Abstract
Since its discovery, nitric oxide (NO) has been observed to play an important role in the physiology of single-celled organisms as well as high-order vertebrates. In this review, we will discuss the involvement of NO in bacterial, plant and human systems. NO originates from a variety of sources, namely bacterial, plant, and mammalian nitric oxide synthases which oxidize L-arginine. Bacterial NO is involved in toxin synthesis, signaling and biofilm formation. Organisms use NO to mediate oxidative stress incurred during the innate immune response. In plants, large amounts of NO hinder plant growth, while lower concentrations regulate normal development. NO and the associated reactive oxygen species (ROS) are effective antibacterial, anti-parasitic, and antifungal agents. Though NO has therapeutic effects in the immune system, the NO response is biphasic and concentration-dependent. NO promotes tumorigenesis within a concentration range, and induces apoptosis of cancerous cells at other concentrations. The biphasic response to NO is also evident in the regulation of chemokine, interleukins, and NF-κB, which can promote or inhibit inflammation. The physiologic response to NO is concentration dependent. NO, by way of non-adrenergic noncholinergic (NANC) nerve transmission, propagates a cascade of molecular signaling that facilitates smooth muscle cell relaxation and increased arterial inflow into the corpora, initiating an erectile response. Additional NO is released through NOS activity in the endothelium in response to cholinergic nerve activity and shear stress, which helps to maintain erection.
Keywords: nitric oxide, nitric oxide synthase family, nitric oxide and erectile function, nitric oxide and bacterial cells, nitric oxide and plants
History of NO
Discovery and major players
Atmospheric NO (nitrous air) was first observed in 1774 by Joseph Priestly [1]. Amyl Nitrite was first synthesized in 1844 by Antoine Balard. Vasoactive properties were first reported by Frederick Guthrie in 1859 [2, 3]. Brunton in 1867 reported on the use of nitrite of amyl as a treatment for angina pectoris [4]. In 1903 Francois-Franck suggested that amyl nitrate is a vasodilator [5]. The metabolic sources and functions of NO in bacteria, humans, and plants is illustrated in the Figure 1.
Figure 1.
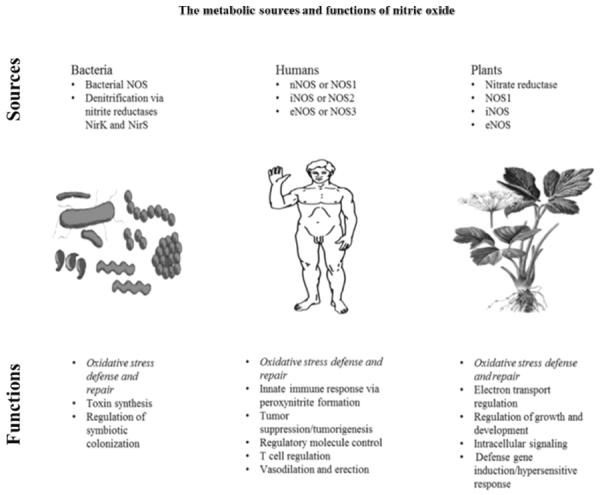
This figure illustrates the role of nitric oxide synthase (NOS) and nitric oxide in bacteria, humans, and plants in regard to oxidative stress, cellular defense and repair, and other cellular processes. There are 3 isoforms of NOS in humans and more are present in plants. It has recently been discovered that NOS and NO play an important role in cardiovascular health and erectile function. The roles of NOS and NO are diverse and important in bacteria, humans, animals, and plants.
Role of NO in bacteria
Bacterial synthesis of NO proceeds from the oxidation of L-arginine via the N-ω̵-hydroxy-L-arginine intermediate. Additionally, denitrification is a facultative pathway shared by many bacteria and archaea whereby nitrate is reduced to nitrite, nitric oxide, nitrous oxide and finally nitrogen gas [6]. The four steps of denitrification are characterized by seven enzymes [7]. Of these enzymes, the reductases Nar or Nap catalyze the reduction of nitrate to nitrite, and the nitrite reductases NirK or NirS reduce soluble NO2- to NO gas [6].
The identification of bacterial proteins homologous to mammalian NOS have shed light on new roles of NO in microbes [8]. Studies on the biological functions of these bacterial NOSs show that NO is involved in toxin biosynthesis, defense against oxidative stress, and regulation of growth responses after radiation exposure [8].
Toxin synthesis and host defense
In certain Streptomyces strains, NOS plays a role in the biosynthesis of thaxtomin, a plant toxin contributing to the virulence in scab-causing pathogens [9, 10]. NO, produced by NOS, is directly linked to the nitration of thaxtomin [11]. Bacillus subtilis, a bacteria that exhibits many multicellular traits such as biofilm formation and swarming motility, uses NOS-derived NO defensively and collectively to protect against host oxidative attack [12]. This is employed against a common form of host defense which is driven by the Fenton reaction, and uses ferrous iron and peroxide to generate hydroxyl radicals that have a deleterious effect on cells [13]. Reduction of the ferric iron by cellular reductants, such as cysteine, maintains the continuity of the Fenton reaction by providing ferrous iron. Using NO, bacterial s-nitrosation of cysteine inhibits recycling of this ferrous iron, thereby preventing oxidative damage to the Bacilli [8]. NO also blocks the detrimental effects of oxidation of DNA and proteins by activating a Bacilli-specific catalase that breaks down peroxide [13]. The same defensive mechanism involving endogenous and exogenous NO also occurs in Bacillus anthracis and is activated upon macrophage-induced oxidative stress which helps ensure the survival of the pathogen [14]. During a host-pathogen interaction, the hosts fight infection by causing indiscriminate oxidative damage, but, like their bacterial assailants, the hosts also produces NO in response to pathogen virulence and oxidative stress [8].
Bacterial NO signaling
Deinococcus radioduransis is a highly resilient bacteria able to survive in stringent conditions, including desiccation, exposure to reactive oxygen species, and significant radiation exposure [15]. However, and significant radiation exposure [15]. However Δnos, a strain of D. radiodurans in which the nos gene has been deleted, displays minimal cell repair after irradiation. The addition of exogenous NO at any stage of damage promotes the growth recovery of the strain [15]. The protective mechanism exhibited by bacterial NOS is further established through the correlation observed between NO generation, levels, and the activation of the obgE gene. The gene codes for GTPases involved in regulation of developmental processes and cell proliferation [16]. D. radiodurans exposed to UV light synthesizes NO. This results in the upregulation of obgE gene which induces cell repair signaling [15]. NO also elicits responses involving regulatory proteins in other bacteria [17].
Symbiosis and NO
NO is involved in signaling pathways of endosymbionts, used primarily as a way to avoid cascades of host derived ROS and RNS from attacking their proteins and lipids [18]. In the squid-vibrio light organ, bacterial symbionts with bacterial heme-containing H-NOX proteins sense host-derived NO and regulate the symbiotic colonization of the light organ [19]. NO is also involved in signaling in plant root nodules containing nitrogen-fixing bacteria. Recent studies indicate that the amount of NO generated by a host's immune response in response to a pathogenic or beneficial microbe is modulated by class 1 hemoglobin genes, which lower concentrations of NO in the presence of nitrogen fixing bacteria [20]. NO also plays a role in signaling involving the symbiotic relationships of a diverse collection of animal hosts [21]. Cellobiose, a cell wall component in plants, also induces production of NO at the host-pathogen interface and the excess of NO in toxin biosynthesis implicate the role of NO in tissue growth [8].
Role of NO in plants
Physiology
NO has been shown to stimulate seed germination in plants. It is also known to play a role in mitochondrial respiration and chloroplast electron transport, where it serves to regulate the terminal transport step and rate of electron transport [22–25]. Treatment of plants exposed to an oxidative stress inducing herbicide along with the NO donor sodium nitroprusside showed a protective effect against ROS [26]. At high doses, NO can retard plant growth, whereas at lower concentrations NO promotes normal growth and development [27]. Application of SNP to roots has shown to stimulate lateral root development whereas applying a NO scavenger to the root has been shown to stimulate primary root growth and hinder lateral root growth [28]. Nitric oxide has also been indicated to be involved with the regulation of fertility. A plant mutant under-producing NO develops faster, whereas plants given NO donors or plants with higher endogenous production of NO exhibit a delayed flowering time [29].
Plants are known to possess many distinct nitric oxide synthases (NOS). The enzymes generally catalyze NO from arginine, similar to what is seen in mammalian cells [30]. Plant iNOS was found to be induced upon virus inoculation and was present in very low numbers in uninfected cells. This enzyme was also shown to be sensitive to inhibitors of animal NOS such as L-NMMA, L-NAME and aminoguanidine [31, 32]. Another NOS, NOS1, has been shown to be required for maximal growth and proper organ development [33]. Distinct NOS enzymes have also been discovered in peroxisomes and apoplasts [34, 35].
Second messengers and defense
Cyclic GMP has been proposed as a common second messenger for NO in plants. Experimentally, NO has been shown to regulate calcium ion channels in stomatal guard cells (cells regulating gas exchange in the leaf through the opening and closing of the stoma) via promotion of calcium release from intracellular stores [36, 37]. The subsequent rise of free calcium in the cytosol was blocked by guanylate cyclase antagonists, implicating a cGMP dependent second messenger system [37].
Defense genes such as pathogenesis-related 1 protein and phenylalanine ammonia lyase have been observed to be induced by the addition of NO donors (SNP). The same genes were observed to be induced by cGMP, further implying a guanylate cyclase pathway [38]. NO plays a key signaling role during the hypersensitive response, a reactive oxygen species generating response resulting in localized cell death limiting nutrient availability to an invading pathogen. NO, in concert with hydrogen peroxide, can induce cell death in this role [39].
Physiologic roles of NO
Immune system
Many cell types express NO within the immune system, such as macrophages, neutrophils, NK cells, mast cells, phagocytic cells and dendritic cells. Other cells involved in immune response also express NO such as endothelial cells, epithelial cells, vascular smooth muscle cells, fibroblasts and many others [40]. The NO response in infection has direct effects on DNA through mutation, the inhibition of DNA repair and synthesis, the S-nitrosylation of proteins, tyrosine nitration, or enzymatic inactivation [40].
The oxidation of proteins and DNA at different sites is often carried out by peroxynitrite and NO2 [41]. The targeting of this nitrogen-reactive species in bacteria such as Salmonella typhimurium shows its efficacy as an antibacterial agent [42]. In viral infections, such as seen in Epstein-Barr virus infections, peroxynitrite formation from NO hinders viral replication and blocks activation of the genome, thereby inhibiting the virus [43]. Anti-parasitic and antifungal activity has also been credited to peroxynitrite synthesis [41].
iNOS
iNOS induction contributes to indirect effects of NO antimicrobial activity [40]. Induced iNOS in macrophages diminishes the growth factor arginine, contributing to growth inhibition or possibly parasitic death [44]. In a similar manner, Nω-hydroxy-L-arginine, a stable intermediate in the NO synthesis pathway, blocks arginase activity, which leads to the killing of Leishmania [45].
Tumors
The ability of interferon-λ to suppress tumor cell growth in mice established the first known function of NO in the immune system [46]. The sulfated polysaccharide fucoidan exhibits cytotoxic properties against tumor cells, which mechanistically stems from the activation of the NOS gene and the increase of NO synthesis [47]. Studies show that the NOS inhibitor L-NAME blocks NO production as well as the cytocidal effects seen in fucoidan. However, contradictory evidence exists regarding the endogenous expression of iNOS in tumor cells inducing DNA-dependent protein kinase that protect the cells from NO cytotoxicity [40]. NO is still considered a “double-edged sword” in oncology due to its involvement in both inhibition and promotion of tumorigenesis [48]. Within a certain concentration range, NO facilitates the survival of tumors, but beyond this critical NO concentration NO has been observed to sensitize the cancerous cells to apoptosis. Therefore, the NO response in tumor cells is biphasic. The duality of the NO response has been exploited in pre-clinical cancer models to hinder tumor growth as well as improve the effectiveness of chemotherapy and radiotherapy [49].
Tissue damage
Though NO has proven therapeutic effects in the immune system, it can also act in tissue-damaging pathways [40]. NO has been implicated in neurodegeneration through its connection to the neurotransmitter glutamate [50]. Deviation in glutamate signaling transduction and release of NO are factors in ischemic stroke [51]. Microglia are brain immune cells responsible for preservation of the neural environment, including the regulation of immune responses, and increased activity of NOS in microglia following transient brain ischemia leads to the production of cytotoxic NO, which has a deleterious effect [52, 53]. The combination of nitric oxide and superoxide yields peroxynitrite, which causes the cleavage of DNA strands. This leads to the activation of a DNA repair protein and consumption of NAD+, the root cause of the observed brain damage [52].
Immune response to NO
The physiologic response to NO in the immune system is varied due to the expression of all known isoforms of NOS in the immune system as well as the ability of NO to easily cross membranes, and be transported with various low molecular weight compounds such as S-nitrothiols at sites distal to NO production. NO can bind a variety of targets, many of which are regulatory molecules [40]. Consequently, nitric oxide possesses both an anti-inflammatory and immunosuppressive effect in the body [54]. In rat models, evidence for this effect has been observed in the attenuation of acute inflammation and adjuvant arthritis. L-arginine was shown to enhance these effects [55–63]. In humans, synthesis of NO in the colon is increased in patients with ulcerative colitis and NOS inhibitors have been shown to suppress an experimental model of induced chronic ileitis [64]. There are a wide range of responses mediated by NO, from vasodilation and edema, sensory nerve modulation, leukocyte activity modulation, to tissue cytotoxicity [65–68]. These varied responses support reports showing evidence of tissue protection through the administration of both NO donors and NOS inhibitors [54, 69, 70].
Immune system regulation
Another function of NO within the immune system is cytokine, chemokine, and growth factor regulation. NO induces both pro and anti-inflammatory responses through these mediators. NO produced by macrophages, T cells, endothelial cells and fibroblasts can cause the up and down regulation of interleukins (e.g. IL-1, IL-6, IL-8, IL-10, IL-12, IL-18), IFN-γ, TNF, growth factors (TGF-β, G-CSF, M-CSF, VEGF), CC chemokines, and macrophage inflammatory proteins. The modulation of these molecules affects signaling cascades, transcription factors, proteins regulating mRNA translation, and cytokine enzymes and precursors [71–78]. Regulation of NF-κB is also determined in a biphasic manner by the concentration of NO present [79].
T cell regulation
NO also modulates T helper cell levels and ratios. Th1 cells are more susceptible to apoptosis than Th2 cells and this apoptotic pathway has been shown to be regulated by NO, likely through its reactions with other ROS within the cell. Th2 ratios in relation to Th1 are increased in the presence of NO through the up regulation of IL-2 in murine lymphocytes and the up regulation of IL-4 in human cells [80–85]. NO also down regulates the expression of P and E selectin, vascular cell adhesion molecule, and intracellular adhesion molecule-1, which inhibits the rolling of leukocytes along the endothelium and prevents migration of helper cells from vessels into tissues. P and E selectins preferentially recruit Th1 cells into inflamed tissues. Suppression of these selectins shows that NO works to inhibit the accumulation of Th1 cells at sites of inflammation via adhesion interference [86–88].
Role of NO in erection
Erection is achieved through an integration of central and peripheral processes which result in physiologic vasodilation, arterial inflow into the paired corpora cavernosa and veno-occlusion in the penis. Nonadrenergic, noncholinergic transmission of NO from the cavernosal nerve terminals initiates a cascade of molecular signaling mediated through the heterodimericheme protein soluble guanylate cyclase. sGC activation results in conversion of intracellular GTP to cGMP. Increased intracellular levels of cGMP activate a cGMP-dependent protein kinase, causing membrane hyperpolarization and uptake of calcium into the endoplasmic reticulum. The decrease in bioavailable intracellular calcium facilitates smooth muscle cell relaxation. As the arterial inflow into the corpora increases, shear stress activates an endothelial nitric oxide synthase, serving as an additional source of NO to facilitate an erection. A cGMP dependent PDE-5 converts cGMP to GDP, blocks membrane hyperpolarization and abolishes the erectile response [89–91].
Early investigations performed with human corporal smooth muscle cell cultures showed that these cells respond identically to vascular smooth muscle cells when cGMP was added to the culture media and suggested a role for EDRF in the process of erection [92, 93]. This notion was further developed with additional experiments in which rat aortas were harvested and hung on a column over a rat cavernosal smooth muscle cell culture. Calcium efflux was measured in the cavernosal culture after human serum had been dripped through the aortas. The results of this study also suggested EDRF was responsible for cavernosal smooth muscle cell relaxation [93]. Dr. Jake Rajfer contributed significantly to these early studies and one of his colleagues at UCLA medical center, Dr. Louis Ignarro, had recently reported that the identity of EDRF released from the vascular smooth muscle was NO [86]. The two investigators began a collaboration to show that NO was the compound responsible for smooth muscle cell relaxation in the corpora cavernosa and erection. Their initial study showed that relaxation of rabbit cavernosal tissue by electrical field stimulation was attenuated by compounds that inhibited NO synthesis [94, 95]. Within the next two years, 3 studies were published showing the role of NO in relaxation of human corporal smooth muscle tissue [93, 96–98]. Rajfer and Ignarro's 1992 article in the New England Journal of Medicine was especially significant because it was the first article to demonstrate that inhibition of phosphodiesterase enhanced the relaxation response of corporal strips to EFS and NO [93, 98]. A study by Dr. Arthur Burnett at Johns Hopkins demonstrated the localization of NO to the penile nerves of the rat using antibodies [89]. Shortly thereafter, the effects of phosphodiesterase inhibition upon erectile responses was reported in vivo [99]. These experiments provided the basis for what is now considered to be the gold standard in treatment of erectile dysfunction, the PDE-5 inhibitor.
Acknowledgments
Research Support: NIH Grant HL 62000 and HL 77421
ABBREVIATIONS
- NO
nitric oxide
- Nar
respiratory nitrate reductase
- Nap
periplasmic nitrate reductases
- NirK, NirS
nitrite reductase genes
- NOS
nitric oxide synthase
- GTPase
large family of enzymes that can bind and hydrolyze guanosine triphosphate
- ROS
reactive oxygen species
- RNS
reactive nitrogen species
- H-NOX
heme nitric oxide/oxygen
- SNP
sodium nitroprusside
- iNOS
inducible nitric oxide synthase
- L-NMMA
L-NG-monomethyl Arginine citrate
- L-NAME
L-NG-Nitroarginine methyl ester (hydrochloride)
- cGMP
cyclic guanosine monophosphate
- NK cells
natural killer cells
- DNA
deoxyribonucleic acid
- DNA-PKcs
DNA-dependent protein kinase
- IL
interleukins
- IFN-γ
interferon-gamma
- TNF
tumor necrosis factors
- TGF-β
transforming growth factor beta
- G-CSF
granulocyte colony-stimulating factor
- M-CSF
macrophage colony-stimulating factor
- VEGF
vascular endothelial growth factor
- CC chemokine
ß-chemokine
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- Th
T helper cell
- NANC
Nonadrenergic, noncholinergic
- sGC
soluble guanylate cyclase
- GTP
guanosine-5'-triphosphate
- eNOS
endothelial nitric oxide synthase
- PDE-5
type-5 phosphodiesterase
- GDP
guanosine diphosphate
- EDRF
endothelium-derived relaxing factor
- EFS
electrical field stimulation
REFERENCES
- 1.Priestley J. Experiments and observations on different kinds of air, 1774. Printed for J. Johnson; London: [Google Scholar]
- 2.Marsh N, Marsh A. Clin. Exp. Pharmacol. Physiol. 2000;27:313–9. doi: 10.1046/j.1440-1681.2000.03240.x. [DOI] [PubMed] [Google Scholar]
- 3.Guthrie F. Quarterly Journal of the Chemical Society of London. 1859;11:245–252. [Google Scholar]
- 4.T LB. The Lancet. 1867;90:97–98. [Google Scholar]
- 5.Fye WB. Circulation. 1986;74:222–9. doi: 10.1161/01.cir.74.2.222. [DOI] [PubMed] [Google Scholar]
- 6.Jones CM, Stres B, Rosenquist M, Hallin S. Mol. Biol. Evol. 2008;25:1955–66. doi: 10.1093/molbev/msn146. [DOI] [PubMed] [Google Scholar]
- 7.Philippot L. Biochim. Biophys. Acta. 2002;1577:355–76. doi: 10.1016/s0167-4781(02)00420-7. [DOI] [PubMed] [Google Scholar]
- 8.Crane BR, Sudhamsu J, Patel BA. Annu. Rev. Biochem. 2010;79:445–70. doi: 10.1146/annurev-biochem-062608-103436. [DOI] [PubMed] [Google Scholar]
- 9.Kers JA, Wach MJ, Krasnoff SB, Widom J, Cameron KD, Bukhalid RA, Gibson DM, Crane BR, Loria R. Nature. 2004;429:79–82. doi: 10.1038/nature02504. [DOI] [PubMed] [Google Scholar]
- 10.Hogenhout SA, Loria R. Curr. Opin. Plant Biol. 2008;11:449–56. doi: 10.1016/j.pbi.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Johnson EG, Sparks JP, Dzikovski B, Crane BR, Gibson DM, Loria R. Chem. Biol. 2008;15:43–50. doi: 10.1016/j.chembiol.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 12.Schreiber F, Beutler M, Enning D, Lamprecht-Grandio M, Zafra O, Gonzalez-Pastor JE, de Beer D. BMC Microbiol. 2011;11:111. doi: 10.1186/1471-2180-11-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gusarov I, Nudler E. Proc. Natl. Acad. Sci. USA. 2005;102:13855–60. doi: 10.1073/pnas.0504307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shatalin K, Gusarov I, Avetissova E, Shatalina Y, McQuade LE, Lippard SJ, Nudler E. Proc. Natl. Acad. Sci. USA. 2008;105:1009–13. doi: 10.1073/pnas.0710950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel BA, Moreau M, Widom J, Chen H, Yin L, Hua Y, Crane BR. Proc. Natl. Acad. Sci. USA. 2009;106:18183–8. doi: 10.1073/pnas.0907262106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Czyz A, Wegrzyn G. Acta Biochim. Pol. 2005;52:35–43. [PubMed] [Google Scholar]
- 17.Spiro S. FEMS Microbiol. Rev. 2007;31:193–211. doi: 10.1111/j.1574-6976.2006.00061.x. [DOI] [PubMed] [Google Scholar]
- 18.Feng FC. Nature Reviews Microbiology. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Dufour YS, Carlson HK, Donohue TJ, Marletta M, Ruby E. Proc. Natl. Acad. Sci. USA. 2010:8375–8380. doi: 10.1073/pnas.1003571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagata M. Mol. Plant Microbe Interact. 2008;21:1175–1183. doi: 10.1094/MPMI-21-9-1175. [DOI] [PubMed] [Google Scholar]
- 21.Ganassi S, Tagliazucchi D, Mola L. Eur. J. Histochem. 2005;49:385–93. doi: 10.4081/967. [DOI] [PubMed] [Google Scholar]
- 22.Caro A, Puntarulo S. Free Radic. Res. 1999;31(Suppl):S205–12. doi: 10.1080/10715769900301521. [DOI] [PubMed] [Google Scholar]
- 23.van Rensen JJ. Photosynth. Res. 2002;73:185–92. doi: 10.1023/A:1020451114262. [DOI] [PubMed] [Google Scholar]
- 24.Yamasaki H, Shimoji H, Ohshiro Y, Sakihama Y. Nitric Oxide. 2001;5:261–70. doi: 10.1006/niox.2001.0353. [DOI] [PubMed] [Google Scholar]
- 25.Beligni MV, Lamattina L. Planta. 2000;210:215–21. doi: 10.1007/PL00008128. [DOI] [PubMed] [Google Scholar]
- 26.Beligni MV, Fath A, Bethke PC, Lamattina L, Jones RL. Plant Physiol. 2002;129:1642–50. doi: 10.1104/pp.002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beligni MV, Lamattina L. Trends Plant Sci. 2001;6:508–9. doi: 10.1016/s1360-1385(01)02156-2. [DOI] [PubMed] [Google Scholar]
- 28.Correa-Aragunde N, Graziano M, Lamattina L. Planta. 2004;218:900–5. doi: 10.1007/s00425-003-1172-7. [DOI] [PubMed] [Google Scholar]
- 29.He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, Fiorani F, Jackson RB, Crawford NM, Pei ZM. Science. 2004;305:1968–71. doi: 10.1126/science.1098837. [DOI] [PubMed] [Google Scholar]
- 30.Shapiro AD. Vitam. Horm. 2005;72:339–98. doi: 10.1016/S0083-6729(05)72010-0. [DOI] [PubMed] [Google Scholar]
- 31.Chandok MR, Ytterberg AJ, van Wijk KJ, Klessig DF. Cell. 2003;113:469–82. doi: 10.1016/s0092-8674(03)00350-7. [DOI] [PubMed] [Google Scholar]
- 32.Ribeiro EA, Jr., Cunha FQ, Tamashiro WM, Martins IS. FEBS Lett. 1999;445:283–6. doi: 10.1016/s0014-5793(99)00138-6. [DOI] [PubMed] [Google Scholar]
- 33.Guo FQ, Okamoto M, Crawford NM. Science. 2003;302:100–3. doi: 10.1126/science.1086770. [DOI] [PubMed] [Google Scholar]
- 34.Barroso JB, Corpas FJ, Carreras A, Sandalio LM, Valderrama R, Palma JM, Lupianez JA, del Rio LA. J. Biol. Chem. 1999;274:36729–33. doi: 10.1074/jbc.274.51.36729. [DOI] [PubMed] [Google Scholar]
- 35.Zhang C, Shapiro AD. BMC Plant Biol. 2002;2:9. doi: 10.1186/1471-2229-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desikan R, Cheung MK, Bright J, Henson D, Hancock JT, Neill SJ. J. Exp. Bot. 2004;55:205–12. doi: 10.1093/jxb/erh033. [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Mata C, Gay R, Sokolovski S, Hills A, Lamattina L, Blatt MR. Proc. Natl. Acad. Sci. USA. 2003;100:11116–21. doi: 10.1073/pnas.1434381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durner J, Wendehenne D, Klessig DF. Proc. Natl. Acad. Sci. USA. 1998;95:10328–33. doi: 10.1073/pnas.95.17.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delledonne M, Xia Y, Dixon RA, Lamb C. Nature. 1998;394:585–8. doi: 10.1038/29087. [DOI] [PubMed] [Google Scholar]
- 40.Bogdan C. Nat. Immunol. 2001;2:907–16. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 41.Jones ML, Ganopolsky JG, Labbe A, Wahl C, Prakash S. Appl. Microbiol. Biotechnol. 2010;88:401–7. doi: 10.1007/s00253-010-2733-x. [DOI] [PubMed] [Google Scholar]
- 42.Bryk R, Griffin P, Nathan C. Nature. 2000;407:211–5. doi: 10.1038/35025109. [DOI] [PubMed] [Google Scholar]
- 43.Kawanishi M. Intervirology. 1995;38:206–13. doi: 10.1159/000150434. [DOI] [PubMed] [Google Scholar]
- 44.Piacenza L, Peluffo G, Radi R. Proc. Natl. Acad. Sci. USA. 2001;98:7301–6. doi: 10.1073/pnas.121520398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iniesta V, Gomez-Nieto LC, Corraliza I. J. Exp. Med. 2001;193:777–84. doi: 10.1084/jem.193.6.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nathan C. FASEB J. 1992;6:3051–64. [PubMed] [Google Scholar]
- 47.Takeda K, Tomimori K, Kimura R, Ishikawa C, Nowling TK, Mori N. Int. J. Oncol. 2011 doi: 10.3892/ijo.2011.1168. in press. [DOI] [PubMed] [Google Scholar]
- 48.Bonavida B, Baritaki S. Nitric Oxide. 2011;24:1–7. doi: 10.1016/j.niox.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 49.Singh S, Gupta AK. Cancer Chemother. Pharmacol. 2011;67:1211–24. doi: 10.1007/s00280-011-1654-4. [DOI] [PubMed] [Google Scholar]
- 50.Strijbos PJ. Crit. Rev. Neurobiol. 1998;12:223–43. doi: 10.1615/critrevneurobiol.v12.i3.40. [DOI] [PubMed] [Google Scholar]
- 51.Gupta YK, Chauhan A. Indian J. Med. Res. 2011;133:15–26. [PMC free article] [PubMed] [Google Scholar]
- 52.Love S. Brain Pathol. 1999;9:119–31. doi: 10.1111/j.1750-3639.1999.tb00214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kraft AD, Harry GJ. Int. J. Environ. Res. Public Health. 2011;8:2980–3018. doi: 10.3390/ijerph8072980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tripathi P, Kashyap L, Singh V. FEMS Immunol. Med. Microbiol. 2007;51:443–52. doi: 10.1111/j.1574-695X.2007.00329.x. [DOI] [PubMed] [Google Scholar]
- 55.Ding AH, Nathan CF, Stuehr DJ. J. Immunol. 1988;141:2407–12. [PubMed] [Google Scholar]
- 56.McCall TB, Boughton-Smith NK, Palmer RM, Whittle BJ, Moncada S. Biochem. J. 1989;261:293–6. doi: 10.1042/bj2610293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moncada S, Palmer RM, Higgs EA. Pharmacol. Rev. 1991;43:109–42. [PubMed] [Google Scholar]
- 58.Nathan CF, Hibbs JB., Jr. Curr. Opin. Immunol. 1991;3:65–70. doi: 10.1016/0952-7915(91)90079-g. [DOI] [PubMed] [Google Scholar]
- 59.Marletta MA. J. Med. Chem. 1994;37:1899–907. doi: 10.1021/jm00039a001. [DOI] [PubMed] [Google Scholar]
- 60.Griffith OW, Stuehr DJ. Annu. Rev. Physiol. 1995;57:707–36. doi: 10.1146/annurev.ph.57.030195.003423. [DOI] [PubMed] [Google Scholar]
- 61.MacMicking J, Xie QW, Nathan C. Annu. Rev. Immunol. 1997;15:323–50. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 62.Stuehr DJ. Biochim. Biophys. Acta. 1999;1411:217–30. doi: 10.1016/s0005-2728(99)00016-x. [DOI] [PubMed] [Google Scholar]
- 63.Bogdan C, Rollinghoff M, Diefenbach A. Immunol. Rev. 2000;173:17–26. doi: 10.1034/j.1600-065x.2000.917307.x. [DOI] [PubMed] [Google Scholar]
- 64.Billiar TR, Harbrecht BG. Gastroenterology. 1997;113:1405–7. doi: 10.1053/gast.1997.v113.agast971131405. [DOI] [PubMed] [Google Scholar]
- 65.Laskin JD, Heck DE, Laskin DL. Trends Endocrinol. Metab. 1994;5:377–82. doi: 10.1016/1043-2760(94)90105-8. [DOI] [PubMed] [Google Scholar]
- 66.Honold J, Pusser NL, Nathan L, Chaudhuri G, Ignarro LJ, Sherman MP. Nitric Oxide. 2000;4:35–46. doi: 10.1006/niox.1999.0267. [DOI] [PubMed] [Google Scholar]
- 67.Wei LH, Morris SM, Jr., Cederbaum SD, Mori M, Ignarro LJ. Arch. Biochem. Biophys. 2000;374:255–60. doi: 10.1006/abbi.1999.1563. [DOI] [PubMed] [Google Scholar]
- 68.Griscavage JM, Wilk S, Ignarro LJ. Proc. Natl. Acad. Sci. USA. 1996;93:3308–12. doi: 10.1073/pnas.93.8.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ignarro LJ, Napoli C, Loscalzo J. Circ. Res. 2002;90:21–8. doi: 10.1161/hh0102.102330. [DOI] [PubMed] [Google Scholar]
- 70.Jacobs AT, Ignarro LJ. J. Biol. Chem. 2001;276:47950–7. doi: 10.1074/jbc.M106639200. [DOI] [PubMed] [Google Scholar]
- 71.Marshall HE, Merchant K, Stamler JS. FASEB J. 2000;14:1889–900. doi: 10.1096/fj.00.011rev. [DOI] [PubMed] [Google Scholar]
- 72.Bogdan C. Trends Cell Biol. 2001;11:66–75. doi: 10.1016/s0962-8924(00)01900-0. [DOI] [PubMed] [Google Scholar]
- 73.Berendji D, Kolb-Bachofen V, Zipfel PF, Skerka C, Carlberg C, Kroncke KD. Mol. Med. 1999;5:721–30. [PMC free article] [PubMed] [Google Scholar]
- 74.Wang S, Wang W, Wesley RA, Danner RL. J. Biol. Chem. 1999;274:33190–3. doi: 10.1074/jbc.274.47.33190. [DOI] [PubMed] [Google Scholar]
- 75.Vodovotz Y, Chesler L, Chong H, Kim SJ, Simpson JT, DeGraff W, Cox GW, Roberts AB, Wink DA, Barcellos-Hoff MH. Cancer Res. 1999;59:2142–9. [PubMed] [Google Scholar]
- 76.Zhang Z, Kolls JK, Oliver P, Good D, Schwarzenberger PO, Joshi MS, Ponthier JL, Lancaster JR., Jr. J. Biol. Chem. 2000;275:15839–44. doi: 10.1074/jbc.M000604200. [DOI] [PubMed] [Google Scholar]
- 77.Uma S, Yun BG, Matts RL. J. Biol. Chem. 2001;276:14875–83. doi: 10.1074/jbc.M011476200. [DOI] [PubMed] [Google Scholar]
- 78.Schindler H, Bogdan C. Int. Immunopharmacol. 2001;1:1443–55. doi: 10.1016/s1567-5769(01)00089-3. [DOI] [PubMed] [Google Scholar]
- 79.Connelly L, Palacios-Callender M, Ameixa C, Moncada S, Hobbs AJ. J. Immunol. 2001;166:3873–81. doi: 10.4049/jimmunol.166.6.3873. [DOI] [PubMed] [Google Scholar]
- 80.Liew FY. Adv. Neuroimmunol. 1995;5:201–9. doi: 10.1016/0960-5428(95)00009-q. [DOI] [PubMed] [Google Scholar]
- 81.Niedbala W, Wei XQ, Piedrafita D, Xu D, Liew FY. Eur. J. Immunol. 1999;29:2498–505. doi: 10.1002/(SICI)1521-4141(199908)29:08<2498::AID-IMMU2498>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 82.Bauer H, Jung T, Tsikas D, Stichtenoth DO, Frolich JC, Neumann C. Immunology. 1997;90:205–11. doi: 10.1046/j.1365-2567.1997.00161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van der Veen RC. Int. Immunopharmacol. 2001;1:1491–500. doi: 10.1016/s1567-5769(01)00093-5. [DOI] [PubMed] [Google Scholar]
- 84.Huang FP, Niedbala W, Wei XQ, Xu D, Feng GJ, Robinson JH, Lam C, Liew FY. Eur. J. Immunol. 1998;28:4062–70. doi: 10.1002/(SICI)1521-4141(199812)28:12<4062::AID-IMMU4062>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 85.Taylor-Robinson AW, Liew FY, Severn A, Xu D, McSorley SJ, Garside P, Padron J, Phillips RS. Eur. J. Immunol. 1994;24:980–4. doi: 10.1002/eji.1830240430. [DOI] [PubMed] [Google Scholar]
- 86.Ignarro LJ, Buga GM, Wood KS, Byrns RE, Chaudhuri G. Proc. Natl. Acad. Sci. USA. 1987;84:9265–9. doi: 10.1073/pnas.84.24.9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Palmer RM, Ferrige AG, Moncada S. Nature. 1987;327:524–6. doi: 10.1038/327524a0. [DOI] [PubMed] [Google Scholar]
- 88.Adams MR, Forsyth CJ, Jessup W, Robinson J, Celermajer DS. J. Am. Coll. Cardiol. 1995;26:1054–61. doi: 10.1016/0735-1097(95)00257-9. [DOI] [PubMed] [Google Scholar]
- 89.Burnett AL, Lowenstein CJ, Bredt DS, Chang TS, Snyder SH. Science. 1992;257:401–3. doi: 10.1126/science.1378650. [DOI] [PubMed] [Google Scholar]
- 90.Lasker GF, Maley JH, Kadowitz PJ. Adv. Pharmacol. Sci. 2010:ii, 730861. doi: 10.1155/2010/730861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Andersson KE, Wagner G. Physiol. Rev. 1995;75:191–236. doi: 10.1152/physrev.1995.75.1.191. [DOI] [PubMed] [Google Scholar]
- 92.Krall JF, Fittingoff M, Rajfer J. Biol. Reprod. 1988;39:913–22. doi: 10.1095/biolreprod39.4.913. [DOI] [PubMed] [Google Scholar]
- 93.Rajfer J. Int. J. Impot. Res. 2008;20:431–6. doi: 10.1038/ijir.2008.10. [DOI] [PubMed] [Google Scholar]
- 94.Ignarro LJ, Bush PA, Buga GM, Wood KS, Fukuto JM, Rajfer J. Biochem. Biophys. Res. Commun. 1990;170:843–50. doi: 10.1016/0006-291x(90)92168-y. [DOI] [PubMed] [Google Scholar]
- 95.Ignarro LJ, Bush PA, Buga GM, Rajfer J. Nature. 1990;347:131–2. doi: 10.1038/347131b0. [DOI] [PubMed] [Google Scholar]
- 96.Holmquist F, Hedlund H, Andersson KE. Acta Physiol. Scand. 1991;141:441–2. doi: 10.1111/j.1748-1716.1991.tb09103.x. [DOI] [PubMed] [Google Scholar]
- 97.Kim N, Azadzoi KM, Goldstein I, Saenz de Tejada I. J. Clin. Invest. 1991;88:112–8. doi: 10.1172/JCI115266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rajfer J, Aronson WJ, Bush PA, Dorey FJ, Ignarro LJ. N. Engl. J. Med. 1992;326:90–4. doi: 10.1056/NEJM199201093260203. [DOI] [PubMed] [Google Scholar]
- 99.Trigo-Rocha F, Aronson WJ, Hohenfellner M, Ignarro LJ, Rajfer J, Lue TF. Am. J. Physiol. 1993;264:H419–22. doi: 10.1152/ajpheart.1993.264.2.H419. [DOI] [PubMed] [Google Scholar]


