletal voxels as a guide, the calculation of the volume is not complete yet. There are voxels in the scanned image that are not indexed in the skeleton because of the erosion involved in the skeletonization process; these voxels have to be recovered. The following steps are performed for every edge in the skeleton:
For each skeletal point in the edge, recover any contiguous eroded voxels in the slice/plane.
Voxels may have been “eaten” away from the ends of the edge. There can be one or two ends. Recover these voxels.
Due to the intrinsic problem of potential voxel sharing between blood vessels (edges) a table is compiled, during voxel recovery, for every distinct, contiguous group of voxels in a plane. Associated with each entry in the table is a list of the edges that pass through that group of voxels. With this technique we can determine when some voxels are shared between two or more vessel segments. In this case the number of shared voxels is divided equally between the segments (Fig. 5A - C).
Fig. (5). Representative μCT image of a Microfil vessel cast of the mouse hindlimbs with corresponding VolCal analysis and accuracy.
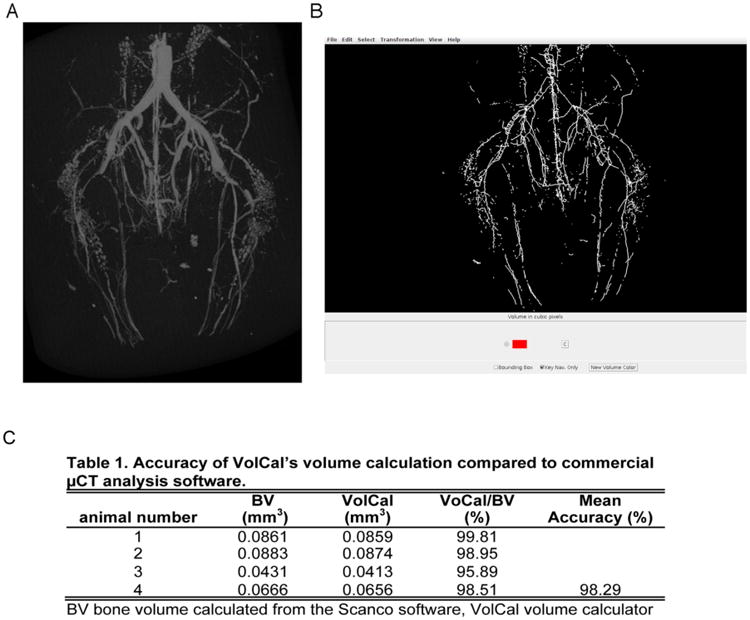
A) Representative μCT image. B) Corresponding VolCal analysis. The analysis for both images is presented in Table 1.
Experimental
Mouse hindlimb ischemia model
We chose to use a mouse strain with a targeted mutation in the Sprouty1 (Spry1) gene for this analysis. Spry1 is a negative regulator of tyrosine receptor kinase signaling [12], but has also been suggested as having an impact on other cytokine signals such as interferons [13] and TGFβ [14]. We have characterized the Spry1 mutant mice previously in bone loss and adipose tissue accumulation [15], mesenchymal stem cell lineage allocation [16], and hemangioblast and endothelial cell differentiation [17]. In addition, we characterized Spry1 as an angiogenesis regulator in vitro [18]. Because of these studies and Spry1 regulation of critical angiogenic cytokine pathways, we used the Spry1+/− and Spry1−/− mice in this analysis. Two-month old heterozygous and homozygous Spry1 knockout littermates (Spry1+/−, Spry1−/−, FVB background, obtained from the Mutant Mouse Regional Resource Center, UC, Davis) underwent surgery on day 0 following the procedure by Couffinhal et al. [19]. Mice were anesthetized by peritoneal injection of 2.5% Avertin (100ul/10g body weight) and placed on a pre-warmed heating pad. An incision in the skin approximately 1 cm long from the knee towards to the medial thigh was made in the left limb. The femoral artery was separated from the femoral vein and nerves. Two ligatures were placed on the femoral artery, one proximal close to groin and one distal close to the popliteal artery. Transection of the femoral artery was made between the distal and proximal ligatures. The wound was closed using 5-0 sutures. Mice were allowed to recover on the heating pad and then subcutaneously injected with days post surgery days post surgery buprenorphine (0.05mg/kg) for analgesia. MRAs were acquired on days 1, 4, 7, 10, and 14.
Experimental protocols were approved by the Institutional Animal and Use Committee of Maine Medical Center and followed the recommendations of current National Institutes of Health and American Physiological Society guidelines for the use and care of laboratory animals.
Magnetic resonance angiograms
Images were acquired with a BRUKER PharmaScan 7 T (BRUKER, Billerica, MA) small animal imaging system. Mice were anaesthetized and anesthesia was maintained with 1-2% isoflurane and a flow of 0.8 L O2/min throughout the experiment. Mice were placed prone into the animal holder and respiration and body temperature was monitored using the SA Instruments set up. T1 weighted MR angiograms of the hindlimb were acquired using a FLASH-TOF-2D pulse sequence with TR 13 ms, TE 4 ms, matrix 256 × 256, FOV 30 × 30 mm, slice thickness 0.4 mm, 120 slices, inter-slice distance 0.25 mm, 2 averages, and total acquisition time 11 min 1 sec.
μCT imaging
For specimen preparation mice were anesthetized with Avertin. The thoracic cavity was opened, rapidly exposing the heart. Prior to instigating perfusion, the right atrium was opened to serve as a drain vent. The animals were perfused with Microfil medium mixture (Microfil MV-122, Flow Tech; Carver, MA) through the left ventricle at 3ml/min, until all organs showed a rich yellow coloration. The specimens were refrigerated overnight, to allow polymerization. Mouse hindlimbs were dissected from the specimens and placed in a 10% EDTA solution for 10 days to decalcify the bone. Hindlimb vasculature was imaged using a high-resolution micro-CT (VivaCT-40, Scanco Medical, Basserdorf, Switzerland) at 12.5 μm resolution, with a voltage of 55 kVp and a current of 145 μA. Resolution was set to high, which created a 2048 × 2048 pixel image matrix. The tomograms were globally thresholded based on X-ray attenuation and used to render binarized 3-D images of the hindlimb vasculature.
VolCal validation using synthetic images
To validate the accuracy of VolCal's pixel recovery after skeletonization, we used synthetic images of blood vessels generated by Galarreta-Valverde et al. (2013) [8]. The images were imported into VolCal and the number of pixels and the volume calculated compared to the total pixel count and volume calculated using the built-in Fiji/ImageJ histogram function.
Results
VolCal correctly displays the vasculature in the mouse hindlimb and achieves 95% accuracy in blood vessel volume calculation
We utilized the mouse hindlimb number of junctions accuracy (%) ischemia model to validate VolCal because it is a well-established and frequently used technique to study arteriogenesis and angiogenesis [12,20]. During the injury and remodeling phases, there are significant changes in blood vessel volume and size, and comprehensive analysis requires a three dimensional analysis of the vascular network. As shown in Figs. (1 and 2), VolCal correctly displays the blood vessel tree of the mouse hindlimbs. This is important for the correct identification of collateral vessel formation in the ischemic hindlimb in order to determine their origin above and insertion below the ischemic area. The ability to turn the 3D image in VolCal and zoom in on certain areas aids in detection and analysis of these blood vessels. To demonstrate the reproducibility of blood vessel volume calculations we analyzed the MRAs of 4 Spry1+/−and 3 Spry 1−/− mice during their recovery from the ischemic hindlimb injury. Fig. (3A) shows that one-day post surgery Spry1−/− mice had slightly higher blood vessel volumes in the ischemic leg compared to Spry1+/− controls (8.76 ± 2.68 vs. 6.78 ± 1.44 (μl). After 8 and 15 days recovery and blood vessel volumes in both groups were equal. On the nonischemic side Spry1−/− mice have a persistent higher blood vessel volume on days 1 and 8 (11.33±2.01 and 12.21=1=1.67 in Spry1−/− vs. 9.22±1.63 and 10.12±1.88 μl in Spry1+/−) that only after full recovery of the ischemic muscle tissue resolves at day 15 (9.82±3.79 in Spry1−/− vs. 10.41±1.91 in Spry1+/−). Because of the activity of Spry1 to suppress receptor tyrosine kinase signaling pathways, loss of Spry1 function in vivo would result in enhanced RTK signaling, and the increased blood vessel volumes in Spry1−/− mice shortly after ischemic insult suggests Spry 1−/− have increased vascular remodeling. These data are similar to enhanced angiogenesis observed in Spry4−/− mice using the hindlimb ischemia model [21]. The consistency of VolCal in volume calculation is also demonstrated in the constant blood vessel volume of the tail veins between all animals ranging from a minimum of 2.4±0.09 μl to a maximum of 3.11±0.43 μ1. To further demonstrate the accuracy of VolCal the total voxel counts for all MRAs was calculated using the built-in Fiji/ImageJ histogram function. Each histogram provided the total count of “on” voxels for that image. These counts were compared to the total voxel count achieved by VolCal. Fig. (3B) shows that the accuracy is above 95% for all animals and scans analyzed, and does not change over time, as the repeated measurements of the same individual during ischemic remodeling shows.
Fig. (3). Quantitative analysis of hindlimb ischemia in Spry+/− and Spry1/− mice.
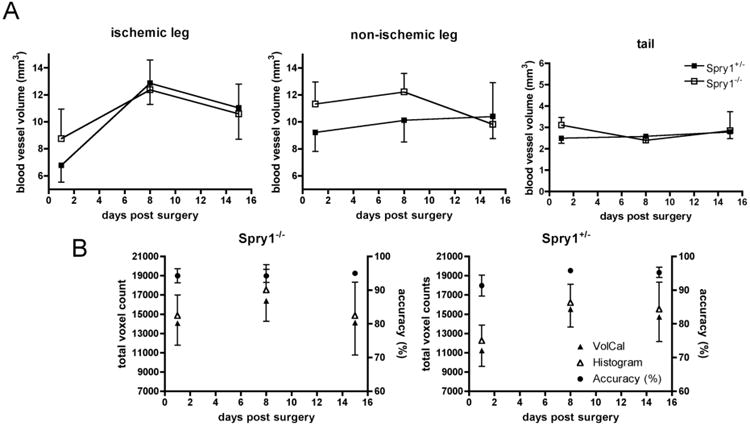
VolCal was used to analyze the recovery from hindlimb ischemia in Spry1 null and their respective Spry1+/− control mice. A) Quantification of blood vessel volumes in the ischemic and nonischemic leg, and tail. On the ischemic side the Spry null mice had a slightly higher blood vessel volume one day post surgery compared to Spry1+/− control mice, suggesting that these mice are either not as susceptible to this kind of injury or that these mice have higher vascularized hindlimbs compared to their wild type control mice. This is further supported by the fact that the non-ischemic leg has a slightly higher blood vessel volume on days 1 and 8 post surgery. The overall volume of the tail vein in both groups of mice is constant. N = 4 Spry1+/− and 3 Spry null mice, Means ± SE. B) Analysis of the total voxel count and accuracy in Spry1−/− and Spry 1+/− mice. The total voxel count (left y axis) was calculated as the sum of the voxels of the ischemic, the non-ischemic and the tail vein from the counts obtained using VolCal (filled triangles) and the Fiji Histogram image processing function (open triangles). Counts were collected post surgery at days 1, 8 and 15 for all animals in the study. The graph shows the expected increase in voxel numbers at day 8 when the maximum of remodeling is achieved. The accuracy of VolCal (right axis) was calculated as the ratio of VolCal to Fiji Histogram counted voxels and expressed as a percentage (filled circles). VolCal achieves, on average, a 95% accuracy that does not change over the duration of the experimental time line chosen.
VolCal correctly displays synthetic blood vessels and achieves higher than 99% accuracy in volume calculation
In vivo MRAs can have background noise and the absolute accuracy of VolCal in pixel recovery cannot be established using in vivo images, because the absolute volume of the blood vessels is not known. Thus, we used synthetic blood vessels that were generated by Galarretta-Valverde et al. (2013) [8] for which the absolute number of pixles is known and thus the absolute volume can be calculated. Fig. (4) shows one example of a synthetic image and its representation in VolCal. The accuracy VolCal achieves in pixel and volume calculation is higher than 99% for all four synthetic images we used (Rvdiametro, ArboDeLado, noestocastico6its, 9ItersSPIE) (Fig. 4C).
Fig. (4). Representative synthetic blood vessel tree image and the corresponding VolCal analysis and accuracy.
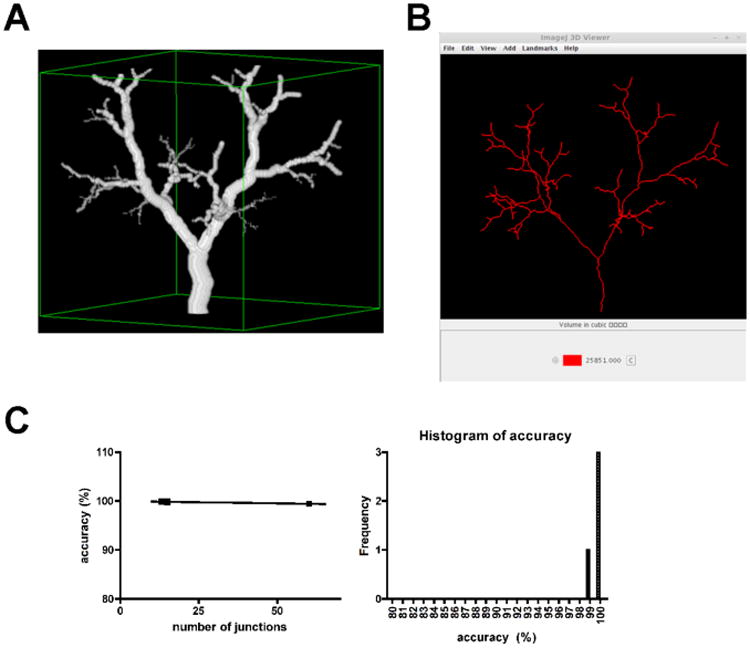
A) Example of a synthetic blood vessel tree. Image (9itersSPIE) is shown with approval of Galarreta-Valverde et al. and taken from reference[8]. B) The synthetic image is correctly displayed in VolCal. C) The pixel and volume analysis of four different synthetic images in VolCal leads to higher than 99% accuracy in pixel recovery and volume calculation.
VolCal can be used for different raw data inputs from MRAs and μCT images
In order to validate the volumes obtained for MRAs, we used (μCT images of Microfil-perfused mouse hindlimbs and compared the volume calculated by commercially available software for bone volume calculation (IPLV6_TRABECULAR, Scanco) with VolCal's calculations. Fig. (5) shows the (μCT image and the corresponding VolCal 3D reconstruction, which shows very good representation of the (μCT image. The comparison of the volume calculations shows that VolCal achieves 98% accuracy (Fig. 5C). This also demonstrates how versatile VolCal is for large volume data analysis in different experimental settings.
VolCal's volume calculation is independent of the number of blood vessel junctions
Due to the intrinsic problem of potential voxel sharing between blood vessels (or edges) in the analysis and our solution to divide the shared voxels by the number of branches we calculated the dependence of the volume on the number of junctions in a chosen blood vessel tree. Fig. (4C, 6D and E) show that the accuracy of VolCal's volume calculation is independent of the number of junctions in the analysis. This analysis obtained an accuracy of close to 95% as well.
Fig. (6). The accuracy of VolCal is independent of the number of blood vessel junctions and the shape of junctions in an MRA.

A) Graphical description of the voxel distribution in edges after skeletonization and the sharing of voxels. The gray boxes show the voxels belonging to 3 different edges associated with a simple blood vessel bifurcation shown in B). In C) the edges overlay the voxels in the different slices to demonstrate the sharing of voxels and the equal division of voxels between the number of edges. D) Relationship of accuracy vs. the number of junctions. The accuracy of VolCal is independent of the number of junctions included in the analysis and thus the equal splitting of the junction area between the number of branches is justified. E) The histogram shows that for most cases VolCal calculates the volume with 95% accuracy. 21 different vessel tree sections were included in this analysis and single data points plotted.
Discussion
We provide a novel approach to the analysis of complex blood vessel trees that is both rapid and accurate in obtaining the volume sets of vessels. We validated the volume quantification with (μCT images of mouse hindlimbs and achieved 95% accuracy compared with commercial available software for total voxel counts and 98% accuracy compared to the “bone volume” calculated with the Scanco (μCT analysis software (also based on voxel counts). The VolCal analysis plug-in allows for large data throughput and straightforward vessel tree comparisons.
As mentioned earlier, other methods for vessel volume calculation exist that feature automated steps [3-5,11, 22-26]. In particular, we note the work by Stefancik et al. [5]. The novel aspect of their methodology is the separation of venal and arterial vessels in the scanned image via heuristic methods. Manual steps are still required to enable the proper identification of vessels. In contrast, VolCal can identify voxels that are shared by vessels and then simply allocates the shared voxels evenly between the vessels. Our validation experiments indicate that there is little loss in accuracy with our approach. Finally, it does not appear that their method produces a final three-dimensional representation whereby a user can select individual segments for quantification.
Limitations of the MRAs
Magnetic resonance imaging is an excellent tool to measure blood flow to various organs and visualize blood vessels. The method we applied in this study is a time-of-flight 2D imaging routine. The limitations of this method are well known, and it does not represent the total volume of a blood vessel but rather the volume that represents the highest blood velocity. We chose part of the femoral artery to calculate the volumes in MRAs and μCT images from the same mice and were careful to use the same length in both images. Based on our comparison of MRA blood vessel volumes and μCT blood vessel volumes we calculated that the error in MRA volume is 12% (MRA: 3.5 ± 0.5 μl vs μCT: 4.0 ± 0.4 μl, n = 3, p = 0.55, t -test).
Limitations of the graphical analysis
We developed an algorithm for recovering eroded voxels and determining how contiguous voxel groups are shared amongst edges. In brief, recovering the original (binary) image's voxels that were removed by skeletonization is primarily a process of examining voxels increasingly outward (in a slice/plane) from a starting point and stopping at those points where the voxel is not set to the “on” color. In this manner, adjacent voxels are determined to be part of a particular line (vessel). However, the process of skeletonization is not perfectly bidirectional, leading to forms of ambiguity in the graphic representation of the original image; these ambiguities arise from vessels sharing voxels, especially at junctures.
The problem becomes one of deciding to which vessel a particular voxel belongs. This is a difficult problem; VolCal solves it by dividing the voxel count evenly between the shared junctions and/or vessels. As mentioned earlier, other techniques have been developed for separating and analyzing vessels in MRA images that could be employed as input to VolCal.
Because of the nature of the algorithms employed, we found that VolCal works best with images that are:
tree-like, e.g., vasculatures
skeletonizable; that is, the skeletonization algorithm can produce an accurate representation of the image's original structure. For this reason, images with jagged edges would often result in the creation of false edges and inaccurate volume measurements.
Our experiment's images were clear and exhibited, distinct tree-like structures which led to the accurate results as reported above.
Potential clinical uses of VolCal
The simplicity and versatility of VolCal suggests multiple uses in routine clinical imaging practices. This includes angiograms for any etiology in any organ system, ranging from cardiac angiograms and the specific volume analysis of coronary arteries to tumor vasculature analysis to determine pharmacological treatment success. VolCal may also aid in determining the decision for intervention for example in aortic aneurysm as the volume assessment and the monitoring of progression can be complicated using the tools currently available.
The use of VolCal as a tool for the quantification of blood vessels need not be restricted to studies involving the ischemic model. It could enhance data analysis from various studies such as for example in cerebrovascular [22,23], pulmonary [24], hepatic [25], and intracranial vessel [26] vascular studies.
In summary, we present a novel tool for the fast, reproducible, and interactive volume analysis of complex blood vessel tree. It is available as a Fiji/ImageJ plug-in (http://fiji.sc/Volume_Calculator).
Acknowledgments
This work was supported in part by the Mouse Transgenic and In Vivo Imaging Core and the Administrative Core of Facility NIH grant P30GM103392 (R.E. Friesel, PI) Phase III COBRE in Vascular Biology.
Footnotes
Conflict of Interest: The authors confirm that this article content has no conflicts of interest.
References
- 1.Bühler K, Felkel P, La Cruz A. Geometric Methods for Vessel Visualization and Quantification--A Survey. Citeseer. 2002 [Google Scholar]
- 2.Lesage D, Angelini ED, Bloch I, Funka-Lea G. A review of 3D vessel lumen segmentation techniques: Models, features and extraction schemes. Med Image Anal. 2009;13(6):819–45. doi: 10.1016/j.media.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Hernández-Hoyos M, Orkisz M, Puech P, Mansard-Desbleds C, Douek P, Magnin IE. Computer-assisted Analysis of Three-dimensional MR Angiograms1. RadioGraphics Radiol Soc N Am. 2002;22(2):421–36. doi: 10.1148/radiographics.22.2.g02mr03421. [DOI] [PubMed] [Google Scholar]
- 4.Duvall CL. Quantitative microcomputed tomography analysis of collateral vessel development after ischemic injury. AJP: Heart Circul Physiol. 2004;287(1):H302–10. doi: 10.1152/ajpheart.00928.2003. [DOI] [PubMed] [Google Scholar]
- 5.Stefancik RM, Sonka M. Highly automated segmentation of arterial and venous trees from three-dimensional magnetic resonance angiography (MRA) Int J Cardiovasc Imag. 2001;17(1):37–47. doi: 10.1023/a:1010656618835. [DOI] [PubMed] [Google Scholar]
- 6.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open- source platform for biological-image analysis. Nat Meth. 2012;9(7):676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Meth. 2012;9(7):671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galarreta-Valverde MA, Macedo MMG, Mekkaoui C, Jackowski MP. Three-dimensional synthetic blood vessel generation using stochastic L-systems. SPIE Med Imag SPIE. 2013:86691I–86691I–6. [Google Scholar]
- 9.Arganda-Carreras I, Fernández-González R, Muñoz-Barrutia A, Ortiz-De-Solorzano C. 3D reconstruction of histological sections: Application to mammary gland tissue. Microsc Res Tech. 2010;73(11):1019–29. doi: 10.1002/jemt.20829. [DOI] [PubMed] [Google Scholar]
- 10.Sowizral K, Rushforth K, Sowizral H. The Java 3D API Specification. 1997 [Google Scholar]
- 11.Schmid B, Schindelin J, Cardona A, Longair M, Heisenberg M. A high-level 3D visualization API for Java and Image J. BMC Bioinformatics. 2010;11(1):274. doi: 10.1186/1471-2105-11-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mason JM, Morrison DJ, Albert Basson M, Licht JD. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 2006;16(1):45–54. doi: 10.1016/j.tcb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Sharma B, Joshi S, Sassano A, et al. Sprouty Proteins Are Negative Regulators of Interferon (IFN) Signaling and IFN-inducible Biological Responses. J Biol Chem. 2012;287(50):42352–60. doi: 10.1074/jbc.M112.400721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin EHH, Basson MA, Robinson ML, McAvoy JW, Lovicu FJ. Sprouty is a negative regulator of transforming growth factor β-induced epithelial-to-mesenchymal transition and cataract. Mol Med. 2012;18:861–73. doi: 10.2119/molmed.2012.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urs S, Henderson T, Le P, Rosen CJ, Liaw L. Tissue-specific expression of Sprouty1 in mice protects against high-fat diet-induced fat accumulation, bone loss and metabolic dysfunction. Br J Nutr. 2012;108(6):1025–33. doi: 10.1017/S0007114511006209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urs S, Venkatesh D, Tang Y, et al. Sprouty1 is a critical regulatory switch of mesenchymal stem cell lineage allocation. FASEB J. 2010;24(9):3264–73. doi: 10.1096/fj.10-155127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, Gong Y, Friesel R. Spry1 is expressed in hemangioblasts and negatively regulates primitive hematopoiesis and endothelial cell function. PLoS ONE. 2011;6(4):e18374. doi: 10.1371/journal.pone.0018374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S, Bui Nguyen TM, Kovalenko D, et al. Sprouty1 inhibits angiogenesis in association with up-regulation of p21 and p27. Mol Cell Biochem. 2010;338(1-2):255–61. doi: 10.1007/s11010-009-0359-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couffinhal T, Silver M, Zheng LUP, Kearney M, Witzenbichler B, Isner JM. Mouse model of angiogenesis. Am J Pathol. 1998;152(6):1667. [PMC free article] [PubMed] [Google Scholar]
- 20.Couffinhal T, Dufourcq P, Barandon L, Leroux L, Duplaa C. Mouse models to study angiogenesis in the context of cardiovascular diseases. Front Biosci. 2009;14:3310–25. doi: 10.2741/3454. [DOI] [PubMed] [Google Scholar]
- 21.Taniguchi K, Sasaki KI, Watari K, et al. PLoS ONE. 5. Vol. 4. Public Library of Science; 2009. Suppression of Sproutys Has a Therapeutic Effect for a Mouse Model of Ischemia by Enhancing Angiogenesis; p. e5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Baz A, Elnakib A, Khalifa F, et al. Precise segmentation of 3-D magnetic resonance angiography. IEEE Trans Biomed Eng. 2012;59(7):2019–29. doi: 10.1109/TBME.2012.2196434. [DOI] [PubMed] [Google Scholar]
- 23.Gao X, Uchiyama Y, Zhou X, Hara T, Asano T, Fujita H. A Fast and Fully Automatic Method for Cerebrovascular Segmentation on Time-of-Flight (TOF) MRA Image. J Digit Imag. 2010;24(4):609–25. doi: 10.1007/s10278-010-9326-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shingrani R, Krenz G, Molthen R. Automation process for morphometric analysis of volumetric CT data from pulmonary vasculature in rats. Comput Methods Programs Biomed. 2010;97(1):62–77. doi: 10.1016/j.cmpb.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauer C, Pock T, Sorantin E, Bischof H, Beichel R. Med Image Anal. 2. Vol. 14. Elsevier B.V; 2010. Segmentation of interwoven 3d tubular tree structures utilizing shape priors and graph cuts; pp. 172–84. [DOI] [PubMed] [Google Scholar]
- 26.Chapman BE, Stapelton JO, Parker DL. Intracranial vessel segmentation from time-of-flight MRA using pre-processing of the MIP Z-buffer: accuracy of the ZBS algorithm. Med Image Anal. 2004;8(2):113–26. doi: 10.1016/j.media.2003.12.002. [DOI] [PubMed] [Google Scholar]


