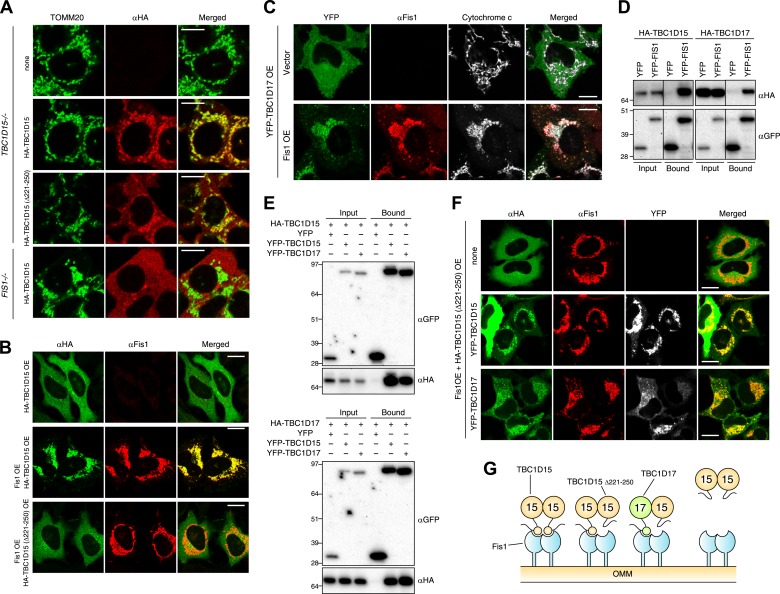
Figure 6. Identification of TBC1D17 as a Fis1 and TBC1D15 binding protein.
(A) TBC1D15−/− cells and those stably expressing HA-TBC1D15 WT and HA-TBC1D15 (Δ221-250), and FIS1−/− cells stably expressing HA-TBC1D15 WT were subjected to immunostaining with anti-TOMM20 and anti-HA antibodies. Scale bars, 10 μm. (B) HA-TBC1D15 WT or HA-TBC1D15 (Δ221-250) with or without Fis1 was transiently overexpressed (OE) in HeLa cells. Cells were subjected to immunostaining with anti-HA and anti-Fis1 antibodies. Scale bars, 20 μm. (C) YFP-TBC1D17 together with pcDNA vector or Fis1 was transiently overexpressed (OE) in HeLa cells. Cells were subjected to immunostaining with anti-Fis1 and anti-Cytochrome c antibodies. Scale bars, 20 μm. (D) YFP or YFP-Fis1 was co-overexpressed with HA-TBC1D15 or HA-TBC1D17 in HEK293 cells. The cell extracts were subjected to pull down assays with GFP-Trap. 5% input and bound fractions were analyzed by immunoblotting with anti-HA (upper panel) and anti-GFP (lower panel) antibodies. (E) YFP, YFP-TBC1D15, or YFP-TBC1D17 was co-overexpressed with HA-TBC1D15 (upper panel) or HA-TBC1D17 (lower panel) in HEK293 cells. The cell extracts were subjected to pull down assays with GFP-Trap. 5% input and bound fractions were analyzed by immunoblotting with anti-GFP and anti-HA antibodies. (F) HA-TBC1D15 (Δ221-250) together with Fis1 and YFP-TBC1D15 WT or YFP-TBC1D17 WT were transiently overexpressed (OE) in HeLa cells. Cells were subjected to immunostaining with anti-HA and anti-Fis1 antibodies. Images of HA and Fis1 staining were merged in the right panels. Scale bars, 20 μm. (G) Schematic model of Fis1, TBC1D15, and TBC1D17 binding. Homo- or hetero-dimer of TBC1D15 can interact with Fis1 dimer on the mitochondrial outer membrane (OMM).