Abstract
Genetic variation in the leucine-rich repeat and Ig domain containing 1 gene (LINGO1) was recently associated with an increased risk of developing essential tremor (ET) and Parkinson disease (PD). Herein, we performed a comprehensive study of LINGO1 and its paralog LINGO2 in ET and PD by sequencing both genes in patients (ET, n=95; PD, n=96) and by examining haplotype-tagging single-nucleotide polymorphisms (tSNPs) in a multicenter North American series of patients (ET, n=1,247; PD, n=633) and controls (n=642). The sequencing study identified six novel coding variants in LINGO1 (p.S4C, p.V107M, p.A277T, p.R423R, p.G537A, p.D610D) and three in LINGO2 (p.D135D, p.P217P, p.V565V), however segregation analysis did not support pathogenicity. The association study employed 16 tSNPs at the LINGO1 locus and 21 at the LINGO2 locus. One variant in LINGO1 (rs9652490) displayed evidence of an association with ET (odds ratio (OR)=0.63; P=0.026) and PD (OR=0.54; P=0.016). Additionally, four other tSNPs in LINGO1 and one in LINGO2 were associated with ET and one tSNP in LINGO2 associated with PD (P<0.05). Further analysis identified one tSNP in LINGO1 and two in LINGO2 which influenced age at onset of ET and two tSNPs in LINGO1 which altered age at onset of PD (P<0.05). Our results support a role for LINGO1 and LINGO2 in determining risk for and perhaps age at onset of ET and PD. Further studies are warranted to confirm these findings and to determine the pathogenic mechanisms involved.
Keywords: Essential tremor, Parkinson disease, LINGO1, LINGO2, Genetic association
Introduction
Essential tremor (ET) and Parkinson disease (PD) are prevalent age-related movement disorders affecting about 3–6% (ET) and 1–2% of individuals over the age of 65 years (PD) [1–4]. While both ET and PD may cause significant motor impairment with tremor, they are regarded as distinct entities based on major differences at the clinical and pathological levels. ET patients display mostly symmetric action tremor which contrasts with asymmetric PD tremor that occurs at rest and is associated with bradykinesia, rigidity, and postural instability. Pathologically, some ET cases have Purkinje cell loss and Purkinje cell axonal dilations (torpedoes) in the cerebellum [5]. Some cases have α-synuclein immunopositive Lewy bodies (LB) in the brainstem [5]. Conversely, in PD there is severe neuronal loss in brainstem nuclei with abundant LB pathology [5, 6]. Despite these differences, clinical evidence indicates an overlap exists between ET and PD with a fourfold increased risk of PD in patients with ET, increased prevalence of ET in relatives of patients with PD and the presence of action tremor often preceding the onset of PD symptoms [7]. Furthermore, imaging studies found signs of dopaminergic deficiency in some ET patients and brainstem LB have been reported in ET cases [8].
The leucine-rich repeat and Ig domain containing 1 gene (LINGO1) has recently been associated with an increased risk of developing ET and PD, providing the first evidence of a genetic link between the two diseases [9]. LINGO1 is a central nervous system-specific component of the Nogo-66 receptor (NgR1)/p75/LINGO1 signaling complex implicated in inhibition of oligodendrocyte differentiation, axonal myelination and regeneration, and neuronal survival [10–16]. Expression of LINGO1 is increased after neuronal damage or cell death and its inhibition promotes functional recovery and axonal sprouting after spinal cord injury [10, 14, 17]. The expression of LINGO1 is higher in the substantia nigra of patients with PD compared to age-matched controls and increases in ventral midbrain neurons in animal models of PD after neurotoxic lesions [10]. Furthermore, reduction of LINGO1 activity was shown to improve survival, growth, and function of dopaminergic neurons both in primary cell cultures and in vivo experimental models of parkinsonism in rodents [10, 18]. These data highlight the functional relevance of LINGO1 as a regulator of neuronal death, which is consistent with LINGO1 variability altering the risk for ET and PD [9, 19, 20].
The leucine-rich repeat and Ig domain containing 2 gene (LINGO2) is a much less well characterized paralog of LINGO1. In contrast to the other LINGO1 paralogs (LINGO3 and LINGO4), LINGO2 expression is detectable in the mouse adult brain and appears to be restricted to neuronal tissue [21, 22]. We recently performed a genome-wide association study in a PD patient-control series that identified single-nucleotide polymorphisms (SNPs) in SNCA and LRRK2 associated with increased disease risk (unpublished findings). Although none of the SNPs in LINGO2 were found to associate with PD after correction for multiple testing, nominal significant P values were observed. Given the high degree of homology between the LINGO1 and LINGO2 proteins (61%), and recently reported association studies, both LINGO1 and its paralog LINGO2 are reasonable candidate genes for ET and PD.
In the present study, we examine the role of LINGO1 and LINGO2 in ET and PD by sequencing both genes in a series of patients with ET (n=95) and PD (n=96), and by performing association studies in ET and PD patient-control series (combined n=2,522) using tagging SNPs (tSNPs) that capture >95% of the genetic variability of LINGO1 and LINGO2. We identified ten rare coding variants (nine novel) in LINGO1 and LINGO2; three of them did not segregate with disease within families. However, we found evidence suggesting LINGO1 and LINGO2 variation influences risk for and onset age of ET and PD, expanding the scope of genetic factors common to both diseases.
Methods
Study population
A total of 1,247 patients with ET, 633 patients with PD, and 642 control subjects of Caucasian origin from North America were included in this study (Mayo Clinic Jacksonville: 150 ET, 438 PD, and 423 controls; Emory University: 214 ET, 195 PD, and 219 controls; Columbia University: 449 ET; Baylor College of Medicine: 228 ET; and University of Saskatchewan: 206 ET). The control groups consisted of unrelated individuals and spouses free of known neurological disease. Demographics for each group are given in Table 1. All patients were examined by a movement disorders neurologist and diagnosed with PD according to published criteria [23] or satisfied clinical criteria for definite or probable ET [24]. All sites obtained local ethics committee approval prior to subject enrollment. Individuals were informed of all aspects pertaining to their participation in the study and gave either written or proxy consent.
Table 1.
Demographic characteristics of patients and controls
| Controls | Essential tremor | Parkinson disease | |
|---|---|---|---|
| No. of patients | 642 | 1,247 | 633 |
| Age | 73±10 (33–101) | 67±15 (9–97) | 71±11 (30–92) |
| Male gender (n, %) | 310 (48%) | 530 (43%) | 359 (57%) |
| Age at disease onset | N/A | 50±20 (4–88) | 62±12 (16–85) |
The sample mean ± SD (range) is given for age and age at disease onset. Age at disease onset was only available in 396 ET and 423 PD cases
DNA sequencing of LINGO1 and LINGO2
Genomic DNA was extracted from peripheral blood lymphocytes using standard protocols. Primer pairs for LINGO1 and LINGO2 (available on request) were used to sequence all coding exons and exon–intron boundaries by polymerase chain reaction (PCR) in 95 randomly selected ET and 96 PD probands from the Mayo Clinic Jacksonville. PCR products were purified from unincorporated nucleotides using Agencourt bead technology (Beverly, MA, USA) with Biomek FX automation (Beckman Coulter, Fullerton, CA, USA). Sequence analysis was performed as previously described [25]. All novel variants were examined for disease segregation when possible in affected and unaffected family members by additional sequencing.
Genetic association analysis
The population frequency of six known coding variants with minor allele frequency (MAF) <10% and six novel LINGO1 and three LINGO2 variants was assessed in the case-control series. Selection of additional tSNPs was based on HapMap Phase II data using Haploview software [26]. The regions containing LINGO1 and LINGO2 exons from any reported transcript (±2.5 kb surrounding noncoding exons or ±10 kb for coding exons) were used for the selection of tSNPs. In total 16 tSNPs across LINGO1 and 21 across LINGO2 loci were selected to capture >95% of the polymorphic variation in these regions (MAF>5% and r2>0.8) in Caucasian population standards. Genotyping of tSNPs and of known and novel coding variants (MAF<10%), identified by sequencing, was performed on a Sequenom MassArray iPLEX platform (San Diego, CA, USA); all primer sequences are available on request. For each variant genotyping error was assessed by deviation from Hardy–Weinberg equilibrium expectation. All genotypes are given on the “+” strand.
Associations for PD and ET were evaluated using logistic regression models adjusted for age and gender; odds ratios (ORs) and 95% confidence intervals (CIs) were estimated. Single SNP associations with age at disease onset were examined using linear regression models adjusted for gender; regression coefficients and 95% CIs were estimated. Due to the alternate association findings in previous reports, both dominant (major allele homozygote vs minor allele homozygote and heterozygote) and recessive (minor allele homozygote vs major allele homozygote and heterozygote) models were considered in all regression analyses. P values<0.05 were considered significant, and no adjustment for multiple testing was performed in this exploratory study.
Results
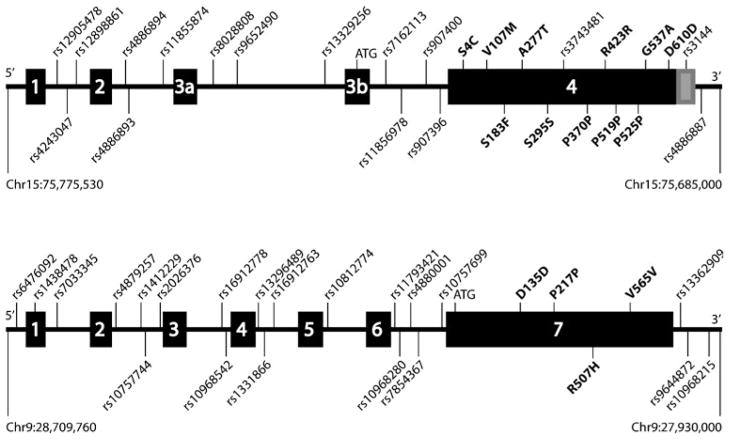
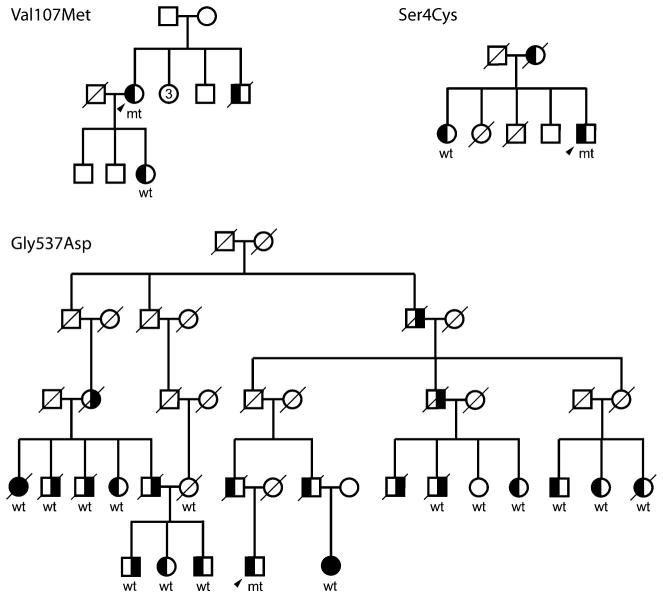
Sequencing analysis in 95 ET and 96 PD patients identified six novel coding variants in LINGO1 (Ser4Cys, Val107Met, Ala277Thr, Arg423Arg, Gly537Ala, and Asp610Asp) and three in LINGO2 (Asp135Asp, Pro217Pro, and Val565Val; Fig. 1, Table 2). In addition five known polymorphisms were detected in LINGO1 (rs2271398, rs2271397, rs2271396, rs3743481, and rs61737308), four of which with a MAF>10%. Three novel variants in LINGO1 (Ser4Cys, Val107Met, and Gly537Ala) did not segregate with disease within families (Fig. 2); two of these variants, Ser4Cys and Gly537Ala, as well as Ala277Thr, Arg423Arg, and Asp610Asp in LINGO1 and Pro217Pro and Arg507His in LINGO2 were observed exclusively in cases and not in controls (Table 2).
Fig. 1.
Schematic representation of the LINGO1 (top) and LINGO2 (bottom) genes indicating the exons (boxes) and the variants studied. Novel (above the genes) and known (below the genes) coding variants are given in bold. For LINGO1 exon arrangement was based on the two transcripts BC068558 and BC011057. The position of the start codon (ATG) was determined according to BC011057
Table 2.
Minor allelic counts and frequency for LINGO1 and LINGO2 coding variants
| rs/ss number | Amino acid change | Controls | Essential tremor | Parkinson disease |
|---|---|---|---|---|
| LINGO1 | ||||
| ss179321698 | S4C | 0 | 1 (0.04%) | 1 (0.08%) |
| ss179321700 | V107M | 1 (0.08%) | 1 (0.04%) | 0 |
| rs9855 | S183F | 0 | 0 | 0 |
| ss179321701 | A277T | 0 | 2 (0.08%) | 1 (0.08%) |
| rs34904447 | S295S | 0 | 0 | 0 |
| rs61737308 | P370P | 18 (1.40%) | 46 (1.84%) | 21 (1.66%) |
| ss179321703 | R423R | 0 | 1 (0.04%) | 0 |
| rs61737307 | P519P | 0 | 0 | 0 |
| rs11853548 | P525P | 0 | 0 | 0 |
| ss179321704 | G537A | 0 | 3 (0.12%) | 1 (0.08%) |
| ss179321705 | D610D | 0 | 2 (0.08%) | 0 |
| LINGO2 | ||||
| ss179321693 | D135D | 3 (0.23%) | 2 (0.08%) | 2 (0.16%) |
| ss179321694 | P217P | 0 | 1 (0.04%) | 0 |
| rs17506843 | R507H | 0 | 2 (0.08%) | 0 |
| ss179321696 | V565V | 1 (0.08%) | 1 (0.04%) | 1 (0.08%) |
Only those variants with a minor allele frequency over 10% were analyzed in this study
Fig. 2.
Segregation analysis of three novel coding LINGO1 variants in three pedigrees, representing males as squares, females as circles, whereas a number inside a symbol indicates the number of additional siblings. Patients with PD have right-half-dark-filled symbols, patients with ET have left-half-dark-filled symbols, deceased individuals are indicated with a diagonal line, and probands with an arrow head
Results of single SNP associations with ET and PD are presented in Table 3. Whereas the original report and one replication study identified the minor allele of rs9652490 associated with an increased risk of ET [19, 20], we previously identified association with ET and PD for the major allele [9]. The association study of LINGO1 tSNPs identified only the previously reported variant (rs9652490) being associated with both ET and PD under a recessive model (ET, OR=0.63, P=0.026; PD, OR=0.54, P=0.016). The risk allele found to be overrepresented in the disease groups was the major allele (T; genotype frequencies are provided in Supplemental Table 1). This association is consistent with our previous report [9] but in disagreement with other studies [19, 20]. The reasons for this alternate association are unclear, but several theoretical hypothesis have been proposed [27].
Table 3.
Associations between tagging SNPs in LINGO1 and LINGO2 with ET and PD
| Essential tremor (n =1247)
|
Parkinson disease (n=633)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| SNP (MA) | Dominant
|
Recessive
|
Dominant
|
Recessive
|
||||
| LINGO1 | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value |
| rs4886887 (A) | 0.91 (0.74, 1.13) | 0.41 | 1.83 (1.11, 3.01) | 0.018 | 0.95 (0.75, 1.21) | 0.70 | 1.41 (0.80, 2.49) | 0.23 |
| rs3144 (G) | 0.94 (0.77, 1.15) | 0.52 | 1.48 (1.05, 2.08) | 0.030 | 1.01 (0.80, 1.26) | 0.96 | 1.25 (0.85, 1.85) | 0.26 |
| rs3743481 (T) | 1.03 (0.84, 1.27) | 0.78 | 1.08 (0.82, 1.42) | 0.59 | 1.05 (0.83, 1.32) | 0.70 | 1.06 (0.78, 1.44) | 0.72 |
| rs907396 (C) | 1.03 (0.83, 1.27) | 0.80 | 1.04 (0.78, 1.41) | 0.78 | 1.00 (0.78, 1.27) | 0.98 | 0.95 (0.67, 1.33) | 0.74 |
| rs907400 (G) | 1.05 (0.84, 1.31) | 0.70 | 1.29 (0.64, 2.58) | 0.48 | 0.98 (0.76, 1.26) | 0.85 | 0.72 (0.30, 1.70) | 0.45 |
| rs11856978 (C) | 1.09 (0.85, 1.40) | 0.51 | 1.45 (0.56, 3.77) | 0.45 | 0.96 (0.72, 1.28) | 0.77 | 1.50 (0.52, 4.28) | 0.45 |
| rs7162113 (T) | 1.10 (0.90, 1.35) | 0.37 | 1.24 (0.77, 1.99) | 0.37 | 0.93 (0.74, 1.18) | 0.56 | 0.89 (0.51, 1.54) | 0.67 |
| rs13329256 (T) | 1.02 (0.76, 1.39) | 0.88 | 2.73 (0.31, 24.12) | 0.37 | 0.81 (0.57, 1.16) | 0.25 | 4.74 (0.52, 42.95) | 0.17 |
| rs9652490a (C) | 0.95 (0.77, 1.16) | 0.61 | 0.63 (0.42, 0.95) | 0.026 | 1.00 (0.79, 1.25) | 0.98 | 0.54 (0.33, 0.89) | 0.016 |
| rs8028808 (T) | 0.97 (0.78, 1.20) | 0.76 | 0.49 (0.29, 0.83) | 0.008 | 1.00 (0.78, 1.27) | 0.97 | 0.59 (0.32, 1.08) | 0.08 |
| rs11855874 (C) | 1.12 (0.89, 1.41) | 0.33 | 1.03 (0.49, 2.15) | 0.95 | 0.89 (0.68, 1.16) | 0.37 | 1.14 (0.52, 2.50) | 0.75 |
| rs4886893 (A) | 0.95 (0.78, 1.17) | 0.66 | 1.10 (0.63, 1.93) | 0.73 | 0.88 (0.70, 1.11) | 0.29 | 1.19 (0.65, 2.19) | 0.57 |
| rs4886894 (C) | 0.86 (0.70, 1.05) | 0.14 | 1.00 (0.70, 1.41) | 0.98 | 0.88 (0.70, 1.10) | 0.27 | 1.08 (0.74, 1.59) | 0.68 |
| rs12898861(A) | 1.15 (0.93, 1.43) | 0.20 | 1.14 (0.88, 1.47) | 0.32 | 1.01 (0.80, 1.29) | 0.91 | 1.24 (0.94, 1.64) | 0.13 |
| rs4243047 (A) | 0.95 (0.77, 1.16) | 0.59 | 0.85 (0.64, 1.14) | 0.27 | 0.90 (0.72, 1.13) | 0.37 | 0.81 (0.58, 1.12) | 0.20 |
| rs12905478 (G) | 0.88 (0.69, 1.13) | 0.31 | 0.36 (0.16, 0.86) | 0.021 | 1.00 (0.76, 1.31) | 0.97 | 0.76 (0.33, 1.73) | 0.51 |
| LINGO2 | ||||||||
| rs10968215 (A) | 1.02 (0.83, 1.25) | 0.87 | 1.14 (0.81, 1.61) | 0.45 | 0.97 (0.77, 1.23) | 0.81 | 0.88 (0.59, 1.32) | 0.53 |
| rs9644872 (C) | 1.19 (0.96, 1.47) | 0.12 | 1.02 (0.79, 1.32) | 0.89 | 0.96 (0.76, 1.22) | 0.75 | 0.91 (0.68, 1.21) | 0.51 |
| rs13362909 (A) | 1.37 (0.99, 1.90) | 0.06 | 1.84 (0.36, 9.25) | 0.46 | 1.10 (0.76, 1.60) | 0.62 | 2.76 (0.55, 14.01) | 0.22 |
| rs10757699 (C) | 1.07 (0.87, 1.31) | 0.55 | 0.97 (0.73, 1.28) | 0.83 | 0.88 (0.70, 1.11) | 0.30 | 0.81 (0.59, 1.12) | 0.21 |
| rs7854367 (A) | 0.84 (0.66, 1.07) | 0.15 | 1.27 (0.74, 2.20) | 0.39 | 0.97 (0.74, 1.26) | 0.79 | 1.57 (0.88, 2.80) | 0.13 |
| rs4880001 (G) | 1.03 (0.83, 1.27) | 0.79 | 1.12 (0.85, 1.47) | 0.43 | 1.04 (0.82, 1.32) | 0.76 | 0.91 (0.66, 1.25) | 0.56 |
| rs10968280 (T) | 0.82 (0.64, 1.04) | 0.11 | 1.85 (0.72, 4.73) | 0.20 | 0.73 (0.55, 0.97) | 0.029 | 1.28 (0.42, 3.87) | 0.66 |
| rs11793421 (G) | 0.92 (0.72, 1.16) | 0.47 | 0.94 (0.39, 2.29) | 0.90 | 1.24 (0.96, 1.61) | 0.10 | 1.97 (0.82, 4.70) | 0.13 |
| rs10812774 (A) | 0.92 (0.73, 1.16) | 0.46 | 0.90 (0.70, 1.16) | 0.40 | 0.89 (0.69, 1.15) | 0.37 | 0.95 (0.72, 1.26) | 0.71 |
| rs16912763 (A) | 0.82 (0.64, 1.06) | 0.12 | 1.31 (0.50, 3.44) | 0.59 | 1.06 (0.80, 1.40) | 0.68 | 0.99 (0.31, 3.10) | 0.98 |
| rs1331866 (T) | 1.08 (0.85, 1.36) | 0.55 | 1.37 (0.59, 3.17) | 0.46 | 0.83 (0.63, 1.09) | 0.18 | 1.41 (0.56, 3.56) | 0.47 |
| rs13296489 (C) | 1.03 (0.81, 1.30) | 0.81 | 0.82 (0.64, 1.05) | 0.12 | 0.95 (0.73, 1.23) | 0.67 | 0.97 (0.74, 1.27) | 0.81 |
| rs10968542 (A) | 1.00 (0.81, 1.24) | 1.00 | 0.95 (0.73, 1.23) | 0.70 | 0.99 (0.78, 1.26) | 0.94 | 0.99 (0.74, 1.33) | 0.95 |
| rs16912778 (G) | 1.22 (0.99, 1.51) | 0.06 | 1.30 (0.99, 1.72) | 0.06 | 0.96 (0.76, 1.22) | 0.76 | 1.25 (0.92, 1.71) | 0.15 |
| rs2026376 (T) | 1.12 (0.84, 1.51) | 0.44 | 0.49 (0.17, 1.47) | 0.21 | 1.03 (0.74, 1.44) | 0.86 | 0.45 (0.12, 1.77) | 0.25 |
| rs10757744 (C) | 1.05 (0.82, 1.33) | 0.72 | 0.65 (0.32, 1.34) | 0.24 | 0.99 (0.76, 1.30) | 0.94 | 0.81 (0.37, 1.77) | 0.59 |
| rs1412229 (T) | 1.08 (0.86, 1.35) | 0.52 | 0.72 (0.55, 0.94) | 0.015 | 0.81 (0.63, 1.03) | 0.09 | 0.87 (0.65, 1.17) | 0.35 |
| rs4879257 (T) | 0.90 (0.73, 1.10) | 0.30 | 0.73 (0.48, 1.11) | 0.14 | 1.09 (0.86, 1.38) | 0.47 | 0.78 (0.48, 1.26) | 0.30 |
| rs7033345 (G) | 0.85 (0.70, 1.04) | 0.12 | 0.76 (0.52, 1.12) | 0.17 | 1.06 (0.85, 1.33) | 0.61 | 0.83 (0.54, 1.28) | 0.39 |
| rs1438478 (C) | 0.81 (0.63, 1.04) | 0.10 | 0.94 (0.44, 2.02) | 0.87 | 0.99 (0.75, 1.30) | 0.93 | 0.73 (0.29, 1.84) | 0.50 |
| rs6476092 (G) | 0.91 (0.74, 1.12) | 0.39 | 0.71 (0.45, 1.11) | 0.13 | 1.09 (0.87, 1.38) | 0.45 | 0.72 (0.43, 1.21) | 0.21 |
Odd ratios (OR), 95% confident intervals (CI), and P values were obtained from logistic regression models adjusted for age and gender MA, minor allele
Includes 428 control subjects, 353 ET, and 426 PD patients previously reported [9]
Additional associations with ET were identified for rs4886887, rs3144, rs8028808, and rs12905478 (Table 3), spanning the entire LINGO1 gene (Fig. 1). Similarly to the previously described association with rs9652490, the major alleles of rs8028808 and rs12905478 were overrepresented in cases resulting in protective ORs for the minor alleles (OR=0.49 and 0.36, respectively). However, for rs4883887 and rs3144, the associations were driven by the minor alleles (OR=1.83 and 1.48, respectively). No additional tSNPs in LINGO1 were found to be significantly associated with PD. The analysis of LINGO2 tSNPs in ET resulted in only one variant (rs1412229) being associated with disease under a recessive model (OR=0.72, P=0.015). Similar results were obtained for rs10968280 and PD under a dominant model (OR=0.73, P=0.029; Table 3).
One variant in LINGO1 (rs907396) was associated with a 5-year younger mean age at ET onset (P=0.019). Association with the age of PD onset identified two variants conferring a later age at onset by approximately 5 years when the minor allele was present in homozygote form (rs4886887, P=0.047; rs3144, P=0.024; Supplemental Table 2). An earlier age at onset for ET by 4 to 5 years was also observed for two variants in LINGO2 (rs10812774 and rs7033345). In contrast, none of the variants in LINGO2 were significantly associated with age at onset of PD, however a trend toward an association (0.05≤P≤0.07) was observed for four variants (rs9644872, rs11793421, rs4879257, and rs6476092; Supplemental Table 2).
Discussion
Progress in the field of neurodegenerative disorders has highlighted the interplay of combined genetic factors in determining risk for complex traits. While variability in several genes may influence the risk for developing one disease, single genes often affect the risk for more than one trait. This diversity is best exemplified by variability in the tau (MAPT) and α-synuclein (SNCA) genes which alters risk of PD (MAPT and SNCA), progressive supranuclear palsy and corticobasal degeneration (MAPT), and multiple system atrophy (SNCA) [28–31]. However, while a growing number of genes have been implicated in both sporadic and familial PD, genetic factors in ET have remained elusive [9, 32]. The LINGO1 SNP rs9652490 was recently shown to associate with ET, a finding that was replicated independently and extended to PD [9, 19, 20]. Taken together with the established role of LINGO1 in neuronal survival and the preliminary evidence implicating LINGO2 in PD, these data support LINGO1 and LINGO2 as candidate genes for ET and PD. In the present study, we examined this hypothesis by performing a comprehensive evaluation of both genes in a multicenter series of North American patients with ET and PD and in control subjects. The sequencing effort identified six novel coding mutations in LINGO1 and three in LINGO2. However, three of these variants did not display segregation with disease in three families including a multi-incident kindred with PD and ET. Identification of additional families will be required to examine segregation and assess pathogenicity of the other six novel variants. Although all nine novel variants were rare (MAF≤0.16%), six of them were found only in patients and not in control subjects, indicating a possible role in pathogenesis that warrants further studies. Interestingly, three of the six variants found only in patients were identified in both ET and in PD, which supports the notion that genetic factors may influence both diseases simultaneously.
The association study using tSNPs identified one variant in LINGO1 which alters the risk for both ET and PD (rs9652490), consistent with our previous report [9]. Interestingly, there is a discrepancy between the results of our two studies and those of others in which the association with disease was driven by the minor allele of rs9652490 [19, 20]. Possible explanations include population-specific differences, although the largest series used in the replication part of the initial report was ethnically similar to our patient-control series (US Caucasians) [19]. Four additional variants in LINGO1 and one in LINGO2 influenced the risk of ET, and one SNP in LINGO2 altered the risk of PD. Furthermore, five variants in LINGO1 and LINGO2 had an effect on age at onset in ET (three variants) and PD (two variants). Despite the fact that these associations would not have withstood adjustment for the number of statistical tests performed and should therefore be considered exploratory, several lines of evidence are supportive including that: (1) LINGO1 is biologically plausible as a candidate gene for neurodegenerative disease; (2) this is the fourth study by three independent groups in Caucasian and Asian populations that consistently nominate variants in LINGO1 as a risk factor for developing ET; and (3) variants which display evidence of association with disease span the entire LINGO1 gene. Factors contributing to our results not withstanding correction for multiple testing may include diagnostic inaccuracy (known to occur in both ET and PD), or the use of genetically heterogeneous, admixed North American populations. Power is unlikely to have played a major role as our combined ET series is the largest studied so far and our PD population is adequately powered to detect associations within the range of expected magnitude. Assuming a disease prevalence of 1%, an allele frequency of 0.25, and an OR of 2, we have >99% power to detect a dominant and 84% for a recessive association in our ET case-control study and a >99% dominant and 72% recessive association in PD.
A better molecular understanding of the pathogenesis for two prevalent movement disorders (ET and PD) will play a significant part in designing future therapeutic strategies aimed at prevention and cure. The results or our study support a role for LINGO1 and LINGO2 in determining risk for and onset age of ET and PD. Further replication studies on large and ethnically diverse populations are warranted to confirm these findings and to pave the way for the functional work that will unravel the pathogenic mechanisms involved.
Supplementary Material
Acknowledgments
The authors wish to thank the patients and families who participated in the study. This work was supported by the Parkinson’s Disease Foundation (International Research Project Grant awarded to CW), the Morris K. Udall Center, National Institute of Neurological Disorders and Stroke P50 NS40256 and The Mayo Foundation Research Committee, Essential Tremor: Clinical and Molecular Genetic Studies (CR program). CW was also supported by the Swiss National Science Foundation/FSBMB (PASMP3-123268/1). ZKW is also partially funded by P01 AG017216, R01 NS057567, R01 AG015866 and CIHR 121849. EDL was funded by R01 NS042859 and R01 NS039422. MJF and ZKW are also partially funded by P01 AG017216 and the Pacific Alzheimer Research Foundation.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10048-010-0241-x) contains supplementary material, which is available to authorized users.
Contributor Information
Carles Vilariño-Güell, Department of Neuroscience, Mayo Clinic, 4500 San Pablo Road, Jacksonville, FL 32224, USA.
Christian Wider, Department of Neuroscience, Mayo Clinic, 4500 San Pablo Road, Jacksonville, FL 32224, USA. Department of Neurology, Centre Hospitalier Universitaire Vaudois (CHUV), Lausanne, Switzerland.
Owen A. Ross, Department of Neuroscience, Mayo Clinic, 4500 San Pablo Road, Jacksonville, FL 32224, USA
Barbara Jasinska-Myga, Department of Neuroscience, Mayo Clinic, 4500 San Pablo Road, Jacksonville, FL 32224, USA. Department of Neurology, Medical University of Silesia, Katowice, Poland.
Jennifer Kachergus, Department of Neuroscience, Mayo Clinic, 4500 San Pablo Road, Jacksonville, FL 32224, USA.
Stephanie A. Cobb, Department of Neuroscience, Mayo Clinic, 4500 San Pablo Road, Jacksonville, FL 32224, USA
Alexandra I. Soto-Ortolaza, Department of Neuroscience, Mayo Clinic, 4500 San Pablo Road, Jacksonville, FL 32224, USA
Bahareh Behrouz, Department of Neuroscience, Mayo Clinic, 4500 San Pablo Road, Jacksonville, FL 32224, USA.
Michael G. Heckman, Biostatistics Unit, Mayo Clinic, Jacksonville, FL, USA
Nancy N. Diehl, Biostatistics Unit, Mayo Clinic, Jacksonville, FL, USA
Claudia M. Testa, Emory Department of Neurology and Center for Neurodegenerative Diseases, Whitehead Biomedical Research Building, Atlanta, GA, USA
Zbigniew K. Wszolek, Department of Neurology, Mayo Clinic, Jacksonville, FL, USA
Ryan J. Uitti, Department of Neurology, Mayo Clinic, Jacksonville, FL, USA
Joseph Jankovic, Department of Neurology, Parkinson’s Disease Center and Movement Disorders Clinic, Baylor College of Medicine, Houston, TX, USA.
Elan D. Louis, GH Sergievsky Center, College of Physicians and Surgeons, Columbia University, New York, NY, USA. Department of Neurology, College of Physicians and Surgeons, Columbia University, New York, NY, USA. Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, NY, USA. Department of Epidemiology, Mailman School of Public Health, Columbia University, New York, NY, USA
Lorraine N. Clark, Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, College of Physicians and Surgeons, Columbia University, New York, NY, USA
Alex Rajput, Division of Neurology, Royal University Hospital, University of Saskatchewan, Saskatoon, SK, Canada.
Matthew J. Farrer, Email: Farrer.Matthew@mayo.edu, Department of Neuroscience, Mayo Clinic, 4500 San Pablo Road, Jacksonville, FL 32224, USA
References
- 1.de Rijk MC, Launer LJ, Berger K, Breteler MM, Dartigues JF, Baldereschi M, Fratiglioni L, Lobo A, Martinez-Lage J, Trenkwalder C, Hofman A. Prevalence of Parkinson’s disease in Europe: a collaborative study of population-based cohorts. Neurologic diseases in the elderly research group. Neurology. 2000;54:S21–S23. [PubMed] [Google Scholar]
- 2.Louis ED, Thawani SP, Andrews HF. Prevalence of essential tremor in a multiethnic, community-based study in northern Manhattan, New York, NY. Neuroepidemiology. 2009;32:208–214. doi: 10.1159/000195691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajput AH, Birdi S. Epidemiology of Parkinson’s disease. Parkinsonism Relat Disord. 1997;3:175–186. doi: 10.1016/s1353-8020(97)00029-1. [DOI] [PubMed] [Google Scholar]
- 4.Sur H, Ilhan S, Erdogan H, Ozturk E, Tasdemir M, Boru UT. Prevalence of essential tremor: a door-to-door survey in Sile, Istanbul, Turkey. Parkinsonism Relat Disord. 2009;15:101–104. doi: 10.1016/j.parkreldis.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Louis ED, Faust PL, Vonsattel JP, Honig LS, Rajput A, Robinson CA, Pahwa R, Lyons KE, Ross GW, Borden S, Moskowitz CB, Lawton A, Hernandez N. Neuropathological changes in essential tremor: 33 cases compared with 21 controls. Brain. 2007;130:3297–3307. doi: 10.1093/brain/awm266. [DOI] [PubMed] [Google Scholar]
- 6.Dickson DW. Alpha-synuclein and the Lewy body disorders. Curr Opin Neurol. 2001;14:423–432. doi: 10.1097/00019052-200108000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Benito-Leon J, Louis ED, Bermejo-Pareja F. Risk of incident Parkinson’s disease and parkinsonism in essential tremor: a population based study. J Neurol Neurosurg Psychiatry. 2009;80:423–425. doi: 10.1136/jnnp.2008.147223. [DOI] [PubMed] [Google Scholar]
- 8.Isaias IU, Canesi M, Benti R, Gerundini P, Cilia R, Pezzoli G, Antonini A. Striatal dopamine transporter abnormalities in patients with essential tremor. Nucl Med Commun. 2008;29:349–353. doi: 10.1097/MNM.0b013e3282f4d307. [DOI] [PubMed] [Google Scholar]
- 9.Vilarino-Guell C, Ross OA, Wider C, Jasinska-Myga B, Cobb SA, Soto-Ortolaza AI, Kachergus JM, Keeling BH, Dachsel JC, Melrose HL, Behrouz B, Wszolek ZK, Uitti RJ, Aasly JO, Rajput A, Farrer MJ. LINGO1 rs9652490 is associated with essential tremor and Parkinson disease. Parkinsonism Relat Disord. 2009 doi: 10.1016/j.parkreldis.2009.08.006. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inoue H, Lin L, Lee X, Shao Z, Mendes S, Snodgrass-Belt P, Sweigard H, Engber T, Pepinsky B, Yang L, Beal MF, Mi S, Isacson O. Inhibition of the leucine-rich repeat protein LINGO-1 enhances survival, structure, and function of dopaminergic neurons in Parkinson’s disease models. Proc Natl Acad Sci USA. 2007;104:14430–14435. doi: 10.1073/pnas.0700901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mi S, Lee X, Shao Z, Thill G, Ji B, Relton J, Levesque M, Allaire N, Perrin S, Sands B, Crowell T, Cate RL, McCoy JM, Pepinsky RB. LINGO-1 is a component of the Nogo-66 receptor/p75 signaling complex. Nat Neurosci. 2004;7:221–228. doi: 10.1038/nn1188. [DOI] [PubMed] [Google Scholar]
- 12.Mi S, Miller RH, Lee X, Scott ML, Shulag-Morskaya S, Shao Z, Chang J, Thill G, Levesque M, Zhang M, Hession C, Sah D, Trapp B, He Z, Jung V, McCoy JM, Pepinsky RB. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci. 2005;8:745–751. doi: 10.1038/nn1460. [DOI] [PubMed] [Google Scholar]
- 13.Li W, Walus L, Rabacchi SA, Jirik A, Chang E, Schauer J, Zheng BH, Benedetti NJ, Liu BP, Choi E, Worley D, Silvian L, Mo W, Mullen C, Yang W, Strittmatter SM, Sah DW, Pepinsky B, Lee DH. A neutralizing anti-Nogo66 receptor monoclonal antibody reverses inhibition of neurite outgrowth by central nervous system myelin. J Biol Chem. 2004;279:43780–43788. doi: 10.1074/jbc.M401803200. [DOI] [PubMed] [Google Scholar]
- 14.Mi S, Hu B, Hahm K, Luo Y, Kam Hui ES, Yuan Q, Wong WM, Wang L, Su H, Chu TH, Guo J, Zhang W, So KF, Pepinsky B, Shao Z, Graff C, Garber E, Jung V, Wu EX, Wu W. LINGO-1 antagonist promotes spinal cord remyelination and axonal integrity in MOG-induced experimental autoimmune encephalomyelitis. Nat Med. 2007;13:1228–1233. doi: 10.1038/nm1664. [DOI] [PubMed] [Google Scholar]
- 15.Shao Z, Browning JL, Lee X, Scott ML, Shulga-Morskaya S, Allaire N, Thill G, Levesque M, Sah D, McCoy JM, Murray B, Jung V, Pepinsky RB, Mi S. TAJ/TROY, an orphan TNF receptor family member, binds Nogo-66 receptor 1 and regulates axonal regeneration. Neuron. 2005;45:353–359. doi: 10.1016/j.neuron.2004.12.050. [DOI] [PubMed] [Google Scholar]
- 16.Carim-Todd L, Escarceller M, Estivill X, Sumoy L. LRRN6A/LERN1 (leucine-rich repeat neuronal protein 1), a novel gene with enriched expression in limbic system and neocortex. Eur J NeuroSci. 2003;18:3167–3182. doi: 10.1111/j.1460-9568.2003.03003.x. [DOI] [PubMed] [Google Scholar]
- 17.Ji B, Li M, Wu WT, Yick LW, Lee X, Shao Z, Wang J, So KF, McCoy JM, Pepinsky RB, Mi S, Relton JK. LINGO-1 antagonist promotes functional recovery and axonal sprouting after spinal cord injury. Mol Cell Neurosci. 2006;33:311–320. doi: 10.1016/j.mcn.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Mi S, Sandrock A, Miller RH. LINGO-1 and its role in CNS repair. Int J Biochem Cell Biol. 2008;40:1971–1978. doi: 10.1016/j.biocel.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 19.Stefansson H, Steinberg S, Petursson H, Gustafsson O, Gudjonsdottir IH, Jonsdottir GA, Palsson ST, Jonsson T, Saemundsdottir J, Bjornsdottir G, Bottcher Y, Thorlacius T, Haubenberger D, Zimprich A, Auff E, Hotzy C, Testa CM, Miyatake LA, Rosen AR, Kristleifsson K, Rye D, Asmus F, Schols L, Dichgans M, Jakobsson F, Benedikz J, Thorsteinsdottir U, Gulcher J, Kong A, Stefansson K. Variant in the sequence of the LINGO1 gene confers risk of essential tremor. Nat Genet. 2009;41:277–279. doi: 10.1038/ng.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan EK, Teo YY, Prakash KM, Li R, Lim HQ, Angeles D, Tan LC, Au WL, Yih Y, Zhao Y. LINGO1 variant increases risk of familial essential tremor. Neurology. 2009;73:1161–1162. doi: 10.1212/WNL.0b013e3181bacfc9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haines BP, Rigby PW. Expression of the Lingo/LERN gene family during mouse embryogenesis. Gene Expr Patterns. 2008;8:79–86. doi: 10.1016/j.modgep.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Homma S, Shimada T, Hikake T, Yaginuma H. Expression pattern of LRR and Ig domain-containing protein (LRRIG protein) in the early mouse embryo. Gene Expr Patterns. 2009;9:1–26. doi: 10.1016/j.gep.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Gelb DJ, Oliver E, Gilman S. Diagnostic criteria for Parkinson disease. Arch Neurol. 1999;56:33–39. doi: 10.1001/archneur.56.1.33. [DOI] [PubMed] [Google Scholar]
- 24.Louis ED, Ford B, Lee H, Andrews H, Cameron G. Diagnostic criteria for essential tremor: a population perspective. Arch Neurol. 1998;55:823–828. doi: 10.1001/archneur.55.6.823. [DOI] [PubMed] [Google Scholar]
- 25.Mata IF, Kachergus JM, Taylor JP, Lincoln S, Aasly J, Lynch T, Hulihan MM, Cobb SA, Wu RM, Lu CS, Lahoz C, Wszolek ZK, Farrer MJ. Lrrk2 pathogenic substitutions in Parkinson’s disease. Neurogenetics. 2005;6:171–177. doi: 10.1007/s10048-005-0005-1. [DOI] [PubMed] [Google Scholar]
- 26.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 27.Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. Am J Hum Genet. 2007;80:531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goris A, Williams-Gray CH, Clark GR, Foltynie T, Lewis SJ, Brown J, Ban M, Spillantini MG, Compston A, Burn DJ, Chinnery PF, Barker RA, Sawcer SJ. Tau and alpha-synuclein in susceptibility to, and dementia in, Parkinson’s disease. Ann Neurol. 2007;62:145–153. doi: 10.1002/ana.21192. [DOI] [PubMed] [Google Scholar]
- 29.Wider C, Vilarino-Guell C, Jasinska-Myga B, Heckman M, Soto-Ortolaza AI, Cobb SA, Aasly JO, Gibson JM, Lynch T, Uitti RJ, Wszolek ZK, Farrer MJ, Ross OA. Association of the MAPT locus with Parkinson’s disease. Eur J Neurol. 2009 doi: 10.1111/j.1468-1331.2009.02847.x. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peuralinna T, Oinas M, Polvikoski T, Paetau A, Sulkava R, Niinisto L, Kalimo H, Hernandez D, Hardy J, Singleton A, Tienari PJ, Myllykangas L. Neurofibrillary tau pathology modulated by genetic variation of alpha-synuclein. Ann Neurol. 2008;64:348–352. doi: 10.1002/ana.21446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scholz SW, Houlden H, Schulte C, Sharma M, Li A, Berg D, Melchers A, Paudel R, Gibbs JR, Simon-Sanchez J, Paisan-Ruiz C, Bras J, Ding J, Chen H, Traynor BJ, Arepalli S, Zonozi RR, Revesz T, Holton J, Wood N, Lees A, Oertel W, Wullner U, Goldwurm S, Pellecchia MT, Illig T, Riess O, Fernandez HH, Rodriguez RL, Okun MS, Poewe W, Wenning GK, Hardy JA, Singleton AB, Gasser T. SNCA variants are associated with increased risk for multiple system atrophy. Ann Neurol. 2009;65:610–614. doi: 10.1002/ana.21685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farrer MJ. Genetics of Parkinson disease: paradigm shifts and future prospects. Nat Rev Genet. 2006;7:306–318. doi: 10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.