Abstract
A pentapeptide open reading frame equipped with a canonical ribosome-binding site is present in the Escherichia coli 23S rRNA. Overexpression of 23S rRNA fragments containing the mini-gene renders cells resistant to the ribosome-inhibiting antibiotic erythromycin. Mutations that change either the initiator or stop codons of the peptide mini-gene result in the loss of erythromycin resistance. Nonsense mutations in the mini-gene also abolish erythromycin resistance, which can be restored in the presence of the suppressor tRNA, thus proving that expression of the rRNA-encoded peptide is essential for the resistance phenotype. The ribosome appears to be the likely target of action of the rRNA-encoded pentapeptide, because in vitro translation of the peptide mini-gene decreases the inhibitory action of erythromycin on cell-free protein synthesis. Thus, the new mechanism of drug resistance reveals that in addition to the structural and functional role of rRNA in the ribosome, it may also have a peptide-coding function.
Full text
PDF
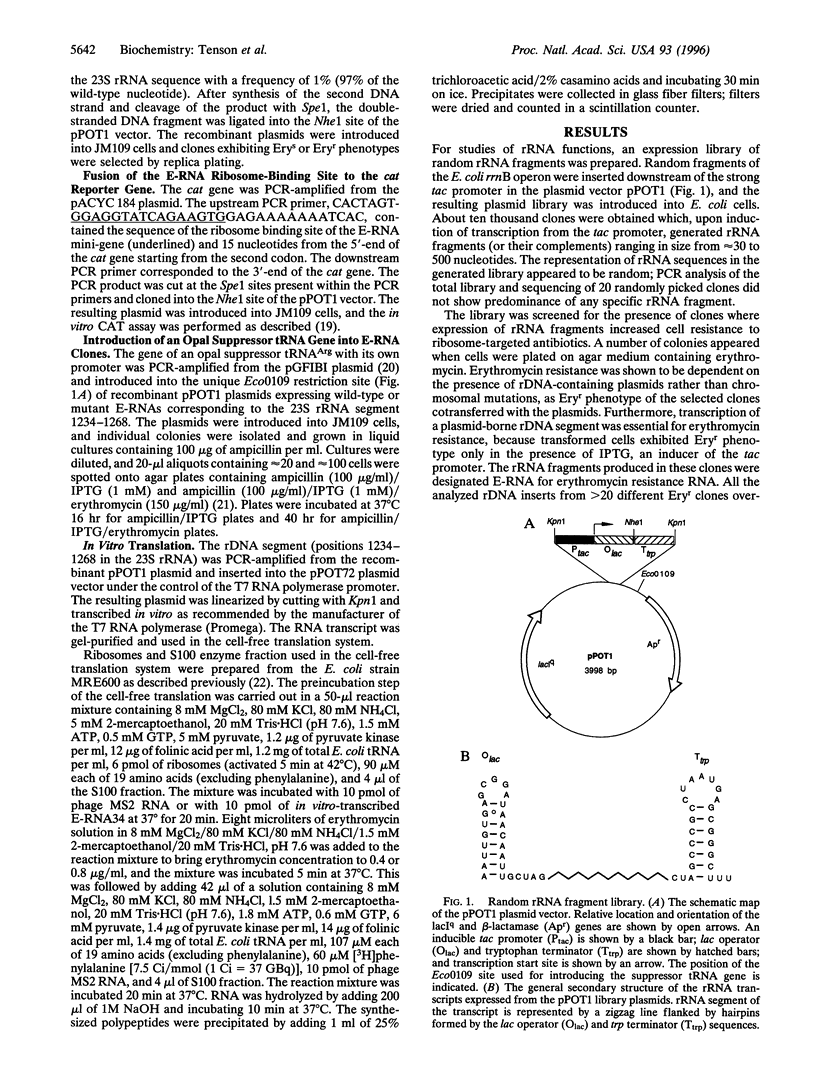
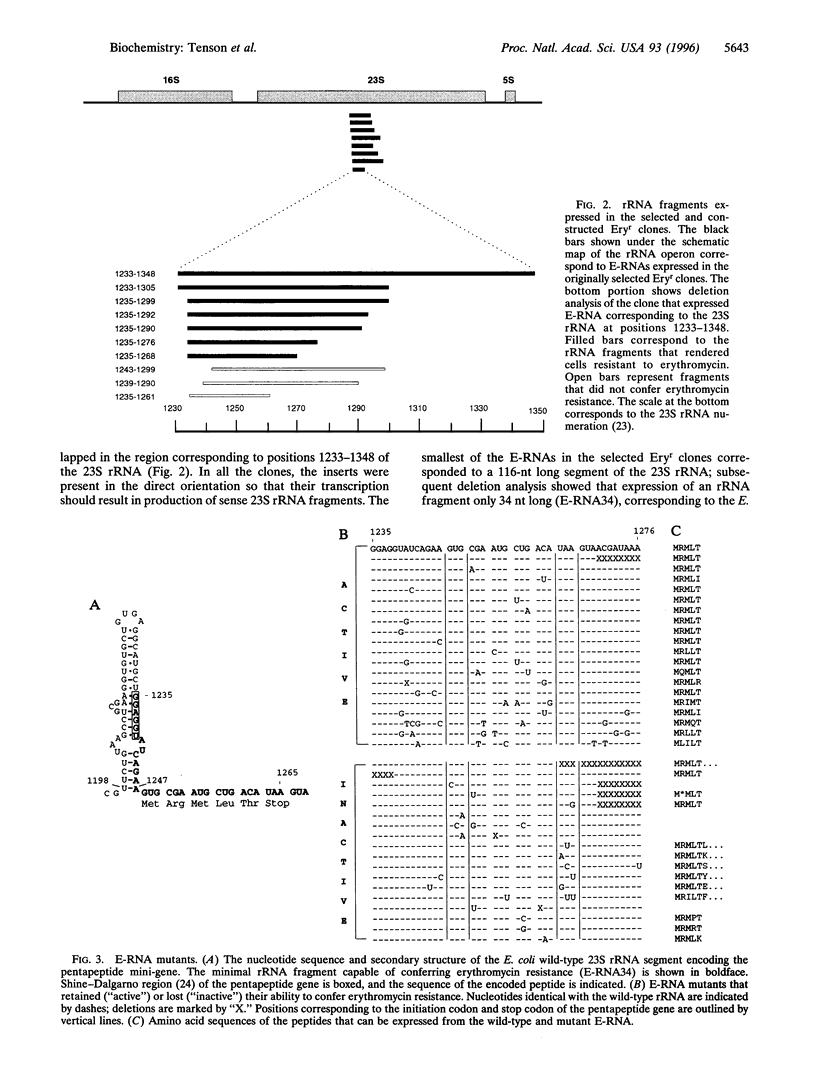
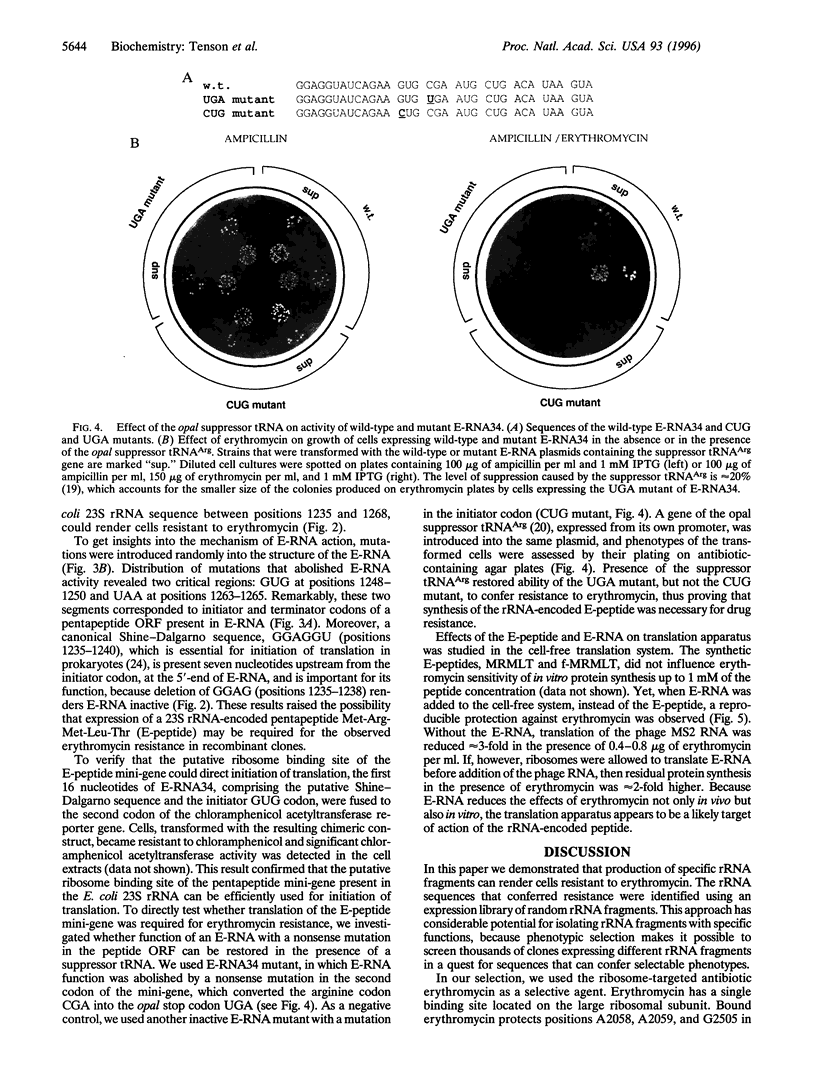
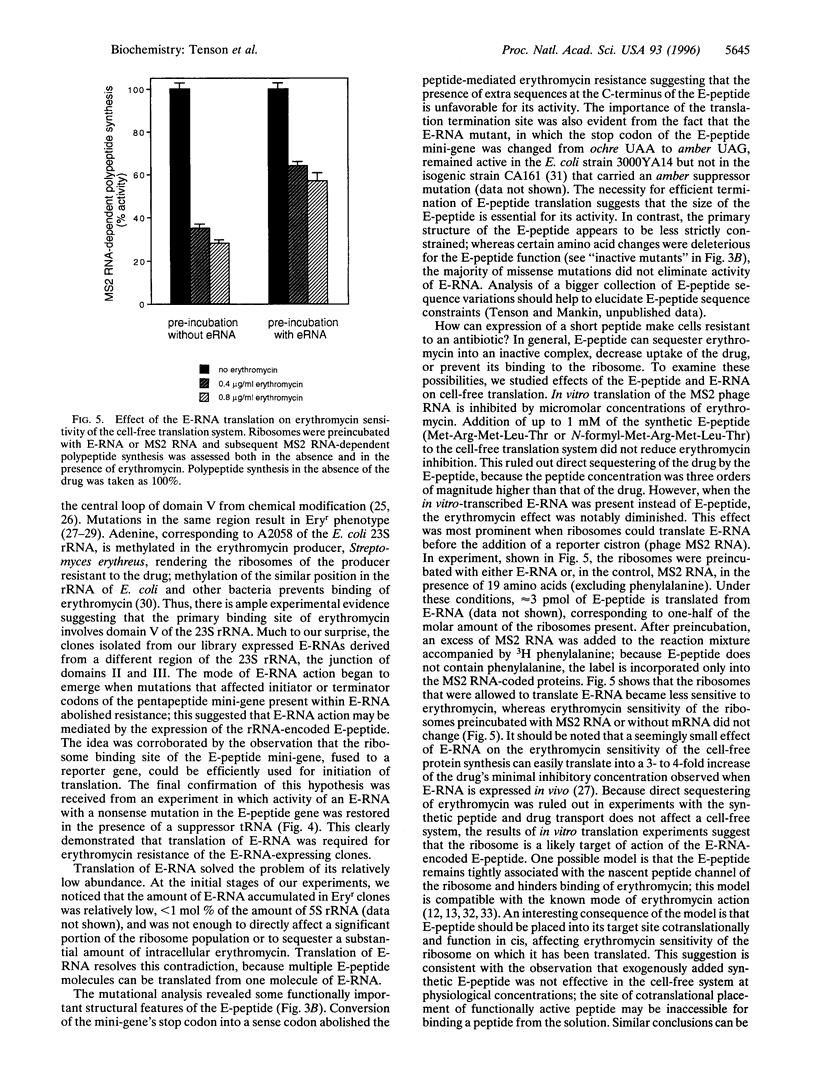

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersson S., Kurland C. G. Elongating ribosomes in vivo are refractory to erythromycin. Biochimie. 1987 Aug;69(8):901–904. doi: 10.1016/0300-9084(87)90218-5. [DOI] [PubMed] [Google Scholar]
- Berg K. L., Squires C. L., Squires C. In vivo translation of a region within the rrnB 16S rRNA gene of Escherichia coli. J Bacteriol. 1987 Apr;169(4):1691–1701. doi: 10.1128/jb.169.4.1691-1701.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Dam M., Douthwaite S., Tenson T., Mankin A. S. Mutations in domain II of 23 S rRNA facilitate translation of a 23 S rRNA-encoded pentapeptide conferring erythromycin resistance. J Mol Biol. 1996 May 31;259(1):1–6. doi: 10.1006/jmbi.1996.0296. [DOI] [PubMed] [Google Scholar]
- Douthwaite S., Aagaard C. Erythromycin binding is reduced in ribosomes with conformational alterations in the 23 S rRNA peptidyl transferase loop. J Mol Biol. 1993 Aug 5;232(3):725–731. doi: 10.1006/jmbi.1993.1426. [DOI] [PubMed] [Google Scholar]
- Douthwaite S., Prince J. B., Noller H. F. Evidence for functional interaction between domains II and V of 23S ribosomal RNA from an erythromycin-resistant mutant. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8330–8334. doi: 10.1073/pnas.82.24.8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emory S. A., Bouvet P., Belasco J. G. A 5'-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992 Jan;6(1):135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- Frankel A. D. Peptide models of the Tat-TAR protein-RNA interaction. Protein Sci. 1992 Dec;1(12):1539–1542. doi: 10.1002/pro.5560011202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geballe A. P., Morris D. R. Initiation codons within 5'-leaders of mRNAs as regulators of translation. Trends Biochem Sci. 1994 Apr;19(4):159–164. doi: 10.1016/0968-0004(94)90277-1. [DOI] [PubMed] [Google Scholar]
- Gu Z., Harrod R., Rogers E. J., Lovett P. S. Anti-peptidyl transferase leader peptides of attenuation-regulated chloramphenicol-resistance genes. Proc Natl Acad Sci U S A. 1994 Jun 7;91(12):5612–5616. doi: 10.1073/pnas.91.12.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Harrod R., Rogers E. J., Lovett P. S. Properties of a pentapeptide inhibitor of peptidyltransferase that is essential for cat gene regulation by translation attenuation. J Bacteriol. 1994 Oct;176(20):6238–6244. doi: 10.1128/jb.176.20.6238-6244.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmayer T. A., Pestov D. G., Roninson I. B. Isolation of dominant negative mutants and inhibitory antisense RNA sequences by expression selection of random DNA fragments. Nucleic Acids Res. 1992 Feb 25;20(4):711–717. doi: 10.1093/nar/20.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R., Moreno F. Genetics of ribosomally synthesized peptide antibiotics. Annu Rev Microbiol. 1992;46:141–163. doi: 10.1146/annurev.mi.46.100192.001041. [DOI] [PubMed] [Google Scholar]
- Lovett P. S. Nascent peptide regulation of translation. J Bacteriol. 1994 Nov;176(21):6415–6417. doi: 10.1128/jb.176.21.6415-6417.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M., Weisblum B. The ermC leader peptide: amino acid alterations leading to differential efficiency of induction by macrolide-lincosamide-streptogramin B antibiotics. J Bacteriol. 1990 Jul;172(7):3772–3779. doi: 10.1128/jb.172.7.3772-3779.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Foss K., Jenkins R. A., Schneider J. Nucleotides that determine Escherichia coli tRNA(Arg) and tRNA(Lys) acceptor identities revealed by analyses of mutant opal and amber suppressor tRNAs. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9260–9264. doi: 10.1073/pnas.87.23.9260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren R. S., Newbury S. F., Dance G. S., Causton H. C., Higgins C. F. mRNA degradation by processive 3'-5' exoribonucleases in vitro and the implications for prokaryotic mRNA decay in vivo. J Mol Biol. 1991 Sep 5;221(1):81–95. [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987 Aug;69(8):879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Kop J., Wheaton V., Brosius J., Gutell R. R., Kopylov A. M., Dohme F., Herr W., Stahl D. A., Gupta R. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 1981 Nov 25;9(22):6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Taylor J. D., Tchen T. T. Increased mitochondrial activities in pigmented (melanized) fish cells and nucleotide sequence of mitochondrial large rRNA. Biochem Biophys Res Commun. 1992 Nov 30;189(1):445–449. doi: 10.1016/0006-291x(92)91578-e. [DOI] [PubMed] [Google Scholar]
- Powers T., Noller H. F. Dominant lethal mutations in a conserved loop in 16S rRNA. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1042–1046. doi: 10.1073/pnas.87.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarma U., Remme J. Novel mutants of 23S RNA: characterization of functional properties. Nucleic Acids Res. 1992 Jun 25;20(12):3147–3152. doi: 10.1093/nar/20.12.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigmund C. D., Ettayebi M., Borden A., Morgan E. A. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 1988;164:673–690. doi: 10.1016/s0076-6879(88)64077-8. [DOI] [PubMed] [Google Scholar]
- Sigmund C. D., Ettayebi M., Morgan E. A. Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 1984 Jun 11;12(11):4653–4663. doi: 10.1093/nar/12.11.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigmund C. D., Morgan E. A. Erythromycin resistance due to a mutation in a ribosomal RNA operon of Escherichia coli. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5602–5606. doi: 10.1073/pnas.79.18.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer E. R., Beckwith J. R., Brenner S. Mapping of suppressor loci in Escherichia coli. J Mol Biol. 1965 Nov;14(1):153–166. doi: 10.1016/s0022-2836(65)80237-6. [DOI] [PubMed] [Google Scholar]
- Skinner R., Cundliffe E., Schmidt F. J. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J Biol Chem. 1983 Oct 25;258(20):12702–12706. [PubMed] [Google Scholar]
- Tenson T., Mankin A. Comparison of functional peptide encoded in the Escherichia coli 23S rRNA with other peptides involved in cis-regulation of translation. Biochem Cell Biol. 1995 Nov-Dec;73(11-12):1061–1070. doi: 10.1139/o95-114. [DOI] [PubMed] [Google Scholar]
- Vester B., Garrett R. A. A plasmid-coded and site-directed mutation in Escherichia coli 23S RNA that confers resistance to erythromycin: implications for the mechanism of action of erythromycin. Biochimie. 1987 Aug;69(8):891–900. doi: 10.1016/0300-9084(87)90217-3. [DOI] [PubMed] [Google Scholar]
- de Boer H. A., Comstock L. J., Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci U S A. 1983 Jan;80(1):21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]





