Abstract
Previous studies have shown that ZBP-89 (Zfp148) plays a critical role in erythroid lineage development, with its loss at the embryonic stage causing lethal anemia and thrombocytopenia. Its role in adult hematopoiesis has not been described. We now show that conditional deletion of ZBP-89 in adult mouse hematopoietic stem/progenitor cells (HSPC) causes anemia and thrombocytopenia that are transient in the steady state, but readily uncovered following chemically induced erythro/megakaryopoietic stress. Unexpectedly, stress induced by bone marrow transplantation of ZBP89−/− HSPC also resulted in a myeloid-to-B lymphoid lineage switch in bone marrow recipients. The erythroid and myeloid/B lymphoid lineage anomalies in ZBP89−/− HSPC are reproduced in vitro in the ZBP-89-silenced multipotent hematopoietic cell line FDCP-Mix A4, and are associated with the upregulation of PU.1 and downregulation of SCL/Tal1 and GATA-1 in ZBP89-deficient cells. Chromatin immunoprecipitation and luciferase reporter assays show that ZBP-89 is a direct repressor of PU.1 and activator of SCL/Tal1 and GATA-1. These data identify an important role for ZBP-89 in regulating stress hematopoiesis in adult mouse bone marrow.
Keywords: Stress hematopoiesis, hematopoietic stem cells, erythroid progenitors, transcription factors, transplantation
Introduction
The hematopoietic system originates from a small population of self-renewing hematopoietic stem cells that differentiate into the various erythroid, myeloid and lymphoid lineages. The hematopoietic lineages tend to be specified in a stepwise process of binary decisions, dependent on particular genetic programs under control of transcription factors. The lineage-specifying and autoregulatory factors PU.1 and GATA1 form a master genetic switch that is responsible for determining the myeloid/lymphoid and erythroid lineages 1, 2, respectively, commonly acting in concert with lineage-restricted factors such as SCL/TAL1 3, 4 and CCAAT/enhancer binding protein α (C/EBPα)5. PU.1 and GATA1 mutually inhibit each other’s expression and transactivation functions, thus favoring one lineage fate over the other 6.
ZBP-89 (Zfp148) belongs to a novel class of GC-rich binding transcription factors phylogenetically conserved in mammals, which contain a characteristic array of four Krüppel type zinc fingers 7. Zebrafish ZBP-89 morphants 8 and ZBP-89 knockout mice 9 die at the embryonic stage with severe anemia and thrombocytopenia. The role of ZBP-89 in adult hematopoiesis has not been explored.
To investigate its role in adult hematopoiesis, we conditionally deleted ZBP-89 in mouse hematopoietic stem/progenitor cells (HSPC). We observed an early drop in red blood cell (RBC) and platelet (PLT) counts in peripheral blood (PB), and a significant reduction in the number of megakaryocyte/erythrocyte progenitors (MEP) in ZBP-89-deficient bone marrow (BM). The defect in the erythro-megakaryocytic lineage was, however, transient under steady state conditions, but was readily uncovered by chemical induction of hematopoietic stress. An unexpected reduction in the myeloid lineage and an increase in B lymphoid lineage were observed in ZBP89−/− BM recipients. Transcriptional profiling revealed a significant increase in PU.1 and a reduction in SCL and GATA1 transcripts in HSPC. Similar anomalous hematopoietic lineage and transcriptional profiles were observed in vitro when ZBP-89 was stably-silenced in the nonleukemic multipotent progenitor cell line FDCP-Mix A4 (A4) 10. Chromatin immunoprecipitation (ChIP) and luciferase reporter assays showed that ZBP-89 binds to the PU.1, SCL and GATA1 promoters, repressing PU.1 and activating SCL and GATA1. The significance of these findings is discussed.
Material and Methods
Mice and cell lines
Mice expressing the targeted ZBP-89 locus with flanking LoxP sites (ZBP-89fl/fl) were produced as described 11. Conditional deletion of ZBP-89 in the hematopoietic system of mice (C57BL/6-CD45.2+ background) was generated as shown in Figure 1A. Transgenic mice expressing an interferon-inducible Cre recombinase (MxCre) (Kuhn et al, 1995) were kindly provided by Dr. Hanno Hock (MGH Cancer Center). Murine erythroleukemia cell line MEL and the non-hematopoietic QM7 cell line, which lacks endogenous ZBP-89 12, were obtained from ATCC and maintained in DMEM medium containing 10% fetal calf serum. The multipotent A4 cell line was maintained in Iscove’s modified Dulbecco’s Medium (IMDM) supplemented with 20% horse serum and Interleukin-3 (IL-3)(100 units/ml, R&D Systems). Congenic C57BL/6-CD45.1 mice (6 to10 weeks old) were used as donors for purification of wild type cells and as recipients for BM transplantation (BMT) experiments. All animal experiments were performed in accordance with legal and ethical requirements demanded by law and approved by the Massachusetts General Hospital Subcommittee on Research Animal Care.
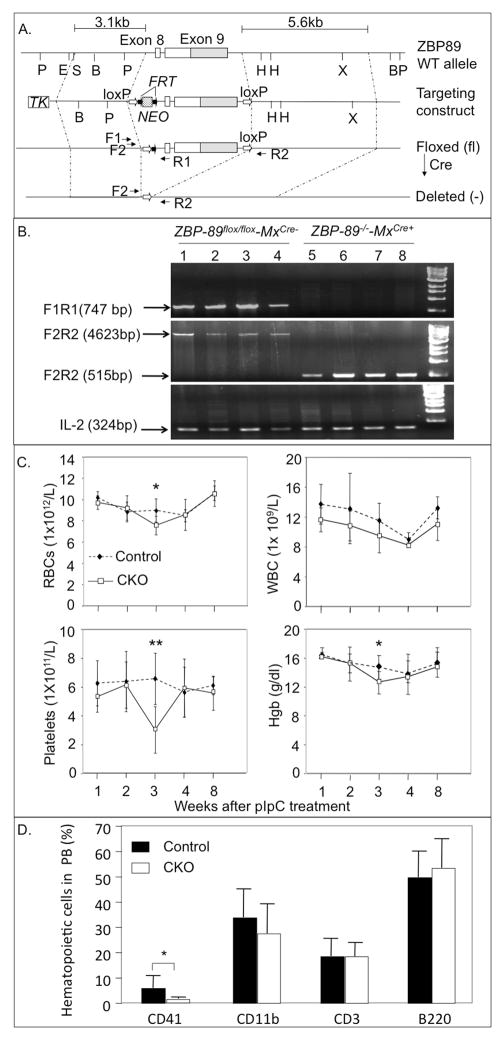
Figure 1. Generation and PB phenotype of ZBP-89 CKO mice.
(A) Strategy for inducible inactivation of ZBP89 in HSPC. Schematic of targeted ZBP-89 exons 8 and 9 (in white), with non-coding region of exon 9 in gray. Restriction sites (P, PstI; E, EcoR1; B, BamH1; H, HindIII; X, Xba1), LoxP sites (open arrows) and FRT sites (closed arrows) are indicated. F1, F2, R1 and R2 represent approximate position and orientation of the primers used in PCR. TK, thymidine kinase; NEO, neomycin. Sizes of the left (3.1kb) and right (5.6kb) vector arms are shown. (B) PCR genotyping showing deletion of the floxed segment of ZBP-89 in single CFU-GM stem cell colonies 8 weeks after pIpC treatment. Lanes 1–4, BM colonies from control (ZBP-89fl/fl-MxCre−) mice, and lanes 5–8 are colonies from ZBP-89 CKO (ZBP-89−/−-MxCre+) mice post pIpC. IL2 is included as internal control. (C) PB white blood cells (WBC)-, RBC-, and platelet counts and hemoglobin (Hgb) level 1–8 weeks after pIpC in (C) PB white blood cells (WBC)-, RBC-, and platelet counts and hemoglobin (Hgb) level 1–8 weeks after pIpC in six control mice (filled diamonds, dotted lines) and in seven ZBP-89 CKO mice (open squares, solid lines). Results are shown as mean ± SD, from 2 independent experiments. *P < 0.05, **P < 0.01. (D) CD41+, CD11b+, CD3+ and B220+ cells in PB from normal (open bars) and ZBP89 CKO mice (black bars) 3 weeks after pIpC injections (mean ± SD, n=6 in each group).
ZBP-89-silenced MEL and A4 cells
21-mers encoding 5 different short hairpin (sh) RNAs (H2 to H6)(Broad Institute, Cambridge, MA) were used for stable silencing of mouse wild-type ZBP-89 in MEL cells. Each shRNA was cloned into the pLK0.1-puromycin plasmid and the plasmid incorporated into lentivirus using the helper and packaging system pΔD8.9, pMD.G (VSV-G). Virus-infected MEL cells were selected by puromycin (5μg/ml), isolated and tested for ZBP-89 silencing by Real-time reverse-transcribed polymerase chain reaction (RT-PCR) and western blotting. The H5 21-mer, which produced maximal silencing of ZBP-89 in MEL (Supplemental Figure 1), was used to stably silence ZBP-89 in A4 cells.
Induction of the Cre transgene in vivo
To induce the MxCre transgene, ZBP-89fl/fl-MxCre+ or ZBP-89fl/fl-MxCre− control mice were injected intraperitoneally with polyinosinic-polycytidylic acid (pIpC, Amersham) at 20 μg/g dissolved in saline on every other day for a total of seven doses. Mouse PB and bone marrow cells were harvested for analysis at different time points after the last pIpC injection.
Recombination analysis
PCR analysis was performed using progenitor colony genomic DNA. To amplify the floxed (non-deleted) allele product and flanking DNA sequence, the forward/reverse primers F1, 5′-AGACCTACGACCCACAGGGTGG-3; R1, 5′-GGCTT CTCTCCACTGTGAGTT-3′; F2, 5′-TGTCCTCTCACCTCTGCACATTCAGCGACAC-3′ (between intron 7 and the reverse primer); R2, 5′-TGCGCCACAGACACACATC AGTCTTCAGATCG-3′ (at the 3′ untranslated region) were used. To amplify the recombined allele product and Cre gene, the following primers were used: MH61, 5′-GACCAGGTTCGTTCACTCATGG-3′; MH63, 5′-AGGCTAAGTGCCTTCTCTACAC-3′. The internal control primers were: IMR0042, 5′-CTAGGCCACAGAATTGAAAGATCT-3′ and IMR0043, 5′-GTAGGTGGAAATTCTAGCATCATCC-3′.
Hematologic analysis and cell culture
Blood samples were obtained from mice by tail puncture and placed in EDTA-coated microtubes. Blood counts were performed with a VetScan HM5 (Abaxis Veterinary Diagnostic Company). Murine colony assays were performed by plating 1×105 BM cells/ml of methocult M3434 (Stem Cell Technologies, Vancouver, BC) in 6-well plates in duplicate and cultured at 37°C for 10 days. Burst-forming unit erythroid (BFU-E), CFU-granulocyte (CFU-G), CFU-granulocyte-macrophage (CFU-GM), granulocyte-erythrocyte-macrophage-megakaryocyte (CFU-GEMM) colony numbers were counted based on colony morphology.
Chemical induction of hemolytic anemia and thrombocytopenia
One week post pIpC, mice were injected subcutaneously with 0.4% phenylhydrazine (PHZ; Sigma-Aldrich) in saline (12μl/g body weight) for 2 consecutive days. Mouse peripheral blood was collected for analysis on day 5 and day 15 after PHZ injection. Bone marrow and spleen cells were harvested from control and ZBP-89 CKO mice (n=3 in each group) on day 5 post PHZ injection. To induce thrombocytopenia, mice were injected intraperitoneally with 150 mg/kg 5-fluorouracil (5FU)(Sigma, St Louis, MO) one week post pIpC, and samples were collected 6 days later.
Hematologic analysis, flow cytometry and cell sorting
PB cell types were identified by flow cytometry following staining with cell-type specific antibodies using LSR II cytometer (BD Biosciences). Single cell suspensions of spleen and BM cells were obtained as detailed elsewhere 13. Surface phenotypes of isolated HSPC were as follows: BM-derived Lineage-negative (Lin−), LSK (Lin− Sca-1+ C-kit+); LT-HSC (LSK CD150+CD48−); multipotent progenitors (MPP) (LSK CD34+FLK3+); common myeloid progenitors (CMP) (Lin−C-kit+Sca1−CD34med CD16/32med); common lymphoid progenitors (CLP) (Lin−C-kitmed Sca1medIL7R+); granule-monocyte progenitors (GMP) (Lin−C-kit+ Sca1−CD34+CD16/32+); MEP (Lin−C-kit+Sca1−CD34−CD16/32−); precursor-B-progenitor B (Pre-Pro-B) (AA4.1+IL-7R+B220MedC-kit+); Pro-B (AA4.1+IL-7+B220MedC-kit−); Pre-B (AA4.1+IL-7+B220hiC-kit−); BM- or spleen-derived Pro- (CD71highTer119low), Basophilic- (CD71highTer119high), polychromatic (CD71lowTer119high) erythroblasts 14. Quantitative analysis of the distinct stages of erythroblast development was conducted as described 15. Briefly, bone marrow or spleen cells were first blocked with rat anti-mouse CD16/32 (BD Science) for 15 minutes, stained with the labeled rat anti-mouse antibodies FITC-ter119, APC-CD44, APC-Cy7 CD45, APC-Cy7 CD11b and APC-Cy7 GR1 (BD sciences), then subsequently stained with 7-AAD (BD science) before cell analysis using flow cytometry. In chimeric mice, PB and HSPC were stained with both Pacific blue-anti-CD45.2, PE-Cy7-anti-CD45.1 and the lineage specific FITC-anti-CD41, PE-anti-CD11b, PE-CY5-anti-CD3 and APC-anti-B220 antibodies, and the dual-positive (Pacific blue-CD45.2+ and lineage-specific cell populations) were gated and quantified. Wild-type or ZBP-89-silenced A4 cells at various passages (12–24 weeks) were immunostained after 1 week in culture with mAbs to the lineage-specific markers CD41, CD11b, CD3 and B220 followed by flow cytometry.
Total RNA isolation and RT-PCR
Total RNA was extracted from fractionated BM progenitors or from A4 cells with the RNAqueous-4PCR kit (AMBION INC, Austin, TX). For each experiment, BM progenitor-derived RNAs from two ZBP-89 CKO or two control mice were pooled. Reverse transcription of BM progenitor- or A4-derived RNA was performed with the High Capacity cDNA Reverse Transcription Kit (AB applied Biosynthesis). RT-PCR was run on Stratagene300 (Stratagene) using Brilliant SYBR Green QPCR Master Mix (Stratagene). Sequences for the primers used are as follows. ZBP89 RTF, 5′-CGGCATAGACGAAATGCAGTC-3′; ZBP89 RTR, 5′-CCTGGTGAGGCAAACTTCGAT-3′; SCLRTF, 5′-CACTAGGCAGTGGGTTCTTTG-3′, SCL RTR, 5′-GGTGTGAGGACCATCA GAAATCT-3′; GAPDH RFF,5′-AGGTCGGTGTGAACGGATTTG-3′; GAPDH RTR,5′-TGTAGACCATGT AGTTGAGGTCA-3′; GATA1RTF, 5′-TGGGGACCTCAGAACCCTTG-3′; GATA1 RTR, 5′-GGCTGCATTTGGGGAAGTG-3′; PU.1RTF,5′-GGACATGGTGTGC GGAGAA-3′, PU.1 RTR, 5′-AGAAAGCCATAGCGATCACTACT-3′; TIF1γ RTF, 5′-AGATAATGCAAGTGCAGTTGGT-3′, TIF1γRTR, 5′-ACGTCAATCTATCACACGTTTCA-3′;C/EBPα RTF,5′-AGGACACGGGGACCATTAG-3′; C/EBPα RTR,5′-TAGACGTGCA CACTGCCATT-3′; FOXO1 RTF, 5′-ATGCTCAATCCAGAGGGAGG-3′; FOXO1 RTR, 5′-ACTCGCAGGCCACTTAGAAAA-3′; Gfi1 RTF, 5′-CCCTTTGCGTGCGAGATGT-3′, Gfi1RTR, 5′-CACTGCCTTGTGTTGCTCCA-3′; C-myb RTF, 5′-CAGAAGAGGAGGACA GAATCATTT; C-myb RTR, 5′-TTCCAGTGGTTCTTGATAGCATTA-3′; FOG1 RTF, 5′-CAGAGCCTTATCCCCTGAGAG-3′; FOG1 RTR, 5′-CGGCTTCTTCAGTTAGGACCT-3′.
Bone marrow repopulation assays
CD45.2+ ZBP-89flox/flox-Mx-Cre− or ZBP-89flox/flox-Mx-Cre+ bone marrow cells mixed with competitor wild-type CD45.1 bone marrow cells in a 1:1 ratio (a total of 2×106 cells) were injected intravenously into the lateral tail vein of congenic age-matched CD45.1+ mouse recipients (n=5 per genotype) that were lethally-irradiated one day before BMT. 6 weeks later, 20 μg/g pIpC were injected into recipient mice every other day (for a total of seven doses), following which PB cells and BM HSPC were harvested at different times and analyzed for expression of CD45.1 and CD45.2 alleles. For secondary BMT, CD45.2+ ZBP-89-excised BM cells were harvested from primary recipients 7 months after the last pIpC dose, and 2×106 cells were injected into 6-week old irradiated CD45.1+ recipient mice (n=5 per genotype). Reconstitution of secondary recipients was analyzed 6 to 36 weeks after BMT.
Chromatin Immunoprecipitation (ChIP) Assays
This was carried out as described 16. Briefly, 5×106 MEL cells were cross-linked with 1% (v/v) formaldehyde, the reaction stopped with glycine, and washed cells pelleted then resuspended in lysis buffer. Following homogenization, the nuclei were sonicated on ice to a DNA size of 200–800 bp. Protein-DNA complexes were immunoprecipitated using an anti-ZBP-89 antibody (SC-19408; Santa Cruz) (or immunoglobulin G (IgG) as a negative control), and the DNA-protein complexes collected by binding to A/G plus-agarose beads. Washed beads were eluted in 1% SDS/0.1mM NaCHO3. DNA was reverse-crosslinked by incubation at 68°C overnight then purified using QIAGEN quick PCR purification kit. PCR was performed using 10% (3μl) of the bound DNA fraction from the chromatin precipitate or 1% (1μl) of the input DNA fraction. The murine SCL 1a/b promoter fragments (−2000 to +1bp) containing the ZBP-89 element were amplified using the forward primer 5′-TCCCAACGTGAGCGCTCAGCC-3′ and the reverse primer 5′-TGTGCGCCGCCGA GATAAGG-3′ for 1a region, and the forward primer 5′-TTCCTCCGTCTTTCCCCATGC-3′ and the reverse primer 5′-AGCACTCTCAACCCGGCCGCC-3′ for 1b region. The murine PU.1 3.4 kb HindIII UTR fragments containing the ZBP-89 element were amplified using the primers F1: 5′-ACCCCGGGGTTGAAGGAACAC-3′, R1; 5′-TCTCCAGAAAGCCTG TTGCTGTCAG-3′; F2, 5′-TAACCCCTGCACATGAAAGCC-3′, and R2, 5′-TCTGGG CAGGGTCAGAGTGCC-3′. The murine 129bp fragment containing the ZBP-89 binding site in G1HE was amplified using the forward primer 5′-TCCCTTATCTATGCCTTCCCA-3′ and the reverse primer 5′-ATGAAGGGTGCCTCTAAGGAC-3′. PCR products were separated in 2.0% agarose gels containing 0.5 μg/ml ethidium bromide.
Site-directed mutagenesis
Mutations in the ZBP-89 binding site of murine SCL 1a promoter were generated by overlapping PCR, changing the wild type ZBP-89 Ia binding site core sequence 5′-cgcttatcgGGGGcggggcc-3′ to 5′-cgcttcgGAAGcGgggcc-3′. PCR reactions were performed using WT forward primer (WTFP): 5′-GGGGTACCTCAGTTAGCGGTGAAGGCTCATG-3′ (tagged KpnI site underlined) and mutant reverse primer: 5′-GGCCCGCCAGCTTCCGATAAGCG-3′, to generate the first fragment, and the mutant forward primer: 5-CGCTTATCGGAAGCTGGCGGGCC-3′, and WT reverse primer (WTRP): 5′-CCGCTCGAGACCCGGCCGCCCGCACACACC-3′ (tagged XhoI site underlined) to generate the second fragment. DNAs from both PCR amplifications were gel extracted and used in overlapping PCR using WTFP and WTRP, and the final product was KpnI/XhoI restricted then inserted into the KpnI/XhoI- digested pGL2 vector. XL1-Blue competent cells were transformed with ligation mix, plated on LB Amp plates, and incubated overnight at 37°C. Colonies were screened and clones confirmed by DNA sequencing. A similar strategy was used in replacing WT ZBP-89 core binding site 5′-ggctccctctCCCCcgtctcttc-3′ in Ib with 5′-ggctccctctCTTCcgtctcttc-3′.
Luciferase Assay
The pXP-214 kb/-0.334 kb/Luc plasmid containing the 3.5kb −15/−14 URE cloned 5′ of the PU.1 minimal promoter was cloned upstream of the Luciferase reporter 17. A construct containing a 1.5kb segment 5′ of SCL promoter 1a in addition to promoter 1a and 1b was cloned upstream of Luciferase reporter gene in the pGL2 vector 18(SCL-pGL2 plasmid). Control and ZBP89-silenced MEL cells were transiently transfected with SCL-pGL2 or pXP-214 kb/-0.334 kb/Luc plasmids. QM7 cells intrinsically lacking ZBP-89 were co-transfected with luciferase reporter constructs prepared using the wild-type G1HE-(124-235)-luc) or G4 mutant G1HE-(124-235)-Luc reporter 12 and an expression plasmid encoding wild type mouse ZBP-89. Transfected cells were harvested, washed twice in PBS, pelleted, lysed with 300μl of lysis buffer (Promega Inc., WI) for 30 minutes at room temperature, and lysate centrifuged for 10 minutes at 13,000 rpm. Luciferase assays were carried out on 20μl of each cell lysate supernatant and light units measured in a Berthold LB9507 luminometer (Berthod, Germany). Relative light units were normalized for transfection efficiency against β-Gal values obtained from 20μl of each supernatant. β-Galactosidase (β-Gal) assays were performed according to the manufacturer’s instructions (Applied Biosystems).
Statistical Analysis
The significance of the difference between groups in the in vitro and in vivo experiments was determined by analysis of variance followed by a one-tailed Student’s t test. Data are expressed as the mean±SD, with P-values of < 0.05 considered significant.
Results
Anemia and thrombocytopenia in ZBP-89 CKO mice
We conditionally inactivated ZBP-89 in the mouse hematopoietic system by breeding a ZBP-89fl/fl C57BL/6 mouse strain 11 with interferon-inducible Mx-Cre transgenic C57BL/6 mice 19(Fig. 1A), followed by administration of poly(I)-poly(C) (pIpC) every other day for a total of 7 injections. PCR of genomic DNA from control (ZBP-89fl/fl-MxCre−) and ZBP-89fl/fl-MxCre+ mice showed undetectable amounts of ZBP-89fl/fl in single BM-derived colony forming-unit-Granulocyte-Macrophage (CFU-GM) colonies transduced with the Cre gene, indicating nearly complete excision of both ZBP-89fl/fl alleles (Fig. 1B).
PB cells obtained weekly in the first 4 weeks and at 2 months after the last pIpC injection showed a significant drop in red blood cells (RBC) and hemoglobin (Hgb) and in platelet counts and in CD41+ megakaryocytes in ZBP-89−/−-MxCre+ mice but not in control (ZBP-89fl/fl-MxCre) mice 3 weeks after the last pIpC injection (Fig. 1C, 1D). No significant changes in peripheral white blood count (WBC), or circulating myeloid (CD11b+), lymphoid T- (CD3+) or B- (B220+) cells were detected (Fig. 1C, D). Consistently, bone marrow colony forming assays showed reduced ZBP-89-deficient burst-forming unit-erythroid (BFU-E) and Granulocyte-Erythroid-Macrophage-Megakaryocyte (GEMM), but not CFU-GM, CFU-M or CFU-G colonies (Supplemental Figure 2A). Peripheral RBC and platelet counts returned to normal at 4 and 8 weeks post pIpC (Fig. 1C), with an associated normalization of bone marrow BFU-E and CFU-GEMM colony counts of ZBP-89CKO mice (Supplemental Figure 2B).
Changes in HSPC and their transcriptional profile
The percentage of ZBP-89-deficient MEP was significantly reduced 3 weeks post pIpC (Fig. 2A), but no change was detected in ZBP-89-deficient LT-HSC, MPP, CMP, CLP, GMP and MEP.
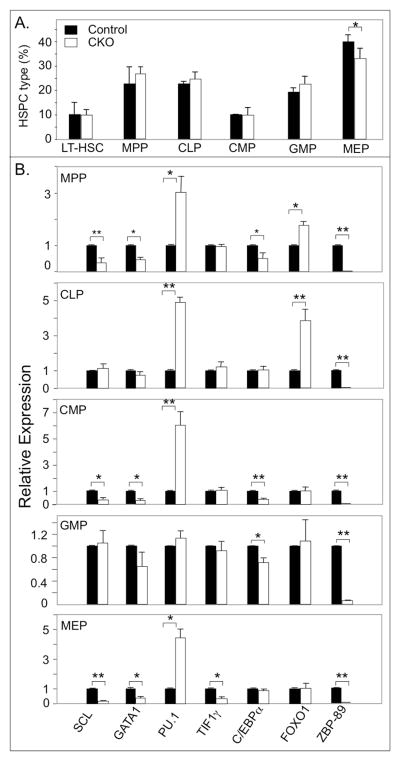
Figure 2. Transcription factor profiles of BM HSPC from ZBP-89 CKO and control mice 3 weeks after the last dose of pIpC.
(A) Histograms (mean ± SD, n=3) showing the percentage of LT-HSC and MPP, CLP, CMP, GMP and MEP in fractionated BM cells from ZBP-89 CKO and in control mice. (B) RT-PCR analysis of transcription factors in BM progenitors derived from ZBP-89 CKO and control mice (bars are colored as in A). Results are from two independent experiments, each representing pooled samples from two mice in each group. * P<0.05; ** P<0.01.
The transcriptional profiles of key transcription factors known to regulate hematopoietic lineage commitment were measured in BM progenitors from ZBP-89-deficient and sufficient animals (Fig. 2B). RT-PCR carried out on mRNA derived from sorted ZBP-89-deficient MPP, CLP, CMP, GMP and MEP revealed a significant induction of PU.1 in MPP, CLP, CMP and MEP, and of FOXO1 in ZBP-89-deficient MPP and CLP. In contrast, SCL/TAL1 and GATA1 levels were significantly reduced by ~60–80% in ZBP-89-deficient MPP, CMP and MEP but not in CLP or GMP. TIF1γ was only suppressed in MEP, while C/EBPα was suppressed in MPP, CMP and GMP (Fig. 2B). As expected, ZBP-89 mRNA levels were minimal in HSPC derived from ZBP-89 CKO mice (Fig. 2B).
ZBP-89 contributes to the erythro- and megakaryopoietic reserve in the adult
In response to erythroid stress in adults, the rate of RBC production rapidly increases from its steady state level 20. We evaluated the response to acute depletion of RBC or platelets in ZBP-89 CKO and control mice. Phenylhydrazine (PHZ) induced a significant (~50%) drop in circulating RBC counts (Fig. 3A) and in bone marrow BFU-E colonies (Supplemental Figure 2C), but not in peripheral blood platelet counts (Fig. 3A) or CFU-GEMM colonies (Supplemental Figure 2C) from ZBP-89 CKO mice. Erythroblast development was examined using two methods 14, 15, both of which showed a significant reduction in pro- and basophilic erythroblasts of ZBP-89 CKO mice (Fig. 3B, Supplemental Figure 3), in parallel with an increase in pro- and basophilic erythroblasts in spleen of ZBP-89 CKO mice (Fig. 3B, Supplemental Figure 3A, B). Polychromatic erythroblasts in spleen were increased ~1.6-fold in ZBP-89 CKO mice when quantified by one method 14 (Fig. 3B) but not the other 15 (Supplemental Figure 3B). There was also a significant increase in spleen size on day 5 after PHZ injection in ZBP-89 CKO mice (Supplemental Figure 4). These changes were insufficient, however, to prevent stress-induced anemia on day 5, but might have contributed to the normalization of RBC, Hb and Hct by day 15 after PHZ injection (Supplemental Figure 5).
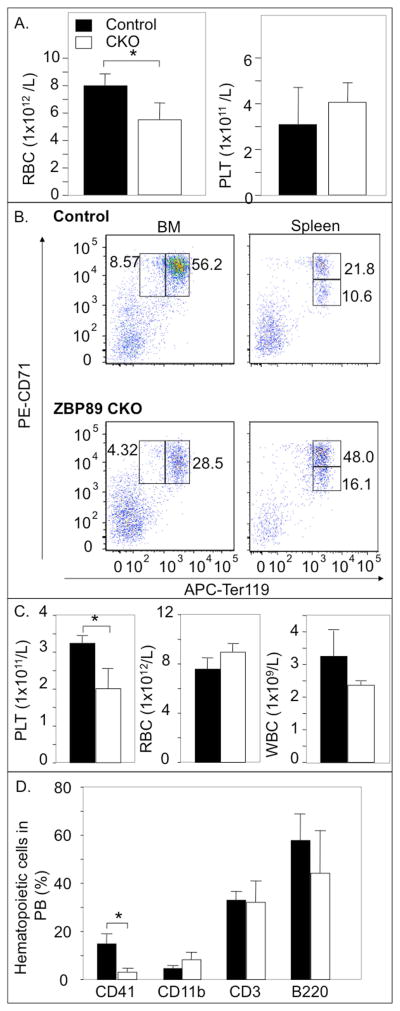
Figure 3. Effect of ZBP-89 CKO on stress erythropoiesis and thrombopoiesis.
(A) Histograms (mean ± SD, n=4) showing the effect of PHZ-induced hemolysis on RBC and platelet (PLT) counts in PB of ZBP-89 CKO mice (white bars here and in C, D) compared to control mice (black bars here and in C, D). (B) Representative FACS analyses of BM proerythroblasts (CD71highTer119low) and basophilic erythroblasts (CD71highTer119high) (left) and in basophilic- and in polychromatic (CD71lowTer119high) erythroblasts in spleen (right) of control and ZBP-89 CKO mice, two weeks after the last pIpC injection and 5 days after PHZ. Numbers for each box reflect percentages of the gated cells, with each representing the mean value from four mice in each group. (C, D) Histograms (mean ± SD, n=4) showing the effect of 5-fluorouracil (5-FU)-induced platelet depletion on circulating platelet-, white blood cells (WBC)- and RBC counts (C), and on surface phenotype, analyzed by FACS from ZBP-89 CKO and control mice, two weeks after the last pIpC treatment and 6 days after 5-FU (D). * P<0.05.
PB platelet count, but not RBC or WBC counts, also significantly decreased in ZBP-89 CKO mice upon acute administration of 5-fluorouracil (5-FU), which depletes immature megakaryocytic progenitors and megakaryocytes in endomitosis 21(Fig. 3C). FACS analysis further confirmed that the megakaryocyte lineage marker CD41 was markedly reduced in PB following 5-FU treatment (Fig. 3D), with no significant changes in PB myeloid (CD11b+), T- (CD3+), or B-cells (B220+).
ZBP-89 deletion results in reciprocal changes in the myeloid- and B-cell lineages in BM repopulation assays
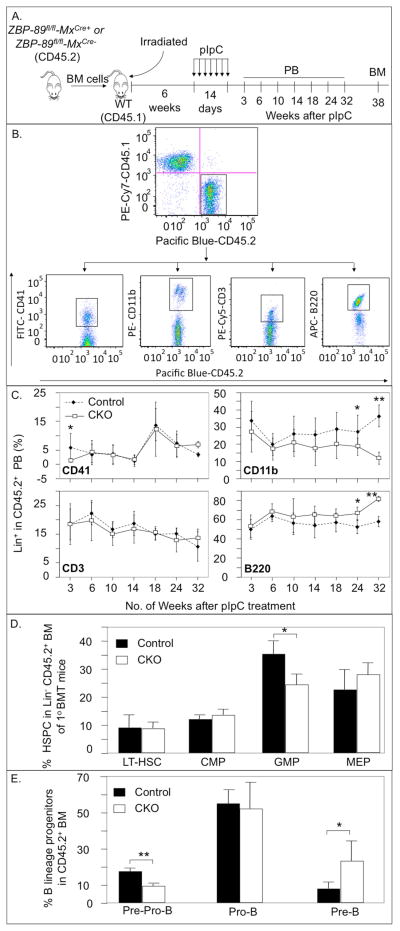
CD45.2+ BM cells from ZBP-89-excised (ZBP-8−fl/fl-MxCre+) and control (ZBP-89fl/fl-MxCre−) mice were transplanted into congenic lethally irradiated CD45.1+ recipients that were then treated with pIpC starting 6 weeks after transplantation (Fig. 4 A). The contribution of ZBP-89-excised CD45.2+ hematopoietic stem cells to PB and BM cells was monitored by surface expression of the CD45.2 allele in HSPC and in PB (Fig. 4 B–E). The proportion of CD45.1+ and of CD45.2+ cells in recipients 3 weeks after primary BMT was equivalent (Fig. 4B, upper panel). Since partially differentiated, yet multi-lineage hematopoietic precursors continue to generate mature cells for up to 3 months post BMT, we measured the percentage of progenitors and differentiated blood cells that descended from donor and competitor precursors up to 8 months after BMT, thus ensuring that all mature blood cells derive from primitive hematopoietic stem cells 22, 23. ZBP-89-excised CD45.2+ cells contributed comparably to peripheral myeloid and lymphoid cells (CD11b+, CD3+ and B220+) in recipient mice for 4.5 months (Fig. 4C). Afterwards, the CD45.2+ CD11b+ PB pool dropped significantly (by ~2/3rd) with a reciprocal increase in the B220+ pool, but with no changes in number of CD3+ cells. At 3 weeks post pIpC, CD45.2+ CD41+ megakaryocytic cell numbers signficantly dropped, but normalized at week 6 post pIpC onwards (Fig. 4C), as in the steady state (Fig. 1C). RBC levels were normal but their origin was not determined in the chimeric mice as RBC lack CD45 expression. A significant reduction in GMP with a corresponding increase in Pre-B and a drop in Pre-Pro-B were observed in the BM, 38 weeks post pIpC (Fig. 4D, E). These changes likely account for the reciprocal alterations in ZBP-89-excised CD45.2+ CD11b+ and CD45.2+ BB20+ cells in peripheral blood. No changes were detected in the LT-HSC, CMP, MEP or Pro-B pools (Fig. 4 D, E). The decrease in the Pre-Pro-B cells and the increase in the Pre-B population (Fig. 4E) may suggest that proliferation of ZBP-89-excised B-cell progenitors in the stages from Pro-B to Pre-B cells is accelerated.
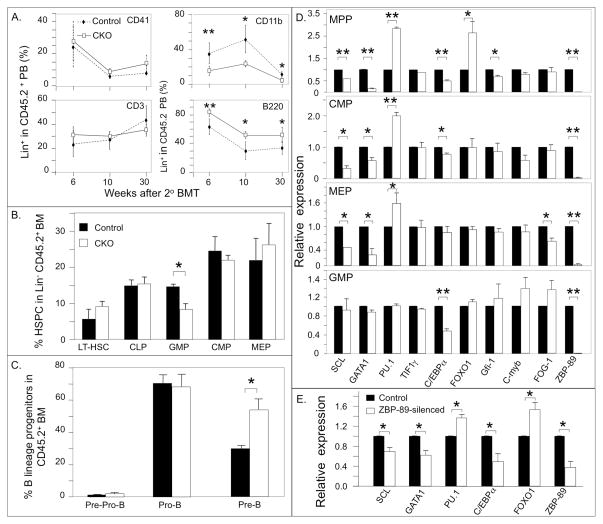
Figure 4.
PB cell counts and percentage of immature hematopoietic lineages in 1° BM transplant recipients. (A) Schematic of the experimental design: BM cells (CD45.2+) from ZBP-89 CKO and control mice were mixed with wild type (WT, CD45.1+) BM cells at a 1:1 ratio and the mixture injected into irradiated wild-type (WT) recipients (CD45.1+). pIpC injections started 6 weeks later and over a 2-week period. PB samples were analyzed at 3–32 weeks after the last pIpC dose and BM was examined at 38 weeks. (B) Isolation of lineage-specific CD45.2+CD45.1− PB cells from control recipients of 1° BM at 3 weeks post pIpC. (C) Percentage of PB megakaryocytes (CD41+), myeloid- (CD11b+), T- (CD3+) and B (B220+)-cells in the CD45.2+ population of ZBP-89−/− and control (ZBP-89fl/fl) mice at the indicated times after pIpC injections. (D) Histograms showing percentage of the different immature hematopoietic lineages in the CD45.2+ BM population of ZBP-89−/− (mean ± SD, n=7) and control mice (mean ± SD, n=8) 38 weeks after pIpC treatment. (E) Histograms (mean ± SD, n=4) showing the percentage of Pre-Pro-B cells, Pro-B cells and Pre-B cells in the CD45.2+ BM population of ZBP-89−/− and control mice 38 weeks after pIpC treatment. * P<0.05, **P<0.01.
We next examined the capacity of ZBP-89-excised CD45.2+ BM HSC for long-term reconstitution of adult hematopoiesis by serial transplantation. Donor CD45.2+ ZBP-89−/− or ZBP-89fl/fl BM (as control) cells were injected into lethally irradiated CD45.1+ recipient mice. The multi-lineage engraftment potential was assessed over an 8-month period. A reduction in PB CD11b+CD45.2+ cells and an increase in B220+CD45.2+ cells were again seen in cells derived from ZBP-89-excised HSPC, beginning earlier (at week 6-post transplantation) (Fig. 5A) vs. 24 weeks following primary BMT (Fig. 4C). Examination of BM hematopoietic progenitors at 7.5 months also showed persistence in the contraction of the GMP pool size and the increase in the Pre-B compartment (Fig. 5 B, C).
Figure 5. Hematopoietic lineages and their transcriptional profiles in 2° BM transplant recipients and in ZBP-89-silenced A4 cells.
(A) Percentages of the different HSPC lineages in CD45.2+ PB at different times after 2° BM transplantation. Results given are mean ± SD (n=5 mice in each group). (B, C) Histograms (mean ± SD, n=4) showing the percentages of the different BM progenitors in CD45.2+ BM from 2nd BMT recipients at 30 weeks post transplantation from control (black bars) and CKO mice (white bars). * P<0.05, ** P<0.01. (D) Histograms (mean ± SD, n=2 independent experiments) showing gene expression profiles in BM progenitors from ZBP-89-deficient cells 36 weeks after 2° BMT (white bars) relative to that in control mice (black bars). For each experiment, RNAs from two mice in each group were pooled for RT-PCR. * P<0.05; ** P<0.01. (E) Histograms (mean ± SD, n=2 experiments) showing gene expression profiles in control and ZBP-89-silenced A4 progenitors. * P<0.05.
Transcriptional deregulation in ZBP-89-excised HSPC
RT-PCR analysis performed on RNA derived from sorted ZBP-89-deficient BM HSPC from secondary BM transplant recipient mice at 36 weeks post transplantation showed persistent upregulation of PU.1 and downregulation of GATA1 and SCL/TAL1 in MPP, CMP and MEP, upregulation of FOXO1 in MPP and downregulation of C/EBPα in MPP, CMP and GMP, and of Gif1 in MPP and FOG-1 in MEP (Fig. 5D). No significant changes in C-myb or Tif-1γ expression were seen.
The hematopoietic lineage anomalies in ZBP-89 CKO mice are reproduced in vitro in ZBP-89-silenced multipotent A4 cells
As previously described 24, immunophenotyping of wild-type A4 cells by flow cytometry showed that the bulk express the B lineage marker B220, the myeloid lineage marker CD11b and the megakaryocytic marker CD41 (Table 1). Stable silencing of ZBP-89 in A4 progenitors caused significant reductions in expression of CD41 and CD1b but increased expression of B220, mirroring the changes observed under BMT stress conditions in vivo. RT-PCR analysis performed on RNA derived from ZBP-89-sufficient and deficient A4 progenitors again showed downregulation of GATA1, SCL/TAL1 and C/EBPα and upregulation of PU.1 and FOXO1 in ZBP-89-silenced cells (Fig. 5E).
Table 1.
Immunophenotype of wild-type and ZBP-89-silenced FDCP-A4 (A4) cells
| Cell Lineage | Surface marker | Percent Positive Cells | P value | |
|---|---|---|---|---|
| A4 | H5-A4 | |||
| Megakaryocytes | CD41 | 80.3±5.1* | 55.1±11.1 | 0.022 |
| Myeloid | CD11b | 92.8±1.5 | 72.0±12.2 | 0.043 |
| B cells | B220 | 47.9±8.1 | 77.2±2.2 | 0.004 |
| T cells | CD3 | 1.6±0.7 | 1.5±1.9 | 0.99 |
Numbers represent the mean ± SD of three independent experiments.
ZBP-89 acts as a direct transcriptional repressor of PU.1 and activator of SCL and GATA1
Induction of PU.1, and suppression of SCL in ZBP-89-silenced BM progenitors, suggested these two factors might be direct downstream targets of ZBP-89, as has been previously shown for GATA112. This was tested (Fig. 6A–F) using ChIP 16 and luciferase reporter assays in WT and ZBP-89-silenced MEL cells or in the non-hematopoietic QM7 cells, which lack endogenous ZBP-89 12, using the known transcriptional regulation of GATA1 by ZBP-89 as a positive control (Fig. 6A–C).
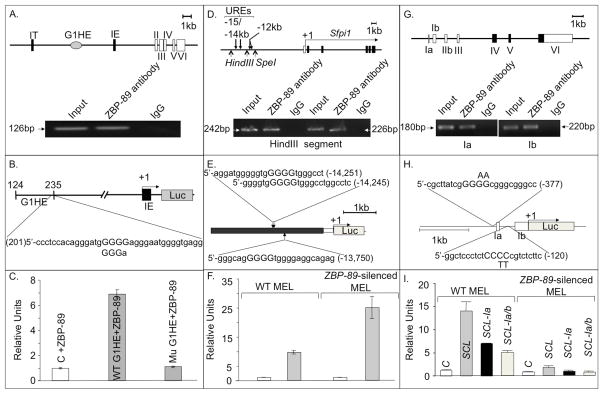
Figure 6. ZBP-89 is a transcriptional regulator of GATA1, PU.1 and SCL.
(A) Schematic of a genomic segment containing mouse GATA1 locus, with two cell-type specific first exons (IT and IE), five coding exons (II–VI), and the G1HE region. The other cis-regulatory elements (double GATA, CACCC and GATA repeats) are not shown. Lower panel, ChIP assays showing specific binding of ZBP-89 to the 5′ region of the GATA1 gene hematopoietic enhancer (G1HE)(which allows GATA1 expression in erythroid lineage). (B) Wt and mutant G1HE (G1HE-(124–235)-luc in which one of the G5 string comprising the ZBP-89 core binding motif is deleted) reporter constructs used (see methods). (C) Histograms (mean±SD, n=2) showing luciferase (Luc) activity in ZBP-89-transfected QM7 cells driven by wild-type (WT) or mutant G1HE (where the ZBP-89 site is mutated). Background luciferase activity was obtained using the control (C) promoter-less vector pGL2. (D) Schematic of a genomic segment containing the nontranslated region of PU.1, the minimal promoter (white bar) and the five coding exons (in black). Transcription start site (+1) and direction (horizontal arrow) are shown. Lower panel, ChIP assays showing specific interaction of ZBP-89 with two PCR products in the −15/−14 URE of PU.1. Normal IgG, used as negative control, and input DNA used as a positive control. (E) pXP-214 kb/-0.334 kb/Luc plasmid containing the 3.5kb −15/−14 URE (in black) cloned upstream of the PU.1 minimal 0.5kb promoter (in white) driving Luc reporter (in gray). Position and sequence of the three predicted ZBP-89 binding sites (core motif capitalized) in −15/14 URE are shown. (F) Histograms (mean±SD, n=2) showing Luc activity in WT and ZBP-89-silenced MEL cells driven by −15/14 URE. In C, F, I, relative units represent Luc activity normalized against β-Gal values. (G) Schematic of a genomic segment containing mouse SCL, comprising 7 exons, with noncoding exons in white. Lower panel, ChIP assays showing specific interaction of ZBP-89 with 1a or 1b promoter regions. (H) Schematic of the construct containing promoters la and lb of SCL cloned upstream of Luc in SCL-pGL2 vector. The two predicted ZBP-89 binding sites are shown. Position and nature of substitution of the two central pyrimidines in the ZBP-89 binding motif of la and lb are indicated above and below the respective sequence. Exons 1a and 1b are shown as white boxes. (I) Histograms (mean ± SD, n=2) showing Luc activity driven by −2kb to +1 DNA region of WT SCL or by SCL in which ZBP-89 consensus-binding sites in promoter 1a (SCL-1a) or 1a+1b (SCL-1a/b) were mutated. Background luciferase activity was obtained using the control promoter-less vector pGL2.
Expression of PU.1 is regulated primarily by a −15/14 kb upstream regulatory region (URE) (Fig. 6D), deletion of which reduces PU.1 expression by 80% in mice 25. C/EBPα binds to URE and prepares an adjacent −12kb enhancer for autoregulatory PU.1 chromatin entry, thus driving myeloid-specific PU.1 expression 26. A Genomatix search 27 identified 3 potential ZBP-89 binding sites in the −15/14 kb URE (Fig. 6E), two of which overlap. ChIP assays confirmed binding of ZBP-89 to URE (Fig. 6D, lower panel). Luciferase reporter assays carried out on WT and ZBP-89-silenced MEL transiently transfected with the pXP-214 kb/-0.5 kb/Luc plasmid 25 (Fig. 6E), revealed a ~2.5 fold induction of PU.1 when ZBP-89 was silenced (Fig. 6F).
Tissue-specific expression of murine (and human) SCL is driven by 2 minimal promoters la and lb 28 (Fig. 6G). Promoter Ia restricts SCL expression to the erythroid lineage 18, and Ib directs SCL expression in CD34+ hematopoietic progenitors 29. A Genomatix search revealed two potential ZBP-89 binding sites in the immediate 5′ regions of 1a and 1b in SCL (Fig. 6H). ChIP assays in WT MEL cells showed that ZBP-89 binds specifically to DNA fragments containing these regions upstream of the respective promoter (Fig. 6G, lower panel). Luciferase reporter assays showed that the -2kbSCL1a1b region drove luciferase expression in WT MEL cells, but minimally in ZBP-89-silenced MEL (the residual activity is likely explained by the incomplete 80% silencing of ZBP-89). Nucleotide substitutions of one or both consensus ZBP-89 binding motifs reduced luciferase reporter activity by 50% and by 60%, respectively (Fig. 6I), suggesting that ZBP-89 binding to the proximal site plays a greater role in SCL induction in the WT MEL cellular context.
Discussion
The role of ZBP-89 in embryonic development of the hematopoietic system has been previously described in zebrafish and mouse embryos 8, 9. Using a conditional knockout model of ZBP-89 in adult mice, we now show that ZBP-89 plays a previously unrecognized role in stress hematopoiesis in the adult.
Excision of ZBP-89 in adult mice resulted in the overt drop in circulating RBC and platelet counts by 3 weeks post pIpC but spared the other hematopoietic lineages, despite the efficient excision of ZBP-89 in the respective hematopoietic progenitors. Anemia and thrombocytopenia were associated with a significant reduction in MEP and in early and late erythroid progenitors in bone marrow, findings that were also observed in the absence of the master erythroid megakaryocytic regulator GATA1 30. This is likely secondary to the higher myeloid potential of CMP and MPP, resulting from reduced GATA1 and increased PU.1 expression in these progenitors 1, 6, 31, 32.
The anemia and thrombocytopenia observed in ZBP-89 CKO mice were however transient, with normalization of RBC and platelet counts in PB 4 and 8 weeks post pIpC, in parallel with normalization of BFU-E and CFU-GEMM BM colonies at 8 weeks post pIpC. This recovery is unlikely to derive from progenitors that escaped Cre-excision of the ZBP-89 gene, since ZBP-89 excision in hematopoietic progenitors is still demonstrable 36 months later (Fig. 5D). A more likely explanation is that loss of ZBP-89 in BM progenitors can be overcome in vivo in the steady state. The redundancy sometimes observed in genetic ablation models is usually explained by the presence of a functionally related gene. Whether ZBP-99 (ZNF281), the other known member of the ZBP-89 family 33, can compensate for loss of ZBP-89 in early BM erythro-megakaryocytic progenitors in the steady state will require further study.
The defect in the erythro-megakaryocytic lineages in ZBP-89 CKO mice can readily be exposed under stress (Fig. 3). Hemolytic stress normally induces expansion of erythroid progenitors in the spleen, driven largely by rapid proliferation of erythroid progenitors 34. The response to stress erythropoiesis in mice, which occurs mainly in the spleen 35, was evident in ZBP-89 CKO mice, but insufficient to prevent overt anemia. Erythropoiesis in the spleen is a molecularly distinct process from BM erythropoiesis, dependent on the glucocorticoid receptor 36, c-Kit 35 and BMP4/Smad5 37, which may explain the differential response to stress in the spleen vs. that in BM of ZBP-89 CKO mice. Impaired development and maturation of the erythroid lineage in BM under stress in ZBP-89 CKO mice has also been observed in GATA1 CKO mice 30 and to a lesser degree in SCL/TAL1 CKO mice 38, suggesting that abnormal stress erythropoiesis in ZBP-89 deficient BM erythroid progenitors is likely caused by suppressed expression of one or both of these factors.
Ablation of ZBP-89 in hematopoietic cells following competitive transplantation of whole BM from CD45.2+ ZBP-89fl/fl-MxCre+ or ZBP-89fl/fl-MxCre− (control) mice and pIpC treatment again lead to the early drop in circulating CD41+ cells at 3 weeks post pIpC, similar to that observed in non transplanted ZBP-89 CKO mice in the steady state. Follow up of primary BMT recipient mice, however, revealed a significant and persistent drop in circulating CD11b+ cells and an increase in B220+ cells by 24 weeks post transplant (Fig. 4C), together with a significant decrease in GMP and an increase in Pre-B BM progenitors (Fig. 4D, E). The reciprocal changes in the progenitor cell pools were also seen earlier (by 6 weeks) following secondary transplantation, which persisted for at least 30 weeks (Fig. 5A). The above lineage anomalies were reproduced in vitro by stable knock down of ZBP-89 in the multipotent A4 cells (Table 1), suggesting that the changes in the hematopoietic progenitors observed in whole animals are cell-autonomous, and directly related to loss of ZBP-89.
Selective transcriptional profiling showed increased PU.1 and suppressed GATA1 and SCL expression in MPP, CMP and MEP from ZBP-89 CKO mice (Fig. 2B and 5D) and in ZBP-89-silenced A4 cells (Fig. 5E). These changes are expected to favor development of the myeloid lineage, based on the binary model of HSC fate decisions. Yet, the measured outcome was a reduction in the myeloid lineage, with a reciprocal increase in the B lymphoid lineage. One potential explanation is the observed concomitant reduction of C/EBPα in ZBP-89-deficient myeloid progenitors (Fig. 5D), which is also observed in ZBP-89-silenced A4 cells (Fig. 5E). The reduction of C/EBPα in GMP compartment could compromise the ability of PU.1 to access the autoregulatory −12kb enhancer element in this pool, which is necessary for its commitment to the myelomonocytic cell lineage 5, 26. This myeloid lineage defect, which is not seen under steady state conditions, is readily uncovered under the replicative stress imposed by serial BMT, which could lead to exhaustion of the GMP pool 39. Formation of a complex of PU.1/E2A/FOXO1 on the −14kb URE in ZBP-89-deficient CLP, where PU.1 is also upregulated, appears sufficient, however, for growth and differentiation of the B cell lineage 26, 40, 41, enhanced in this case by the concomitant upregulation of FOXO1 in this population.
CHIP and luciferase reporter assays show that ZBP-89 acts directly to repress PU.1 while activating both SCL and GATA1, consistent with the lineage anomalies induced in vitro by loss of ZBP-89 in A4 cells. The reciprocal regulation of C/EBPα and FOXO1 by ZBP-89 in hematopoietic progenitors that is seen both in vivo (Fig. 2B, 5D), and in vitro (Fig. 5E) may also be direct (both promoters have potential ZBP-89 binding motifs) or indirect, driven by modulated expression of one or more of the other regulated genes.
In conclusion, we have provided evidence that ZBP-89 plays an important role in stress hematopoiesis in adult mice. Identification of ZBP-89 as an important modulator of key lineage-determining genes provides new insights into the genetic programs that underlie lineage decisions in adult hematopoiesis under steady state and stress conditions.
Supplementary Material
Supplemental Figure 1. Silencing of ZBP-89 in MEL cells. Wild type MEL cells were transfected with five shRNAs targeting different loci in murine ZBP-89. (A) Schematic diagram of the mouse ZBP-89 shRNA clones (H1–H5) targeting different coding regions of ZBP-89. (B) Western blots of cell lysates from clones infected with various shRNAs and probed with anti-ZBP-89 or anti-actin antibodies (for loading control). Maximal knockdown of mouse ZBP-89 expression was achieved in ZBP-89 clone H5.
Supplemental Figure 2. Effects of conditional loss of ZBP-89 on hematopoietic colony formation in vitro. Colony counts were performed at 3 weeks (A) and on 2 months (B) post pIpC-treated bone marrow cells. (C) Colony counts performed on day 5 following PHZ treatment. Histograms show mean + SD of four (A, B) or three (C) independent experiments each carried out in duplicate. BFU-E, blast forming units erythroid; CFU-GEMM, colony forming units granulocyte-erythroid--macrophagemegakaryocyte colonies; CFU-GM, granulocyte-monocyte colonies; CFU-M, monocyte colonies; CFU-G, granulocyte colonies. * P value < 0.05; ** P value < 0.01; ns, not significant.
Supplemental Figure 3. Erythroblast populations in BM and spleen of ZBP-89 CKO mice. (A) Gating of bone marrow and spleen erythroblasts of a Control and a ZBP-89 CKO mice on 5 day after PHZ injection; CD44 is plotted versus FSC of TER119+ cells. Gated erythroblast populations are: I, Pro (CD44 hi Ter119 Low); II, Basophilic (Baso); III, Polychromatic (Poly); IV, Orthochromatic (Ortho); V, Reticulocytes; VI, RBC. (B) The proportion of cells at each stage of erythroblast development form Control and ZBP-89 CKO (3 mice in each group) is normalized based on total nucleated erythroblasts as 100%. Values represent mean + SD.
Supplemental Figure 4. Effect of stress erythropoiesis on spleen size in ZBP-89 CKO mice. Spleen weights/body weight (g) of Control (Cre- zbp89fl/fl) and CKO mice on day 5 post PHZ-induced hemolysis. Histograms represent mean + SD with three mice used in each group. * P< 0.05.
Supplemental Figure 5. Changes in peripheral blood erythrocytes, hemoglobin and hematocrit following PHZ-induced hemolysis. Results shown represent mean ± SD from 7 Control or ZBP-89 CKO mice. * P< 0.05; **P<0.01.
Acknowledgments
We thank Dr. Hanno Hock (MGH Cancer Center) for providing MX1-Cre transgenic mice, Dr. Daniel Tenen (BIDMC, Boston, MA) and Dr. Kinuko Ohneda (Takasaki University of Health and Welfare, Takasaki, Japan) for plasmids and Dr. Ursula Just (University of Kiel, Germany) for FDCP-Mix A4 cells. This work was supported by NIH grants DK081920 (to MAA) and DK55732 (to JLM) from the National Institutes of Health (NIDDK).
Footnotes
Author contributions. MAA conceived and designed experiments; XL, DP, JLM performed experiments; MAA and DTS analyzed data; MAA wrote the paper.
Disclosure of potential conflicts of interest. The authors indicate no potential conflicts of interest.
See www.StemCells.com for supporting information available online.
References
- 1.Iwasaki H, Mizuno S, Wells RA, et al. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity. 2003;19:451–462. doi: 10.1016/s1074-7613(03)00242-5. [DOI] [PubMed] [Google Scholar]
- 2.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nature reviews Immunology. 2007;7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 3.Fujiwara T, O’Geen H, Keles S, et al. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Molecular cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu M, Riva L, Xie H, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Molecular cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeamans C, Wang D, Paz-Priel I, et al. C/EBPalpha binds and activates the PU.1 distal enhancer to induce monocyte lineage commitment. Blood. 2007;110:3136–3142. doi: 10.1182/blood-2007-03-080291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arinobu Y, Mizuno S, Chong Y, et al. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell stem cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Merchant JL, Iyer GR, Taylor BR, et al. ZBP-89, a Kruppel-like zinc finger protein, inhibits epidermal growth factor induction of the gastrin promoter. Molecular and cellular biology. 1996;16:6644–6653. doi: 10.1128/mcb.16.12.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Xiong JW, Shelley CS, et al. The transcription factor ZBP-89 controls generation of the hematopoietic lineage in zebrafish and mouse embryonic stem cells. Development. 2006;133:3641–3650. doi: 10.1242/dev.02540. [DOI] [PubMed] [Google Scholar]
- 9.Woo AJ, Moran TB, Schindler YL, et al. Identification of ZBP-89 as a novel GATA-1-associated transcription factor involved in megakaryocytic and erythroid development. Molecular and cellular biology. 2008;28:2675–2689. doi: 10.1128/MCB.01945-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Just U, Stocking C, Spooncer E, et al. Expression of the GM-CSF gene after retroviral transfer in hematopoietic stem cell lines induces synchronous granulocyte-macrophage differentiation. Cell. 1991;64:1163–1173. doi: 10.1016/0092-8674(91)90271-y. [DOI] [PubMed] [Google Scholar]
- 11.Essien BE, Grasberger H, Romain RD, et al. ZBP-89 Regulates Expression of Tryptophan Hydroxylase I and Mucosal Defense Against Salmonella Typhimurium in Mice. Gastroenterology. 2013;144:1466–1477. e1469. doi: 10.1053/j.gastro.2013.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohneda K, Ohmori S, Ishijima Y, et al. Characterization of a functional ZBP-89 binding site that mediates Gata1 gene expression during hematopoietic development. The Journal of biological chemistry. 2009;284:30187–30199. doi: 10.1074/jbc.M109.026948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurumurthy S, Xie SZ, Alagesan B, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Socolovsky M, Nam H, Fleming MD, et al. Ineffective erythropoiesis in Stat5a(−/−) 5b(−/−) mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Zhang J, Ginzburg Y, et al. Quantitative analysis of murine terminal erythroid differentiation in vivo: novel method to study normal and disordered erythropoiesis. Blood. 2013;121:e43–49. doi: 10.1182/blood-2012-09-456079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez S, Liu J, Javed A, et al. The vitamin D response element in the distal osteocalcin promoter contributes to chromatin organization of the proximal regulatory domain. The Journal of biological chemistry. 2004;279:43581–43588. doi: 10.1074/jbc.M408335200. [DOI] [PubMed] [Google Scholar]
- 17.Okuno Y, Huang G, Rosenbauer F, et al. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Molecular and cellular biology. 2005;25:2832–2845. doi: 10.1128/MCB.25.7.2832-2845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bockamp EO, McLaughlin F, Murrell AM, et al. Lineage-restricted regulation of the murine SCL/TAL-1 promoter. Blood. 1995;86:1502–1514. [PubMed] [Google Scholar]
- 19.Kuhn R, Schwenk F, Aguet M, et al. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 20.Peschle C, Magli MC, Cillo C, et al. Kinetics of erythroid and myeloid stem cells in post-hypoxia polycythaemia. British journal of haematology. 1977;37:345–352. doi: 10.1111/j.1365-2141.1977.tb01005.x. [DOI] [PubMed] [Google Scholar]
- 21.Chenaille PJ, Steward SA, Ashmun RA, et al. Prolonged thrombocytosis in mice after 5-fluorouracil results from failure to down-regulate megakaryocyte concentration. An experimental model that dissociates regulation of megakaryocyte size and DNA content from megakaryocyte concentration. Blood. 1990;76:508–515. [PubMed] [Google Scholar]
- 22.Jordan CT, Lemischka IR. Clonal and systemic analysis of long-term hematopoiesis in the mouse. Genes & development. 1990;4:220–232. doi: 10.1101/gad.4.2.220. [DOI] [PubMed] [Google Scholar]
- 23.Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 24.Ford AM, Healy LE, Bennett CA, et al. Multilineage phenotypes of interleukin-3-dependent progenitor cells. Blood. 1992;79:1962–1971. [PubMed] [Google Scholar]
- 25.Iwasaki H, Somoza C, Shigematsu H, et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106:1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leddin M, Perrod C, Hoogenkamp M, et al. Two distinct auto-regulatory loops operate at the PU.1 locus in B cells and myeloid cells. Blood. 2011;117:2827–2838. doi: 10.1182/blood-2010-08-302976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cartharius K, Frech K, Grote K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 28.Begley CG. The SCL transcription factor and differential regulation of macrophage differentiation by LIF, OSM and IL-6. Stem Cells. 1994;12 (Suppl 1):143–149. discussion 149–151. [PubMed] [Google Scholar]
- 29.Bockamp EO, Fordham JL, Gottgens B, et al. Transcriptional regulation of the stem cell leukemia gene by PU.1 and Elf-1. The Journal of biological chemistry. 1998;273:29032–29042. doi: 10.1074/jbc.273.44.29032. [DOI] [PubMed] [Google Scholar]
- 30.Gutierrez L, Tsukamoto S, Suzuki M, et al. Ablation of Gata1 in adult mice results in aplastic crisis, revealing its essential role in steady-state and stress erythropoiesis. Blood. 2008;111:4375–4385. doi: 10.1182/blood-2007-09-115121. [DOI] [PubMed] [Google Scholar]
- 31.Walsh JC, DeKoter RP, Lee HJ, et al. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17:665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- 32.Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes & development. 1998;12:2403–2412. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Law DJ, Du M, Law GL, et al. ZBP-99 defines a conserved family of transcription factors and regulates ornithine decarboxylase gene expression. Biochemical and biophysical research communications. 1999;262:113–120. doi: 10.1006/bbrc.1999.1180. [DOI] [PubMed] [Google Scholar]
- 34.Mide SM, Huygens P, Bozzini CE, et al. Effects of human recombinant erythropoietin on differentiation and distribution of erythroid progenitor cells on murine medullary and splenic erythropoiesis during hypoxia and post-hypoxia. In Vivo. 2001;15:125–132. [PubMed] [Google Scholar]
- 35.Broudy VC, Lin NL, Priestley GV, et al. Interaction of stem cell factor and its receptor c-kit mediates lodgment and acute expansion of hematopoietic cells in the murine spleen. Blood. 1996;88:75–81. [PubMed] [Google Scholar]
- 36.Bauer A, Tronche F, Wessely O, et al. The glucocorticoid receptor is required for stress erythropoiesis. Genes & development. 1999;13:2996–3002. doi: 10.1101/gad.13.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lenox LE, Perry JM, Paulson RF. BMP4 and Madh5 regulate the erythroid response to acute anemia. Blood. 2005;105:2741–2748. doi: 10.1182/blood-2004-02-0703. [DOI] [PubMed] [Google Scholar]
- 38.Hall MA, Slater NJ, Begley CG, et al. Functional but abnormal adult erythropoiesis in the absence of the stem cell leukemia gene. Molecular and cellular biology. 2005;25:6355–6362. doi: 10.1128/MCB.25.15.6355-6362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrison DE, Astle CM, Delaittre JA. Loss of proliferative capacity in immunohemopoietic stem cells caused by serial transplantation rather than aging. The Journal of experimental medicine. 1978;147:1526–1531. doi: 10.1084/jem.147.5.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 41.Xie H, Ye M, Feng R, et al. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Silencing of ZBP-89 in MEL cells. Wild type MEL cells were transfected with five shRNAs targeting different loci in murine ZBP-89. (A) Schematic diagram of the mouse ZBP-89 shRNA clones (H1–H5) targeting different coding regions of ZBP-89. (B) Western blots of cell lysates from clones infected with various shRNAs and probed with anti-ZBP-89 or anti-actin antibodies (for loading control). Maximal knockdown of mouse ZBP-89 expression was achieved in ZBP-89 clone H5.
Supplemental Figure 2. Effects of conditional loss of ZBP-89 on hematopoietic colony formation in vitro. Colony counts were performed at 3 weeks (A) and on 2 months (B) post pIpC-treated bone marrow cells. (C) Colony counts performed on day 5 following PHZ treatment. Histograms show mean + SD of four (A, B) or three (C) independent experiments each carried out in duplicate. BFU-E, blast forming units erythroid; CFU-GEMM, colony forming units granulocyte-erythroid--macrophagemegakaryocyte colonies; CFU-GM, granulocyte-monocyte colonies; CFU-M, monocyte colonies; CFU-G, granulocyte colonies. * P value < 0.05; ** P value < 0.01; ns, not significant.
Supplemental Figure 3. Erythroblast populations in BM and spleen of ZBP-89 CKO mice. (A) Gating of bone marrow and spleen erythroblasts of a Control and a ZBP-89 CKO mice on 5 day after PHZ injection; CD44 is plotted versus FSC of TER119+ cells. Gated erythroblast populations are: I, Pro (CD44 hi Ter119 Low); II, Basophilic (Baso); III, Polychromatic (Poly); IV, Orthochromatic (Ortho); V, Reticulocytes; VI, RBC. (B) The proportion of cells at each stage of erythroblast development form Control and ZBP-89 CKO (3 mice in each group) is normalized based on total nucleated erythroblasts as 100%. Values represent mean + SD.
Supplemental Figure 4. Effect of stress erythropoiesis on spleen size in ZBP-89 CKO mice. Spleen weights/body weight (g) of Control (Cre- zbp89fl/fl) and CKO mice on day 5 post PHZ-induced hemolysis. Histograms represent mean + SD with three mice used in each group. * P< 0.05.
Supplemental Figure 5. Changes in peripheral blood erythrocytes, hemoglobin and hematocrit following PHZ-induced hemolysis. Results shown represent mean ± SD from 7 Control or ZBP-89 CKO mice. * P< 0.05; **P<0.01.