Abstract
Background and Objectives
Neither the pathogenesis of port wine stain (PWS) birthmarks nor tissue effects of pulsed dye laser (PDL) treatment of these lesions is fully understood. There are few published reports utilizing gene expression analysis in human PWS skin. We aim to compare gene expression in PWS before and after PDL, using DNA microarrays that represent most, if not all, human genes to obtain comprehensive molecular profiles of PWS lesions and PDL-associated tissue effects.
Materials and Methods
Five human subjects had PDL treatment of their PWS. One week later, three biopsies were taken from each subject: normal skin (N); untreated PWS (PWS); PWS post-PDL (PWS + PDL). Samples included two lower extremity lesions, two facial lesions, and one facial nodule. High-quality total RNA isolated from skin biopsies was processed and applied to Affymetrix Human gene 1.0ST microarrays for gene expression analysis. We performed a 16 pair-wise comparison identifying either up- or down-regulated genes between N versus PWS and PWS versus PWS + PDL for four of the donor samples. The PWS nodule (nPWS) was analyzed separately.
Results
There was significant variation in gene expression profiles between individuals. By doing pair-wise comparisons between samples taken from the same donor, we were able to identify genes that may participate in the formation of PWS lesions and PDL tissue effects. Genes associated with immune, epidermal, and lipid metabolism were up-regulated in PWS skin. The nPWS exhibited more profound differences in gene expression than the rest of the samples, with significant differential expression of genes associated with angiogenesis, tumorigenesis, and inflammation.
Conclusion
In summary, gene expression profiles from N, PWS, and PWS + PDL demonstrated significant variation within samples from the same donor and between donors. By doing pair-wise comparisons between samples taken from the same donor and comparing these results between donors, we were able to identify genes that may participate in formation of PWS and PDL effects. Our preliminary results indicate changes in gene expression of angiogenesis-related genes, suggesting that dysregulation of angiogenic signals and/or components may contribute to PWS pathology.
Keywords: gene expression analysis, microarray, port wine stain, pulsed dye laser
INTRODUCTION
Each year 400,000 children are born with port wine stain (PWS) birthmarks worldwide [1]. They and their families are confronted with the psychological and physical consequences of these lesions. The pathogenesis of these lesions is unknown. Lasers, including the flash-lamp-pumped pulsed dye laser (PDL), utilize the principle of selective photothermolysis to target vascular lesions while sparing the epidermis and superficial blood vessels [2]. Laser is the mainstay of treatment for PWS, but most patients do not achieve complete removal even after undergoing many treatments [3]. While laser energy causes damage to targeted vessel walls resulting in lesion lightening, studies have demonstrated that vessels recur and/or new vessels develop as part of the normal wound healing response [4,5]. Our group has studied the presence of angiogenesis mediators following PDL utilizing immunohistochemistry (IHC) [6], and we are now utilizing gene expression analysis for evaluation. A comprehensive literature search shows that gene expression studies on human skin have been conducted, but this methodology has not been utilized to fully evaluate gene expression in PWS or to understand cutaneous effects of PDL treatment.
There are few published studies on the comparison of gene expression between vascular birthmarks and normal skin utilizing microarray analysis or quantitative Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR). A study evaluating fibroblasts (no other cell types) of Sturge–Weber syndrome (SWS) patients found fibronectin (FN1) gene and protein expression upregulated in PWS as compared to normal skin [7]. In a recent follow up study, the same group proposed that somatic mutations in fibroblasts derived from normal and PWS skin of patients with SWS may contribute to disease pathology; they used proteomic analysis of skin-derived fibroblasts from normal and SWS donors and identified small changes (ratios >1.2 and <0.8) in cell proliferation and oxidative stress responses in SWS-associated fibroblasts [8]. Infantile hemangiomas (IH) demonstrated upregulation in ANGPT 1 and 2, Homeobox (Hox) D3, HES/HEY genes (NOTCH receptors), insulin-like growth factor 2 (IGF2), and VEGFA [9–14]. A murine model showed that overexpression of AKT1 in endothelial cells resulted in the development of vascular malformations [15].
Mutations in the RASA1 gene have also been linked to vascular malformations, including hereditary malformations, arteriovenous fistulas, and Parkes–Weber syndrome, although mutations were found in only approximately 30% of capillary malformations tested [16–18].
In this study, we compare gene expression in complete skin samples taken from PWS before and after PDL, using DNA microarrays that represent most, if not all, human genes to obtain comprehensive molecular profiles of PWS lesions and PDL-associated tissue effects.
MATERIALS AND METHODS
The study was approved by the Institutional Review Board at the University of California, Irvine. Five human subjects with PWS participated in this study. Verbal and written informed consent was obtained for all subjects. Each subject had one area of their PWS treated with PDL (7 mm; 1.5 millisecond pulse duration; 9 J/cm2; cryogen spray cooling of 30 millisecond with a 30 millisecond delay). Clinically significant purpura was noted in treated areas for all subjects. The samples included two lower extremity lesions, two facial lesions, and one facial nodule (Table 1). At Day 7, 3 mm punch biopsies were performed on each subject to collect normal skin (N), an area of untreated PWS (PWS), and an area of PWS treated with PDL (PWS + PDL). For the facial nodule subject, three biopsies were performed at Day 7 on N, an untreated PWS nodule (nPWS), and a treated PWS nodule (nPWS + PDL). The N was taken from the corresponding area on the contralateral side of the body from the location of the PWS. Day 7 was the chosen date for measurement as previous literature showed peak levels of VEGF at Day 7 following incisional biopsies in human skin and following laser irradiation in a transgenic rodent model [19,20].
TABLE 1.
Location of PWS on Donors
| Subject number | Age | Gender | Location of PWS |
|---|---|---|---|
| 1 | 22 | Male | Lower extremity |
| 2 | 30 | Female | Lower extremity |
| 3 | 21 | Male | Facial |
| 4 | 43 | Male | Facial |
| 5 | 34 | Male | Facial (nodule) |
All of the collected tissue was immediately placed in RNAlater™ (Qiagen, Valencia, CA) and stored at −20°C prior to RNA isolation. Total RNA was extracted from the skin biopsy samples using the RNeasy purification kit (Qiagen) following manufacturer guidelines. The skin biopsies were homogenized in lysis buffer with β-mercaptoethanol using a FastPrep FP120A homogenizer (Qbiogen, Carlsbad, CA). A small aliquot of each RNA sample was quantified with a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE) and then monitored for integrity on a 2100 Bioanalyzer (Agilent, Santa Clara, CA).
The RNA integrity number (RIN) was the determinant of the suitability of RNA samples for further processing. The RINs of all our samples were greater than ≥7. Following quality assurance, the RNA samples were prepared at the UCI Genomics High Throughput Facility for microarray analysis using the standard Nugen protocol (Nugen, San Carlos, CA). Each sample of RNA was reverse transcribed to cDNA using the Ovation® Pico WTA System (Nugen). This cDNA was then converted into sense transcript cDNA (ST-cDNA) via the WT-Ovation™ Exon Module (Nugen) in preparation for fragmentation and labeling using the Encore™ Biotin Module (Nugen). Once labeled, targets were then hybridized to GeneChip® Human Gene 1.0ST microarrays (Affymetrix, Santa Clara, CA) for gene expression analysis; these microarrays interrogate over 28,000 known (full-length) and predicted (hypothetical) human genes.
Raw array expression data was summarized using Expression Console® Software (Affymetrix) to generate CHP data files and also Probe Logarithmic Intensity Error (PLIER) normalized for further analysis. All data sets passed quality control evaluation and were included in subsequent analyses. Patterns of gene expression varied significantly as judged by unsupervised hierarchical clustering and principal component analyses of gene expression data sets from samples within each sample class (N, PWS, PDL). To allow for this variation, we compared gene expression between N versus PWS, and PWS versus PWS + PDL samples from each donor separately to identify changes in gene expression between these conditions. Thus, for each individual subject, there were two comparison groups. In N versus PWS, the N sample data was set as the control reference point and the gene expression in PWS sample was evaluated based on its change from the baseline N sample. In PWS versus PWS + PDL, the PWS sample was set as the control reference point and the gene expression in PWS + PDL sample was evaluated based on its change from the baseline PWS sample.
Data sets from the four individual PWS donors were initially filtered to remove all probe sets with expression values ≤10 (below reliable levels for detection). The remaining probe sets were used to identify genes associated with PWS or PDL treatment by calculating ratios of expression values between PWS and N samples and PWS + PDL and PWS samples respectively and retaining probe sets with a ratio ≥2 for upregulated genes and ≤0.5 for down-regulated genes. Finally, the resulting gene lists were filtered to remove duplicates and “confounding” genes, defined as those encoding non-proteins, keratin-associate protein (KRTAP) genes, and non-annotated genes. The KRTAP genes were excluded because their expression is believed to be an artifact of the sampling site, being expressed exclusively in hair follicles, rather than associated with disease pathology.
Initially, data for Subject 5 (nPWS) was included in the above described analysis. However, the nPWS data demonstrated obvious differences compared to the other four subjects. This was noted by the lab personnel who were blinded assessors and were not aware of the clinical features of the samples. Due to the marked difference in gene expression profiles for Subject 5, these data were analyzed separately using the same strategy to compare: N versus nPWS and nPWS versus nPWS + PDL profiles.
Donor-specific gene lists from the four donors with standard PWS lesions were then compared to identify genes present in multiple subjects. Shared genes and genes with “co-ordinate” expression between both N versus PWS and PWS versus PWS + PDL were identified. If any gene is associated with the pathology of PWS, one would predict that its expression would be either up- or down-regulated compared to the normal sample. As the treatment is expected to resolve the pathology (partially or completely), then it is reasonable to predict that gene expression would exhibit the opposite trend after receiving treatment. Therefore, genes associated with pathogenesis that are up-regulated in N versus PWS might be down-regulated in PWS versus PWS + PDL, exhibiting a “co-ordinate” expression pattern. Finally, we also looked for changes in specific genes previously associated with vascular malformations, laser effects or angiogenesis, including AKT1, FN1, heat shock proteins (HSP), HES/ HEY transcription factors, hypoxia-inducible factor 1, alpha subunit (HIF1A), HOX, MMPs, RASA1, serine proteinase inhibitors (SERPIN), tissue inhibitors of MMPs (TIMPs), transforming growth factor beta (TGFB), VEGF, and VEGF receptors (VEGFR).
RESULTS
Differential gene expression of PWS was analyzed between samples of the same donor as well as between donors. While there was significant variation in gene expression between the four evaluated subjects with PWS, we were able to identify differentially expressed genes using our pairwise comparison approach. Table 2 shows the most represented functional classes of these differentially expressed genes and demonstrates that those associated with immune, epidermal, and lipid metabolism are up-regulated in PWS skin. Lipid metabolism related gene expression dropped significantly following PDL.
TABLE 2.
Representation of Functional Classes in Genes Exhibiting Changes in Expression Between PWS and Laser Treated Samples in Multiple Donors
| Gene function | N vs. PWS UP | N vs. PWS DOWN | PWS vs. PWS + PDL UP | PWS vs. PWS + PDL DOWN |
|---|---|---|---|---|
| Epidermal | 4 | 1 | 3 | 0 |
| Immune | 6 | 0 | 6 | 2 |
| Lipid metabolism | 4 | 0 | 1 | 7 |
| Signaling | 1 | 0 | 3 | 1 |
N, normal skin; PWS, port wine stain; PWS + PDL, port wine stain treated with pulsed dye laser; UP, up-regulated in PWS or PWS + PDL; DOWN, down-regulated in PWS or PWS + PDL.
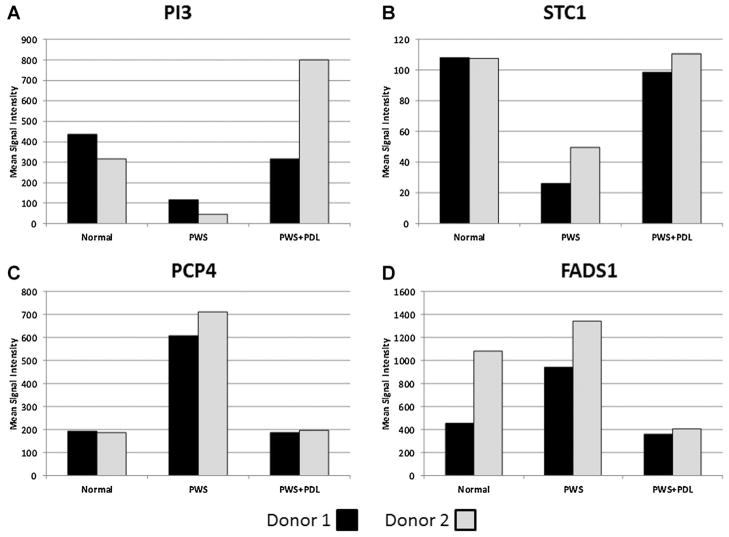
We then went on to find individual genes that exhibited “co-ordinate” regulation between N versus PWS and PWS versus PWS + PDL for Subjects 1–4 (Table 3). Peptidase inhibitor 3 (PI3), a skin-derived PI3, demonstrated the greatest differential expression between PWS and N, being down-regulated in PWS but highly upregulated following PDL. Stanniocalcin 1 (STC1) had a similar profile. Three genes were significantly elevated in PWS compared to N but reduced following PDL: purkinje cell protein 4 (PCP4), fatty acid desaturase 1 (FADS1), and, solute carrier family 45, member 4 (SLC45A4). Expression profiles for the top four genes listed in Table 3 are shown in Figure 1.
TABLE 3.
Genes That Exhibit Co-Ordinate Expression Between PWS and Laser Treated Lesions
| Gene name | Gene symbol | Subject | Ratio | Subject | Ratio | SET | Functional class |
|---|---|---|---|---|---|---|---|
| Peptidase inhibitor 3, skin-derived | PI3 | 2 | 6.85 | 1 | 3.69 | N vs. PWS DOWN | Anti-inflammatory |
| Peptidase inhibitor 3, skin-derived | PI3 | 2 | 17.41 | 1 | 2.69 | PWS vs. PWS + PDL UP | |
| Stanniocalcin 1 | STC1 | 1 | 4.14 | 2 | 2.16 | N vs. PWS DOWN | Metabolism |
| Stanniocalcin 1 | STC1 | 1 | 3.78 | 2 | 2.23 | PWS vs. PWS + PDL UP | |
| Purkinje cell protein 4 | PCP4 | 2 | 3.80 | 1 | 3.13 | N vs. PWS UP | Signaling |
| Purkinje cell protein 4 | PCP4 | 2 | 3.59 | 1 | 3.23 | PWS vs. PWS + PDL DOWN | |
| Fatty acid desaturase 1 | FADS1 | 1 | 4.69 | 2 | 2.09 | N vs. PWS UP | Lipid metabolism |
| Fatty acid desaturase 1 | FADS1 | 4 | 3.28 | 2 | 2.60 | PWS vs. PWS + PDL DOWN | |
| Solute carrier family 45, member 4 | SLC45A4 | 2 | 2.99 | 1 | 2.59 | N vs. PWS UP | Transporter |
| Solute carrier family 45, member 4 | SLC45A4 | 2 | 3.22 | 3 | 3.22 | PWS vs. PWS + PDL DOWN | |
| CD163 molecule | CD163 | 2 | 2.75 | 1 | 2.75 | N vs. PWS UP | Immune |
| CD163 molecule | CD163 | 2 | 2.74 | 4 | 2.50 | PWS vs. PWS + PDL UP |
N, normal skin; PWS, port wine stain; PWS + PDL, port wine stain treated with pulsed dye laser; UP, up-regulated in PWS or PWS + PDL; DOWN, down-regulated in PWS or PWS + PDL.
Fig. 1.
Expression profiles of genes exhibiting co-ordinate expression profiles between PWS and laser treated lesions. Affymetrix GeneChip data are shown as normalized average intensity values for each gene in pairs of subjects as follows: (A) Peptidase Inhibitor 3, Skin-Derived (PI3), (B) Stanniocalcin 1 (STC1), (C) Purkinje Cell Protein 4 (PCP4), (D) Fatty Acid Desaturase 1 (FADS1). Panels A–C; subjects 1 (black bar) and 2 (gray bar), panel D; subjects 2 (black bar) and 4 (gray bar).
Table 4 summarizes the angiogenesis-related genes with significant differential expression and their ratios in the individual subjects with PWS. In comparing N to PWS, ANGPT-like 7 (ANGPTL7) and serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 (SERPINA3) were over-expressed in the PWS skin. PWS + PDL samples demonstrated an upregulation of TIMP1 and VEGFA. No significant changes were seen in the expression of AKT1, FN1, HSP family, HES/ HEY transcription factors, HIF1A, HOX family, MMPs, RASA1, TGFB family, or VEGFRs.
TABLE 4.
Summary of Angiogenesis-Related Genes
| Gene name | Gene symbol | SET | Subject 1 | Subject 2 | Subject 3 | Subject 4 |
|---|---|---|---|---|---|---|
| Angiopoietin-like 7 | ANGPTL7 | N vs. PWS UP | 2.03 | NC | 3.76 | NC |
| Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 | SERPINA3 | N vs. PWS UP | NC | 3.07 | 2.16 | 2.12 |
| Tissue inhibitor of matrix metalloproteinases 1 | TIMP1 | PWS vs. PWS + PDL UP | 2.32 | NC | NC | 2.77 |
| Vascular endothelial growth factor A | VEGFA | PWS vs. PWS + PDL UP | NC | 5.93 | NC | NC |
NC, no change; N, normal skin; PWS, port wine stain; PWS + PDL, port wine stain treated with pulsed dye laser; UP, up-regulated in PWS or PWS + PDL; DOWN, down-regulated in PWS or PWS + PDL.
The nPWS from Subject 5 was analyzed individually because its expression pattern was unique and divergent from the PWS samples, showing differential expression of genes associated with angiogenesis, tumorigenesis, and inflammation. Tables 5 and 6 summarize the findings in the nPWS. Compared to N, ANGPT1, ANGPT2, TGFB3, Fibroblast growth factor (FGF) 7, Thrombospondin 1 (THBS1), FN1, and TIMP 3 were up-regulated in nPWS. Interestingly, MMP12, and VEGFA were found to be greatly down-regulated in nPWS. IGF2 was up-regulated in nPWS + PDL compared to nPWS, whereas PI3 and heat shock 27 kDa protein family, member (HSPB; a heat shock protein) 7 were down-regulated. There were a number of genes that were down-regulated in nPWS compared to N but then up-regulated following treatment. These include dermicidin (DCD), secretoglobulin, family 2A, member 2 (SCGB2A2), prolactin-induced protein (PIP), mucin 7, secreted (MUC7), cysteine-rich secretory protein 3 (CRISP3), and HOXA9. Table 6 specifies the ratios of either up- or down-regulation between comparison groups. No significant changes were seen in AKT1, HES/HEY transcription factors, HIF1A, RASA1, SERPIN family, or VEGFRs.
TABLE 5.
Summary of Nodular PWS Gene Expression
| Gene name | Gene symbol | Ratio | SET | Functional class |
|---|---|---|---|---|
| Angiopoietin 1 | ANGPT1 | 7.21 | N vs. nPWS UP | Pro-angiogenesis |
| Angiopoietin 2 | ANGPT2 | 3.80 | N vs. nPWS UP | Pro-angiogenesis |
| Transforming growth factor, beta 3 | TGFB3 | 2.17 | N vs. nPWS UP | Pro-angiogenesis |
| Fibroblast growth factor 7 | FGF7 | 2.70 | N vs. nPWS UP | Pro-angiogenesis |
| Thrombospondin 1 | THBS1 | 3.30 | N vs. nPWS UP | Anti-angiogenesis |
| Fibronectin 1 | FN1 | 2.70 | N vs. nPWS UP | Angiogenesis, cell movement |
| Tissue inhibitor of matrix metalloproteinases 2 | TIMP3 | 2.30 | N vs. nPWS UP | Tissue remodeling |
| Matrix metalloproteinase 12 | MMP12 | 23.89 | N vs. nPWS DOWN | Tissue remodeling |
| Vascular endothelial growth factor A | VEGFA | 2.13 | N vs. nPWS DOWN | Pro-angiogenesis |
| Insulin-like growth factor 2 | IGF2 | 2.02 | nPWS vs. nPWS + PDL UP | Growth factor |
| Peptidase inhibitor 3, skin-derived | PI3 | 3.78 | nPWS vs. nPWS + PDL DOWN | Anti-inflammatory |
| Heat shock 27 kDa protein family, member 7 (cardiovascular) | HSPB7 | 2.50 | nPWS vs. nPWS + PDL DOWN | Cardiomyopathy |
N, normal skin; nPWS, nodular port wine stain; nPWS + PDL, nodular port wine stain treated with pulsed dye laser; UP, up-regulated in PWS or PWS + PDL; DOWN, down-regulated in PWS or PWS + PDL.
TABLE 6.
Genes That Exhibit Co-Ordinate Expression Between Nodular PWS and Laser Treated Nodule
| Gene name | Gene symbol | Ratio | SET | Function |
|---|---|---|---|---|
| Dermicidin | DCD | 40.62 | N vs. nPWS DOWN | Tumorigenesis |
| Dermicidin | DCD | 277.56 | nPWS vs. nPWS + PDL UP | |
| Secretoglobulin, family 2A, member 2 | SCGB2A2 | 14.02 | N vs. nPWS DOWN | Modulation of inflammation & tissue repair |
| secretoglobulin, family 2A, member 2 | SCGB2A2 | 52.62 | nPWS vs. nPWS + PDL UP | |
| Prolactin-induced protein | PIP | 15.77 | N vs. nPWS DOWN | Tumorigenesis |
| Prolactin-induced protein | PIP | 38.88 | nPWS vs. nPWS + PDL UP | |
| Mucin 7, secreted | MUC7 | 20.75 | N vs. nPWS DOWN | Anti-microbial |
| Mucin 7, secreted | MUC7 | 30.66 | nPWS vs. nPWS + PDL UP | |
| Cysteine-rich secretory protein 3 | CRISP3 | 5.42 | N vs. nPWS DOWN | Tumorigenesis |
| Cysteine-rich secretory protein 3 | CRISP3 | 5.42 | nPWS vs. nPWS + PDL UP | |
| Homeobox A9 | HOXA9 | 8.46 | N vs. nPWS DOWN | Endothelial cells |
| Homeobox A9 | HOXA9 | 2.08 | nPWS vs. nPWS + PDL UP |
N, normal skin; nPWS, nodular port wine stain; nPWS + PDL, nodular port wine stain treated with pulsed dye laser; UP, up-regulated in PWS or PWS + PDL; DOWN, down-regulated in PWS or PWS + PDL.
DISCUSSION
Our gene expression analyses clearly distinguished PWS (Subjects 1–4) and nPWS (Subject 5), and several classes of gene functions were identified in our analysis of PWS and the effects of PDL treatment of these lesions. Significant gene expression pattern variation was observed amongst the donors, although some patterns were common. As PWS are well known to differ in vessel sizes and depths as well as treatment response, this may be expected. It is interesting to note that there was more similar gene expression between PWS in the same anatomic area than between PWS on different anatomic areas (face and leg). This may suggest that what we currently classify as PWS may not be all the same lesion or may simply be a result of different gene expression in skin tissue of various locations. Further research is required to evaluate this possibility. Future work will also be required to determine how genes identified in the current analysis relate to the development and maintenance of PWS.
Table 3 summarizes the genes with “co-ordinate” expression patterns for the PWS samples. PI3, also known as elafin, encodes a small secreted protein with anti-inflammatory activity [21]. STC1 is a hormone that regulates calcium and potassium metabolism [22]. PCP4 participates in calcium-dependent signaling through interaction with calmodulin [23]. FADS1 encodes a rate-limiting enzyme for fatty-acid conversion [24]. The significance and relevance of salt and lipid metabolism to PWS pathology is unclear and merits further exploration. SLCs are regarded as transporters, and there are individual families within the SLCs. There is currently limited literature on the SLC45 family, and none specific to SLC45A4. Other members of the SLC45 family have been associated with cutaneous melanoma and prostate cancer [25,26]. CD163 is a scavenger receptor expressed by dermal macrophages [27]. Differential expression of immune system-associated genes may reflect activation of inflammatory pathways as a result of tissue remodeling in PWS lesions.
In Subjects 1–4, we noted several genes related to angiogenesis, which were either up- or down-regulated (Table 4). There were two genes notably up-regulated in PWS: ANGPTL7 and SERPINA3. ANGPTL7’s function is still poorly understood, but other members of its family are potent regulators of angiogenesis; this family is structurally related to the ANGPTs, which are well-known to participate in angiogenesis [28]. SERPINA3, also known as alpha1-antichymotrypsin (ACT), is part of the serine protease inhibitor (SERPIN) family and has been reported to play a pivotal role in repair of skin wounds following mechanical injury [29]. Pigment endothelium-derived factor (PEDF, now SERPINF1) also belongs to the SERPIN family and has been shown to act as a broad-spectrum angiogenesis inhibitor [30]. Further research will be needed to identify the role of SERPINA3 in PWS.
We found two angiogenesis-related genes up-regulated in PWS following PDL (Table 4): TIMP1 and VEGFA. TIMPs prevent the destruction of tissue, and VEGFA is a well-known and potent angiogenesis factor [31]. We hypothesize that the up-regulation of TIMP1 may be the body’s way of protecting against the effects of the PDL, and that the up-regulation of VEGFA is part of a pro-angiogenic wound healing response, which may limit the efficacy of PDL treatments. Heger et al. [5] among others proposed that the upregulation of VEGF is involved in the angiogenic response to PDL therapy and that the antagonism of this response through VEGF inhibition may be considered as an adjuvant modality. If further studies confirm these results, anti-VEGF agents may be considered as adjunctive treatments to improve PDL effects. Our group and others have reported on increased efficacy with combined PDL and anti-angiogenic agent treatments [32].
The nPWS of Subject 5 exhibited a different expression profile from the other four PWS samples (Table 5). nPWS generally occur in adult patients and nodules often become more numerous with age [33]. Overall, the nPWS showed up-regulation of many angiogenesis-associated genes. The roles of ANGPT1, ANGPT2, and TGFB3 in promoting angiogenesis are well-established [31,34] and ANGPT1 and ANGPT2 are both up-regulated in IH [13]. TGFB2 belongs to the TGFB family, whose members are known inducers of angiogenesis; their proposed mechanism of action is through initiation of VEGF pathways [34]. The entire FGF family is associated with angiogenesis [31,35,36], while FGF7 (also known as keratinocyte growth factor) is a potent epithelial cell-specific growth factor that, in this setting, may be promoting tissue repair [37,38]. We also noted up-regulation of THBS1, a known potent inhibitor of angiogenesis [31,39]. FN1 protein participates in the regulation of angiogenesis, vasculogenesis, cell movement, and growth [7,40] but its exact role is unclear. It is interesting to note that the only other published PWS gene expression study (performed on fibroblasts of SWS patients) reported up-regulation of FN1 protein [7]. In our study, only the nPWS exhibited an up-regulation of FN1 gene compared to N (2.7-fold increase). However, none of our subjects had SWS and we evaluated the whole skin tissue, not just the fibroblasts. FN1 gene expression is likely restricted to fibroblasts, which constitute <5% of skin cells. In the PWS samples (as opposed to nPWS), FN1 mRNA levels may be below our level of detection due to the “dilution” of dermal fibroblast gene expression by the other skin components. TIMP3 was up-regulated in the nPWS compared to N. At first glance, up-regulation of TIMP3, an anti-angiogenic agent, may seem incongruous to our other results. However, the role of TIMPs in angiogenesis is complex due to their interaction and balance with the MMPs [31]. Indeed, MMP12 expression was down-regulated in nPWS compared to N. MMPs are enzymes that digest the extracellular matrix to remodel tissue, allowing for the growth of blood vessels and tumors [31]. Taken together, these observations suggest a dysregulation of angiogenic signals and/or components that may contribute to PWS pathology.
Treatment of the nPWS with PDL resulted in up-regulation of IGF2 and down-regulation of PI3 and HSPB7. IGF2 is involved in human growth and development, and its role in vascular lesions is still not well-described or characterized. PI3 had the most significant change of all genes in the PWS, showing down-regulation in PWS compared to N and then up-regulation following PDL. However, the nPWS showed no significant change in PI3 when compared to N, and then down-regulation after PDL. The difference in expression pattern behavior between nPWS and PWS merits further exploration. HSPB7 is extensively described in association with cardiomyopathies and heart failure as a genetic marker [41–43], but its role in other disease pathology has not been fully explored. The related gene HSPB1 is released primarily by endothelial cells and promotes angiogenesis through direct interaction with VEGF [44].
For the nPWS sample, it is notable that all of the “coordinate” gene expression patterns involved down-regulation in N versus nPWS and up-regulation in nPWS versus nPWS + PDL. The PWS samples did not exhibit this consistency. Table 6 summarizes the genes with “co-ordinate” expression patterns for the nPWS. DCD is primarily associated with antimicrobial peptide activity in the skin, likely participating in innate immunity and/or stress responses [45]. SCGB2A2, formerly known as mammaglobin 1, is best known as a breast cancer marker [46–48]. While its biological activity is not fully defined, the members of the secretoglobin family have been implicated in inflammation and tissue repair and are found in mammalian secretions, including fluids of the lung, lacrimal gland, salivary gland, prostate, and uterus [49]. While the detection of this gene raises the possibility of contamination of the sample with salivary gland tissue, a more likely source is the specialized myoepithelium found in exocrine glands, including sweat, mammary, lacrimal, and salivary glands [50]. SCGB2A2 is known to be found in skin (unpublished results PH/AZ), also localized to sweat glands [51]. Loss of expression of this gene may, therefore, be a consequence of loss of sweat glands in nPWS and the increase following PDL treatment may reflect regrowth of these skin-associated organs. PIP has been linked to both breast and prostate cancer [52], but it also exhibited immunosuppressive activity in a mouse model of allergic contact dermatitis [53]. The finding that PIP expression is significantly reduced in the nPWS and then restored following PDL treatment suggests an inflammatory component to nPWS. MUC7 is a small salivary mucin with microbicidal activity [54,55]. Similar to SCGB2A2, MUC7 expression may indicate a return of normal salivary gland structures and warrants further investigation. Like PIP, CRISP3 is found up-regulated in prostate cancer [56]. Finally, HOXA9 regulates endothelial cell activation, and aberrant expression may be associated with vascular lesions [57].
Previous studies using specific cell types isolated from vascular lesions have identified a number of genes whose expression was altered in the lesion compared to normal skin including AKT1, HES/HEY transcription factors, HIF1A, and RASA1 [10,15–18,58]. None of our comparisons showed any significant difference in the expression of these genes. This may reflect the relatively small changes reported by other groups or be a consequence of different methodologies. Some groups have isolated a certain cell type while we evaluated full skin samples.
There are limitations associated with our study. Microarray analysis provides a detailed transcriptional profile of each sample. However, the number and nature of samples can impact selectivity and sensitivity. PWS are lesions with many variables including vessel size, number, and depth. In this preliminary study, we evaluated five subjects but we will confirm and expand on our results using using qRT-PCR and IHC in a larger number of subjects. We also intend to evaluate gene expression at additional time points post-treatment.
In summary, gene expression profiles from N, PWS, and PWS + PDL demonstrated significant variation within samples from the same donor and between donors. By doing pair-wise comparisons between samples taken from the same donor and comparing these results between donors, we were able to identify genes that may participate in formation of PWS and PDL effects. Our preliminary results indicate changes in gene expression of angiogenesis-related genes, suggesting that dysregulation of angiogenic signals and/or components may contribute to PWS pathology.
Acknowledgments
Contract grant sponsor: American Society for Laser Medicine and Surgery; Contract grant sponsor: National Institutes of Health; Contract grant number: HD065536; Contract grant sponsor: National Institutes of Health Laser Microbeam and Medical Program (LAMMP, a P41 Biotechnology Resource); Contract grant number: RR001192.
The American Society for Laser Medicine and Surgery provided research funds which contributed to this work (K.M.K. and T.C.).
Footnotes
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest and none were reported.
References
- 1.Jacobs AH, Walton RG. The incidence of birthmarks in the neonate. Pediatrics. 1976;58(2):218–222. [PubMed] [Google Scholar]
- 2.Anderson RR, Parrish JA. Selective photothermolysis: Precise microsurgery by selective absorption of pulsed radiation. Science. 1983;220(4596):524–527. doi: 10.1126/science.6836297. [DOI] [PubMed] [Google Scholar]
- 3.Chen JK, Ghasri P, Aguilar G, van Drooge AM, Wolkerstorfer A, Kelly KM, Heger M. An overview of clinical and experimental treatment modalities for port wine stains. J Am Acad Dermatol. 2012;67(2):289–304. e229. doi: 10.1016/j.jaad.2011.11.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi B, Jia W, Channual J, Kelly KM, Lotfi J. The importance of long-term monitoring to evaluate the microvascular response to light-based therapies. J Invest Dermatol. 2008;128(2):485–488. doi: 10.1038/sj.jid.5700991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heger M, Beek JF, Moldovan NI, van der Horst CM, van Gemert MJ. Towards optimization of selective photothermolysis: Prothrombotic pharmaceutical agents as potential adjuvants in laser treatment of port wine stains. A theoretical study. Thromb Haemost. 2005;93(2):242–256. doi: 10.1160/TH04-05-0291. [DOI] [PubMed] [Google Scholar]
- 6.Laquer VT, Dao BM, Pavlis JM, Nguyen AN, Chen TS, Harris RM, Rugg EL, Kelly KM. Immunohistochemistry of angiogenesis mediators before and after pulsed dye laser treatment of angiomas. Lasers Surg Med. 2012;44(3):205–210. doi: 10.1002/lsm.22005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comi AM, Hunt P, Vawter MP, Pardo CA, Becker KG, Pevsner J. Increased fibronectin expression in Sturge–Weber syndrome fibroblasts and brain tissue. Pediatr Res. 2003;53(5):762–769. doi: 10.1203/01.PDR.0000058921.54071.19. [DOI] [PubMed] [Google Scholar]
- 8.Kadam SD, Gucek M, Cole RN, Watkins PA, Comi AM. Cell proliferation and oxidative stress pathways are modified in fibroblasts from Sturge–Weber syndrome patients. Arch Dermatol Res. 2012;304(3):229–235. doi: 10.1007/s00403-012-1210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen SL, Dosanjh A, Young DM, Boudreau N, Hoffman WY. Hemangiomas and homeobox gene expression. J Craniofac Surg. 2006;17(4):767–771. doi: 10.1097/00001665-200607000-00031. [DOI] [PubMed] [Google Scholar]
- 10.Adepoju O, Wong A, Kitajewski A, Tong K, Boscolo E, Bischoff J, Kitajewski J, Wu JK. Expression of HES and HEY genes in infantile hemangiomas. Vasc Cell. 2011;3:19. doi: 10.1186/2045-824X-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu Y, Wylie-Sears J, Boscolo E, Mulliken JB, Bischoff J. Genomic imprinting of IGF2 is maintained in infantile hemangioma despite its high level of expression. Mol Med. 2004;10(7–12):117–123. doi: 10.2119/2004-00045.Bischoff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberger S, Adini I, Boscolo E, Mulliken JB, Bischoff J. Targeting NF-kappaB in infantile hemangioma-derived stem cells reduces VEGF-A expression. Angiogenesis. 2010;13(4):327–335. doi: 10.1007/s10456-010-9189-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Y, Varughese J, Brown LF, Mulliken JB, Bischoff J. Increased Tie2 expression, enhanced response to angiopoietin-1, and dysregulated angiopoietin-2 expression in hemangioma-derived endothelial cells. Am J Pathol. 2001;159(6):2271–2280. doi: 10.1016/S0002-9440(10)63077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenberger S, Boscolo E, Adini I, Mulliken JB, Bischoff J. Corticosteroid suppression of VEGF-A in infantile hemangioma-derived stem cells. N Engl J Med. 2010;362(11):1005–1013. doi: 10.1056/NEJMoa0903036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perry B, Banyard J, McLaughlin ER, Watnick R, Sohn A, Brindley DN, Obata T, Cantley LC, Cohen C, Arbiser JL. AKT1 overexpression in endothelial cells leads to the development of cutaneous vascular malformations in vivo. Arch Dermatol. 2007;143(4):504–506. doi: 10.1001/archderm.143.4.504. [DOI] [PubMed] [Google Scholar]
- 16.Wooderchak-Donahue W, Stevenson DA, McDonald J, Grimmer JF, Gedge F, Bayrak-Toydemir P. RASA1 analysis: Clinical and molecular findings in a series of consecutive cases. Eur J Med Genet. 2012;55(2):91–95. doi: 10.1016/j.ejmg.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Breugem CC, Alders M, Salieb-Beugelaar GB, Mannens MM, Van der Horst CM, Hennekam RC. A locus for hereditary capillary malformations mapped on chromosome 5q. Hum Genet. 2002;110(4):343–347. doi: 10.1007/s00439-002-0700-z. [DOI] [PubMed] [Google Scholar]
- 18.Eerola I, Boon LM, Watanabe S, Grynberg H, Mulliken JB, Vikkula M. Locus for susceptibility for familial capillary malformation (‘port-wine stain’) maps to 5q. Eur J Hum Genet. 2002;10(6):375–380. doi: 10.1038/sj.ejhg.5200817. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi T, Ishida Y, Kimura A, Takayasu T, Eisenmenger W, Kondo T. Forensic application of VEGF expression to skin wound age determination. Int J Legal Med. 2004;118(6):320–325. doi: 10.1007/s00414-004-0468-x. [DOI] [PubMed] [Google Scholar]
- 20.Bui AK, Teves KM, Indrawan E, Jia W, Choi B. Longitudinal, multimodal functional imaging of microvascular response to photothermal therapy. Opt Lett. 2010;35(19):3216–3218. doi: 10.1364/OL.35.003216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alam SR, Newby DE, Henriksen PA. Role of the endogenous elastase inhibitor, elafin, in cardiovascular injury: From epithelium to endothelium. Biochem Pharmacol. 2012;83(6):695–704. doi: 10.1016/j.bcp.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Paulitschke V, Kunstfeld R, Mohr T, Slany A, Micksche M, Drach J, Zielinski C, Pehamberger H, Gerner C. Entering a new era of rational biomarker discovery for early detection of melanoma metastases: Secretome analysis of associated stroma cells. J Proteome Res. 2009;8(5):2501–2510. doi: 10.1021/pr8010827. [DOI] [PubMed] [Google Scholar]
- 23.Wei P, Blundon JA, Rong Y, Zakharenko SS, Morgan JI. Impaired locomotor learning and altered cerebellar synaptic plasticity in pep-19/PCP4-null mice. Mol Cell Biol. 2011;31(14):2838–2844. doi: 10.1128/MCB.05208-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ameur A, Enroth S, Johansson A, Zaboli G, Igl W, Johansson AC, Rivas MA, Daly MJ, Schmitz G, Hicks AA, Meitinger T, Feuk L, van Duijn C, Oostra B, Pramstaller PP, Rudan I, Wright AF, Wilson JF, Campbell H, Gyllensten U. Genetic adaptation of fatty-acid metabolism: A human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am J Hum Genet. 2012;90(5):809–820. doi: 10.1016/j.ajhg.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatzinasiou F, Lill CM, Kypreou K, Stefanaki I, Nicolaou V, Spyrou G, Evangelou E, Roehr JT, Kodela E, Katsambas A, Tsao H, Ioannidis JP, Bertram L, Stratigos AJ. Comprehensive field synopsis and systematic meta-analyses of genetic association studies in cutaneous melanoma. J Natl Cancer Inst. 2011;103(16):1227–1235. doi: 10.1093/jnci/djr219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helgeson BE, Tomlins SA, Shah N, Laxman B, Cao Q, Prensner JR, Cao X, Singla N, Montie JE, Varambally S, Mehra R, Chinnaiyan AM. Characterization of TMPRSS2: ETV5 and SLC45A3: ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68(1):73–80. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- 27.Bechetoille N, Vachon H, Gaydon A, Boher A, Fontaine T, Schaeffer E, Decossas M, Andre-Frei V, Mueller CG. A new organotypic model containing dermal-type macrophages. Exp Dermatol. 2011;20(12):1035–1037. doi: 10.1111/j.1600-0625.2011.01383.x. [DOI] [PubMed] [Google Scholar]
- 28.Comes N, Buie LK, Borras T. Evidence for a role of angiopoietin-like 7 (ANGPTL7) in extracellular matrix formation of the human trabecular meshwork: Implications for glaucoma. Genes Cells. 2011;16(2):243–259. doi: 10.1111/j.1365-2443.2010.01483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffmann DC, Textoris C, Oehme F, Klaassen T, Goppelt A, Romer A, Fugmann B, Davidson JM, Werner S, Krieg T, Eming SA. Pivotal role for alpha1-antichymotrypsin in skin repair. J Biol Chem. 2011;286(33):28889–28901. doi: 10.1074/jbc.M111.249979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orgaz JL, Benguria A, Sanchez-Martinez C, Ladhani O, Volpert OV, Jimenez B. Changes in the gene expression profile of A375 human melanoma cells induced by overexpression of multifunctional pigment epithelium-derived factor. Melanoma Res. 2011;21(4):285–297. doi: 10.1097/CMR.0b013e32834495c3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen A, Hoang V, Laquer V, Kelly KM. Angiogenesis in cutaneous disease: Part I. J Am Acad Dermatol. 2009;61(6):921–942. doi: 10.1016/j.jaad.2009.05.052. quiz 943-924. [DOI] [PubMed] [Google Scholar]
- 32.Tremaine AM, Armstrong J, Huang YC, Elkeeb L, Ortiz A, Harris R, Choi B, Kelly KM. Enhanced port-wine stain lightening achieved with combined treatment of selective photothermolysis and imiquimod. J Am Acad Dermatol. 2012;66(4):634–641. doi: 10.1016/j.jaad.2011.11.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klapman MH, Yao JF. Thickening and nodules in port-wine stains. J Am Acad Dermatol. 2001;44(2):300–302. doi: 10.1067/mjd.2001.111353. [DOI] [PubMed] [Google Scholar]
- 34.Fang S, Pentinmikko N, Ilmonen M, Salven P. Dual action of TGF-beta induces vascular growth in vivo through recruitment of angiogenic VEGF-producing hematopoietic effector cells. Angiogenesis. 2012;15(3):511–519. doi: 10.1007/s10456-012-9278-9. [DOI] [PubMed] [Google Scholar]
- 35.Pfarrer C, Weise S, Berisha B, Schams D, Leiser R, Hoffmann B, Schuler G. Fibroblast growth factor (FGF)-1, FGF2, FGF7 and FGF receptors are uniformly expressed in trophoblast giant cells during restricted trophoblast invasion in cows. Placenta. 2006;27(6–7):758–770. doi: 10.1016/j.placenta.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Berisha B, Welter H, Shimizu T, Miyamoto A, Meyer HH, Schams D. Expression of fibroblast growth factor 1 (FGF1) and FGF7 in mature follicles during the periovulatory period after GnRH in the cow. J Reprod Dev. 2006;52(2):307–313. doi: 10.1262/jrd.17077. [DOI] [PubMed] [Google Scholar]
- 37.Beer HD, Bittner M, Niklaus G, Munding C, Max N, Goppelt A, Werner S. The fibroblast growth factor binding protein is a novel interaction partner of FGF-7, FGF-10 and FGF-22 and regulates FGF activity: Implications for epithelial repair. Oncogene. 2005;24(34):5269–5277. doi: 10.1038/sj.onc.1208560. [DOI] [PubMed] [Google Scholar]
- 38.Finch PW, Rubin JS. Keratinocyte growth factor/fibroblast growth factor 7, a homeostatic factor with therapeutic potential for epithelial protection and repair. Adv Cancer Res. 2004;91:69–136. doi: 10.1016/S0065-230X(04)91003-2. [DOI] [PubMed] [Google Scholar]
- 39.Bagavandoss P, Wilks JW. Specific inhibition of endothelial cell proliferation by thrombospondin. Biochem Biophys Res Commun. 1990;170(2):867–872. doi: 10.1016/0006-291x(90)92171-u. [DOI] [PubMed] [Google Scholar]
- 40.Hamill KJ, Hopkinson SB, Hoover P, Todorovic V, Green KJ, Jones JC. Fibronectin expression determines skin cell motile behavior. J Invest Dermatol. 2012;132(2):448–457. doi: 10.1038/jid.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stark K, Esslinger UB, Reinhard W, Petrov G, Winkler T, Komajda M, Isnard R, Charron P, Villard E, Cambien F, Tiret L, Aumont MC, Dubourg O, Trochu JN, Fauchier L, Degroote P, Richter A, Maisch B, Wichter T, Zollbrecht C, Grassl M, Schunkert H, Linsel-Nitschke P, Erdmann J, Baumert J, Illig T, Klopp N, Wichmann HE, Meisinger C, Koenig W, Lichtner P, Meitinger T, Schillert A, Konig IR, Hetzer R, Heid IM, Regitz-Zagrosek V, Hengstenberg C. Genetic association study identifies HSPB7 as a risk gene for idiopathic dilated cardiomyopathy. PLoS Genet. 2010;6(10):e1001167. doi: 10.1371/journal.pgen.1001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cappola TP, Li M, He J, Ky B, Gilmore J, Qu L, Keating B, Reilly M, Kim CE, Glessner J, Frackelton E, Hakonarson H, Syed F, Hindes A, Matkovich SJ, Cresci S, Dorn GW., II Common variants in HSPB7 and FRMD4B associated with advanced heart failure. Circ Cardiovasc Genet. 2010;3(2):147–154. doi: 10.1161/CIRCGENETICS.109.898395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matkovich SJ, Van Booven DJ, Hindes A, Kang MY, Druley TE, Vallania FL, Mitra RD, Reilly MP, Cappola TP, Dorn GW., II Cardiac signaling genes exhibit unexpected sequence diversity in sporadic cardiomyopathy, revealing HSPB7 polymorphisms associated with disease. J Clin Invest. 2010;120(1):280–289. doi: 10.1172/JCI39085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee YJ, Lee HJ, Choi SH, Jin YB, An HJ, Kang JH, Yoon SS, Lee YS. Soluble HSPB1 regulates VEGF-mediated angiogenesis through their direct interaction. Angiogenesis. 2012;15(2):229–242. doi: 10.1007/s10456-012-9255-3. [DOI] [PubMed] [Google Scholar]
- 45.Paulmann M, Arnold T, Linke D, Ozdirekcan S, Kopp A, Gutsmann T, Kalbacher H, Wanke I, Schuenemann VJ, Habeck M, Burck J, Ulrich AS, Schittek B. Structure-activity analysis of the dermcidin-derived peptide DCD-1L, an anionic antimicrobial peptide present in human sweat. J Biol Chem. 2012;287(11):8434–8443. doi: 10.1074/jbc.M111.332270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zehentner BK, Carter D. Mammaglobin: A candidate diagnostic marker for breast cancer. Clin Biochem. 2004;37(4):249–257. doi: 10.1016/j.clinbiochem.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Zafrakas M, Petschke B, Donner A, Fritzsche F, Kristiansen G, Knuchel R, Dahl E. Expression analysis of mammaglobin A (SCGB2A2) and lipophilin B (SCGB1D2) in more than 300 human tumors and matching normal tissues reveals their co-expression in gynecologic malignancies. BMC Cancer. 2006;6:88. doi: 10.1186/1471-2407-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tassi RA, Calza S, Ravaggi A, Bignotti E, Odicino FE, Tognon G, Donzelli C, Falchetti M, Rossi E, Todeschini P, Romani C, Bandiera E, Zanotti L, Pecorelli S, Santin AD. Mammaglobin B is an independent prognostic marker in epithelial ovarian cancer and its expression is associated with reduced risk of disease recurrence. BMC Cancer. 2009;9:253. doi: 10.1186/1471-2407-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jackson BC, Thompson DC, Wright MW, McAndrews M, Bernard A, Nebert DW, Vasiliou V. Update of the human secretoglobin (SCGB) gene superfamily and an example of ‘evolutionary bloom’ of androgen-binding protein genes within the mouse Scgb gene superfamily. Hum Genomics. 2011;5(6):691–702. doi: 10.1186/1479-7364-5-6-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raubenheimer EJ. The myoepithelial cell: Embryology, function, and proliferative aspects. Crit Rev Clin Lab Sci. 1987;25(2):161–193. doi: 10.3109/10408368709105881. [DOI] [PubMed] [Google Scholar]
- 51.Sjodin A, Guo D, Hofer PA, Henriksson R, Hedman H. Mammaglobin in normal human sweat glands and human sweat gland tumors. J Invest Dermatol. 2003;121(2):428–429. doi: 10.1046/j.1523-1747.2003.12374.x. [DOI] [PubMed] [Google Scholar]
- 52.Baniwal SK, Little GH, Chimge NO, Frenkel B. Runx2 controls a feed-forward loop between androgen and prolactin-induced protein (PIP) in stimulating T47D cell proliferation. J Cell Physiol. 2012;227(5):2276–2282. doi: 10.1002/jcp.22966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugiura S, Fujimiya M, Ebise H, Miyahira Y, Kato I, Sugiura Y, Kimura T, Uehara M, Sato H, Sugiura H. Immunosuppressive effect of prolactin-induced protein: A new insight into its local and systemic role in chronic allergic contact dermatitis. Br J Dermatol. 2010;162(6):1286–1293. doi: 10.1111/j.1365-2133.2010.09756.x. [DOI] [PubMed] [Google Scholar]
- 54.Gomes GP, Assis MA, Fonseca JS, de Souza PE, Zenobio EG, Oliveira DD, Soares RV. Genetic polymorphism of MUC7 in individuals with aggressive or chronic periodontitis. J Oral Sci. 2011;53(4):445–449. doi: 10.2334/josnusd.53.445. [DOI] [PubMed] [Google Scholar]
- 55.Habte HH, de Beer C, Lotz ZE, Roux P, Mall AS. Anti-HIV-1 activity of salivary MUC5B and MUC7 mucins from HIV patients with different CD4 counts. Virol J. 2010;7:269. doi: 10.1186/1743-422X-7-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ribeiro FR, Paulo P, Costa VL, Barros-Silva JD, Ramalho-Carvalho J, Jeronimo C, Henrique R, Lind GE, Skotheim RI, Lothe RA, Teixeira MR. Cysteine-rich secretory protein-3 (CRISP3) is strongly up-regulated in prostate carcinomas with the TMPRSS2-ERG fusion gene. PLoS ONE. 2011;6(7):e22317. doi: 10.1371/journal.pone.0022317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bandyopadhyay S, Harris DP, Adams GN, Lause GE, McHugh A, Tillmaand EG, Money A, Willard B, Fox PL, Dicorleto PE. HOXA9 methylation by PRMT5 is essential for endothelial cell expression of leukocyte adhesion molecules. Mol Cell Biol. 2012;32(7):1202–1213. doi: 10.1128/MCB.05977-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daleprane JB, Schmid T, Dehne N, Rudnicki M, Menrad H, Geis T, Ikegaki M, Ong TP, Brune B, Abdalla DS. Suppression of hypoxia-inducible factor-1alpha contributes to the antiangiogenic activity of red propolis polyphenols in human endothelial cells. J Nutr. 2012;142(3):441–447. doi: 10.3945/jn.111.150706. [DOI] [PubMed] [Google Scholar]