Abstract
Bdellovibrio and like organisms (BALO) are obligate predators of Gram-negative bacteria, belonging to the α- and δ-proteobacteria. BALO prey using either a periplasmic or an epibiotic predatory strategy, but the genetic background underlying these phenotypes is not known. Here we compare the epibiotic Bdellovibrio exovorus and Micavibrio aeruginosavorus to the periplasmic B. bacteriovorus and Bacteriovorax marinus. Electron microscopy showed that M. aeruginosavorus, but not B. exovorus, can attach to prey cells in a non-polar manner through its longitudinal side. Both these predators were resistant to a surprisingly high number of antibiotic compounds, possibly via 26 and 19 antibiotic-resistance genes, respectively, most of them encoding efflux pumps. Comparative genomic analysis of all the BALOs revealed that epibiotic predators have a much smaller genome (ca. 2.5 Mbp) than the periplasmic predators (ca. 3.5 Mbp). Additionally, periplasmic predators have, on average, 888 more proteins, at least 60% more peptidases, and one more rRNA operon. Fifteen and 219 protein families were specific to the epibiotic and the periplasmic predators, respectively, the latter clearly forming the core of the periplasmic ‘predatome', which is upregulated during the growth phase. Metabolic deficiencies of epibiotic genomes include the synthesis of inosine, riboflavin, vitamin B6 and the siderophore aerobactin. The phylogeny of the epibiotic predators suggests that they evolved by convergent evolution, with M. aeruginosavorus originating from a non-predatory ancestor while B. exovorus evolved from periplasmic predators by gene loss.
Keywords: microbial predators, Bdellovibrio and like organisms, epibiotic predators, periplasmic predators, comparative genomics
Introduction
The genomes of predatory bacteria were shown to possess characteristic proteins and pathways, clearly distinguishing them from non-predatory bacterial genomes (Pasternak et al., 2013). However, how and why predators differ from each other is still very much an open question. Among solitary predators, the two main predation strategies are periplasmic, where cells enter the prey periplasm, and epibiotic, where they attach to the prey from outside. The main goal of the present study is to detail how epibiotic and periplasmic predators differ in behavior, phenotype, and genome content and structure, thus deepening our understanding of predators and characterizing the various signatures associated with employing each predatory strategy. The model predators for this comparison are the Bdellovibrio and like organisms (BALO), predatory Gram-negative bacteria belonging to the δ-proteobacteria and α-proteobacteria clades. Although phylogenetically separate, these two subgroups share common behaviors as both are obligate predators of Gram-negative bacteria with strain-dependent prey ranges and complex life cycles composed of two metabolically and spatially separated phases. First, during a free-living ‘attack' phase (AP), a solitary, flagellum-propelled and chemotaxis-directed hunter cell searches for a Gram-negative prey bacterium; and second, a growth phase (GP) is initiated by an AP cell attaching to, and eventually penetrating (in periplasmic species), a prey cell (Sockett, 2009). During GP, the predatory cell engages in prey breakdown for the benefit of synthesizing progeny (Sockett, 2009). The AP phase is conserved among all BALO species: high motility powered by a polar flagellum, no DNA replication or cell division capability, and a relatively short lifespan unless finding a prey cell; the GP phase of different BALOs, however, displays strategies that diverge into two very different behavioral groups, that is, the ‘periplasmic' and the ‘epibiotic' predators. Most δ-proteobacterial species (also called d-BALOs and belonging to the order Bdellovibrionales; Jurkevitch and Davidov, 2007), including Bdellovibrio bacteriovorus, Bacteriovorax marinus, Bacteriolyticum stolpii and Peredibacter starrii, use periplasmic predation. This behavior is characterized by a defined sequence of events during which an individual predatory cell secretes hydrolytic enzymes that help perforate and modify the prey cell's wall (Lerner et al., 2012), enabling the predator to settle within the prey periplasm. At the same time, the infected prey cell, now called a bdelloplast, is killed as respiration comes to a halt and the outer membrane is altered to form an osmotic barrier (Thomashow and Rittenberg, 1978). The invading predator appears to connect to the prey's cytoplasmic membrane, and initiates growth using the cytoplasm of the prey as a food source (Abram et al., 1974). Nutrient transfer may be facilitated by growth stage-specific pumps (Lambert et al., 2010) and the inclusion in the prey cell wall of an outer membrane protein originating from the predator (Barel et al., 2005). The predatory cell develops as a polynucleoid filament, the length of which is regulated by prey cell size (Kessel and Shilo, 1976). Finally, this filament septates into individual attack-phase cells that grow a flagellum, induce the formation of pores in the cell wall and burst into the external medium to engage in another cycle (Fenton et al., 2010a). In contrast to this intracellular life style, the d-BALO B. exovorus (formerly Bdellovibrio sp. strain JSS) and the α-proteobacterium (a-BALO) Micavibrio aeruginosavorus present an epibiotic strategy. Here, predation is extracellular as the predators remain attached to the cell wall but do not penetrate the prey cell, consuming it from the outside before dividing into two daughter cells via binary fission (Koval et al., 2012). Finally, the growing cell divides by binary division. Much less is known on the physiology, ecology and molecular mechanisms involved in epibiotic predation and how they differ from the more ‘classical' periplasmic predators. The isolation, culture and sequencing of epibiotic taxa now enable their comparison to the periplasmic predators. In this study, we characterized the growth parameters of the epibiotic B. exovorus JSS and M. aeruginosavorus EPB, sequenced their genomes de novo and compared them with those of the periplasmic predators Bdellovibrio bacteriovorus HD100 and Bacteriovorax marinus SJ to reveal fundamental differences between the two predatory strategies and their underlying molecular basis.
Materials and methods
Bacterial strains, media and growth conditions
B. exovorus strain JSS was isolated from sewage in London, Ontario (Canada) using Caulobacter crescentus as prey (Koval et al., 2012). M. aeruginosavorus strain EPB was isolated from saline soil in Israel using Pseudomonas corrugata as prey (Davidov et al., 2006). The prey strains were cultured in PYE and NB, respectively. The predators were grown in double-layer agar to obtain plaques or in liquid medium and stored as previously described (Jurkevitch, 2005). To obtain synchronized growth of M. aeruginosavorus strain EPB, a prey to predator ratio of ca. 1:2 was obtained by mixing ten-times concentrated predatory cells from a 2-day-old lytic culture with prey bacteria (OD540=10) and adding 2.7 v/v times of HEPES buffer amended with 2 mM Ca2+, and 1 mM Mg2+ adjusted to pH 7.8. The tubes were then shaken at 250 r.p.m., 30 °C. In order to enrich for predators attached to prey bacteria in both synchronized and unsynchronized cultures, predator and prey cultures of M. aeruginosavorus EPB with P. corrugata or of B. exovorus JSS with C. crescentus were centrifuged at 2000 r.p.m. for 2 min and 1500 r.p.m. for 3 min, respectively. The pelleted cells were then re-suspended in 200 μl standard HEPES buffer.
Prey range
The ability of the two epibiotic predators M. aeruginosavorus EPB and B. exovorus JSS to use Agrobacterium tumefaciens, Bordetella sp., Caulobacter crescentus, Enterobacter agglomerans, Escherichia coli 999 (deficient in diaminopimelic acid synthesis), Escherichia coli ML35, Pseudomonas aeruginosa, P. corrugata, P. fluorescens, P. putida, P. stutzeri, and Variovorax paradoxus was assessed using the double-layer agar plating method (Jurkevitch, 2005). Plates were incubated for up to 10 days at 30 °C and checked for plaque formation. In addition, the growth kinetics of M. aeruginosavorus EPB on the different prey was measured in a 48-well plate where 750 μl of P. corrugata (OD540=1) was mixed with 5 μl of a fresh lytic culture of M. aeruginosavorus EPB in six replicates, and incubated at 30 °C under constant shaking in a Tecan I Control Infinite 200′ plate reader (Tecan, Salzburg, Austria). OD readings were performed for each well at 1-h intervals. Prey without predator was used as controls. Predation efficiency was evaluated by comparing the drop in OD after 6 h of incubation and the slope of the predation curve.
Antibiotic resistance
Resistance of the epibiotic predators to antibiotics (Supplementary Table S1) was assessed using double-layer agar plates. B. exovorus JSS and M. aeruginosavorus EPB were mixed with C. crescentus and P. corrugata, respectively, and incubated at 30 °C for 24 h to initiate growth. Then, 100 μl of 0.2 μm-filtered antibiotic solutions was added to sterile disks, which were subsequently left to dry and placed on the double-layered plates. Resistance was measured as the width of the predation inhibition zone, that is, the inhibition of prey clearing around the disk after 3 days of incubation at 30 °C. Controls included plates without predator to check for prey cell lysis in the presence of antibiotics-imbibed disks.
Transmission electron cryo-microscopy
Synchronized cultures of M. aeruginosavorus EPB cells prepared as above were sampled for 30 and 70 min after mixing predator and prey to obtain active predator–prey complexes and dividing predators, respectively. One hundred μl of B. exovorus JSS was cultured in 10 ml of OD540=1 of C. crescentus for 24 h, resulting in an unsynchronized culture with predator cells at different stages of their lifecycle. All cells for electron microscopy were concentrated by centrifugation. The pellet was drawn into cellulose capillary tubes, mounted on 100-μm-deep aluminum discs (Wohlwend, Sennwald, Switzerland) and covered with a flat disc. The sandwiched sample was frozen in a HPM010 high-pressure freezing machine (Bal-Tec, Balzers, Liechtenstein). Cells were subsequently freeze-substituted in an AFS2 freeze substitution device (Leica Microsystems, Vienna, Austria) in anhydrous acetone containing 2% glutaraldehyde and 0.2% tannic acid and osmium tetroxide for 3 days at −90 °C and then warmed up to −30 °C over 24 h. Samples were washed three times with acetone, incubated for 1 h at room temperature with 2% osmium tetroxide, washed three times with acetone and infiltrated for 5–7 days at room temperature in a series of increasing concentration of Epon in acetone. After polymerization at 60 °C, 60–80 nm sections were stained with uranyl acetate and lead citrate and examined in a Tecnai Spirit electron microscope (FEI, Eindhoven, Holland) operating at 120 kV, utilizing a 2 k × 2 k Eagle CCD camera (FEI).
Protease activity
Proteolytic activity was assessed as in Dori-Bachash et al. (2008). B. exovorus JSS and M. aeruginosavorus EPB were co-cultured with their respective prey cells, C. crescentus and P. corrugata, for up to 48 h. Attack-phase cells were re-suspended in 10 ml standard HEPES for 24 h, and ruptured by sonication. All the samples were filtered through a 0.45-μm and then 0.2-μm filters to eliminate bacterial cells. The filtrates were lyophilized and re-suspended in 2 ml HEPES without calcium or magnesium salts. Two hundred fifty microliters of a 10% skim milk suspension was mixed with 167 μl PBS in tubes, and then 83 μl of a sample solution was added. Phenylmethylsulfonyl fluoride, a serine protease inhibitor, was added in order to identify the activity of serine proteases. The tubes were incubated at 30 °C for 24 h. The optical absorbance (600 nm) was measured against a control inoculated with water.
De novo genome sequencing of epibiotic predators and comparative genome analysis
Attack phase cells of B. exovorus JSS and M. aeruginosavorus EPB were obtained from standard lytic cultures prepared using C. crescentus and P. corrugata as prey, respectively. Attack cells were twice filtered through 0.45 μm filters (Sartorius, Goettingen, Germany) to eliminate remnants of the prey populations, and concentrated by centrifugation. Aliquots were spread on rich-media (NB or PYE) plates, and incubated at 30 °C for 3 days to check for any contaminant by observing prey colony formation. Predator DNA was isolated from these cultures with a commercial kit (Promega, Fitchburg, MA, USA) and used for whole genome paired-ends sequencing with the Genome Analyzer IIx machine (Illumina, San-Diego, CA, USA) at Tel Aviv University Genome High-Throughput Sequencing Laboratory. Both genomes were assembled by sequentially applying the Abyss (Simpson et al., 2009) and Minimus (Sommer et al., 2009) DNA sequence assemblers. The few resulting contigs were ordered and joined into a single chromosomal sequence by identifying genes and repeats present on the ends of the contigs. The resulting genomes were further analyzed, and corrected when needed, by using the reads pairing data. Directed PCR reactions were used to confirm uncertain short regions and to order the repeats in the B. exovorus JSS CRISPR region.
Genomic analysis
For both genomes, ORF prediction was performed with Prodigal (Hyatt et al., 2010), sequence similarity searches with BLAST (Altschul et al., 1997), and protein domain searches using HMMPFAM (Eddy, 1998) and CDD (Marchler-Bauer et al., 2011). Metabolic reconstruction was performed with Asgard (Alves and Buck, 2007), KAAS (Moriya et al., 2007) and extensive manual curation using BLAST. tRNA and rRNA predictions were performed with tRNA-scan (Lowe and Eddy, 1997) and RNAmmer (Lagesen et al., 2007), respectively. Prophage sequences were detected by Prophinder (Lima-Mendez et al., 2008), CRISPR loci by CRISPRFinder (Grissa et al., 2007), signal peptides by SignalP (Petersen et al., 2011) and genomic islands by IslandViewer (Langille and Brinkman, 2009). Venn diagrams were prepared by Venny (Oliveros, 2007) and synteny visualizations by ACT (Carver et al., 2005) using DoubleACT (www.hpa-bioinfotools.org.uk/pise/double_act.html). Directed PCR reactions were used to confirm uncertain short regions and to order the repeats in the B. exovorus JSS CRISPR region. Comparative proteomics was done with MBGD (Uchiyama, 2007): all protein sequences from the genomes of B. bacteriovorus HD100, B. exovorus JSS, B. marinus SJ, M. aeruginosavorus EPB and M. aeruginosavorus ARL13 were BLASTed all-against-all, and proteins that were sufficiently similar (BLAST e-value<10−4, score>200 and identity>50%) were considered a single protein cluster. Clusters that had representatives in the proteomes of both periplasmic predators and no representatives in the proteomes of all three epibiotic predators were considered ‘periplasmic clusters', and vice versa for ‘epibiotic clusters'.
Results
Growth of epibiotic predators
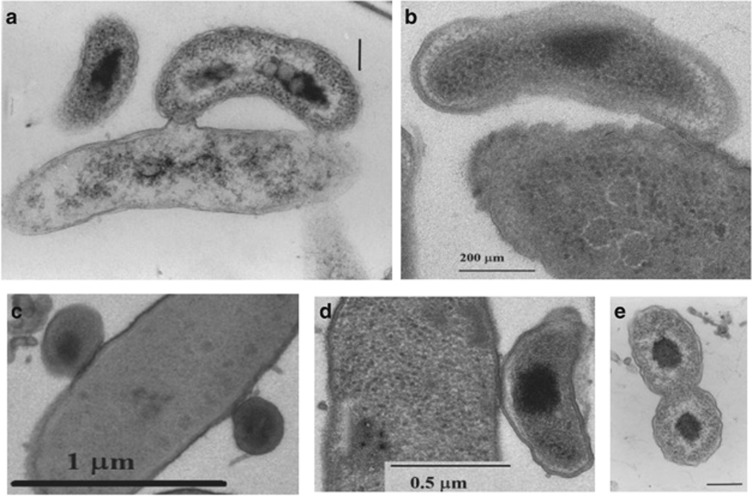
Epibiotic predation and a subsequent binary division pattern were observed in B. exovorus JSS and M. aeruginosavorus EPB growing on Caulobacter crescentus and Pseudomonas corrugata as prey, respectively (Figures 1a and b). In both predators, the prey cell did not round up and no bdelloplast was formed as usually happens during periplasmic predation. M. aeruginosavorus EPB or B. exovorus JSS AP cells were attached to nearly all P. corrugata or C. crescentus prey cells within 30 and 60 min, respectively. An attachment of more than one predatory cell to the prey cell wall was commonly observed with EPB (Figure 1c) but more rarely with JSS. Both predators displayed polar attachment to their prey cells using the proximal (non-flagellated) pole, but M. aeruginosavorus EPB cells were also seen to sometimes attach in a non-polar manner through the longitudinal side of the attack cell (Figure 1d). In old (24 h) cultures, M. aeruginosavorus EPB cells could be observed dividing unbound to prey cells (Figure 1e). EPB preyed most efficiently on P. corrugata but other species were also consumed, including Bordetella sp., Escherichia coli ML 35, P. putida WCS358, P. aeruginosa, P. fluorescens and Enterobacter agglomerans (Supplementary Figure S1). Slight growth of a few plaques of B. exovorus JSS on P. putida was observed, but on no other prey in addition to that occurring with the original C. crescentus prey. These data support the prey range studies in Koval et al. (2012). B. exovorus JSS and M. aeruginosavorus EPB were resistant to about two-thirds of the 23 antibiotics tested, including ampicillin, kanamycin, chloramphenicol, carbapenems and polymixins. Both were sensitive to amikacin, gentamicin and tetracycline; M. aeruginosavorus EPB (but not B. exovorus JSS) was sensitive to all cephalosporins and fluoroquinolones tested, as well as to aztreonam; B. exovorus JSS (but not M. aeruginosavorus EPB) was sensitive to minocycline (Supplementary Figure S2). Although BALOs are defined as obligate predators, host-independent mutants that are able to grow axenically on a rich medium are commonly obtained from B. bacteriovorus (Shilo and Bruff, 1965, Barel and Jurkevitch, 2001). We were unsuccessful at isolating such mutants from M. aeruginosavorus EPB and from B. exovorus JSS.
Figure 1.
(a) A dividing B. exovorus JSS preying on Caulobacter crescentus; (b) M. aeruginosavorus EPB dividing, attached to a Pseudomonas corrugata prey cell; (c) two M. aeruginosavorus EPB attack-phase predators simultaneously attached to a Pseudomonas corrugata prey cell; (d) M. aeruginosavorus EPB attached to a Pseudomonas corrugata prey cell by its longitudinal side; (e) unattached M. aeruginosavorus EPB cells dividing. Bars in (a) and (e) equal 200 nm.
Comparative analysis of the genomes of epibiotic and periplasmic predators
(i) Genome structure
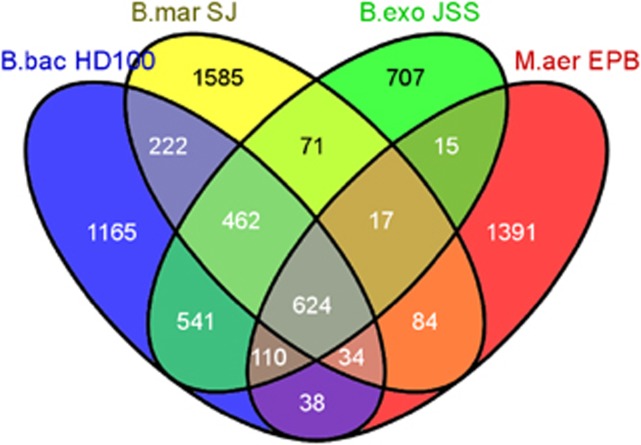
M. aeruginosavorus EPB and B. exovorus JSS genomes were sequenced and assembled from 37.16 and 34.01 million reads, respectively, with an average length of 36 bp (paired end). Circular chromosomes were assembled, and no extra-chromosomal elements were detected. These two genomes were compared to the genomes of the periplasmic predators Bacteriovorax marinus SJ (Crossman et al., 2013), B. bacteriovorus HD100 (Rendulic et al., 2004) and of the epibiotic predator M. aeruginosavorus ARL13 (Wang et al., 2011). A detailed view of the various genomic elements is given in Table 1 and Supplementary Table S2. Genome-wide synteny comparisons are given in Supplementary Figure S3, showing the very high level of conservation between the genomes of HD100 and JSS, and the much smaller conservation between the genomes of EPB and JSS and between HD100 and SJ. A comparison of the genomes of the epibiotic and periplasmic predators revealed that a periplasmic predator has, on average, 1.1 million more base pairs, 888 more protein-coding genes (PCG), at least 60% more peptidase-coding genes, and one more rRNA operon than an epibiotic predator (Table 1). After clustering all the PCG amino-acid sequences encoded by all the genomes using BLAST, a Venn diagram was generated (Figure 2): 17 PCG (in 15 clusters) were present in both epibiotic genomes and absent from all the periplasmic ones (Supplementary Table S3). The PCG of three of these clusters contain signal peptides, that is, they are probably secreted outside the cell; two others contain transmembrane helixes, that is, they are probably membrane proteins. No preference in phase expression (that is, over- or underexpression in the transcriptome) could be seen in this ‘epibiotic predatory' PCG set (Wang et al., 2011). Conversely, there were 257 PCG (in 219 clusters) that were present in the genomes of the periplasmic predators and absent from those of the epibiotic predators (Supplementary Table S4). Of these clusters, 52 (23.7%) are uncharacterized; 20 (9.1%) encode peptidases; 13 (5.9%) encode proteins involved in ABC transport; 12 are components of the purine metabolism—3 participate in the breakdown of adenine to urate and 9 are found in the pathway forming inosine monophosphate from ADP-ribose; 4 encode proteins involved in riboflavin synthesis, and 3 in chemotaxis. Also, 73 (33.3%) of the PCG in these clusters contained a signal peptide and 209 (95.4%) were preferentially transcribed (mean±s.d. 78±170 times) during the growth phase compared to the attack phase, that is, they are GP-specific proteins according to B. bacteriovorus HD100 transcriptomic data (Karunker et al., 2013).
Table 1. Genome characteristics of the five predatory organisms compared in this study: M. aerugnisavorus strains ARL-13 and EPB, B. exovorus JSS, B. bacteriovorus HD100 and B. marinus SJ.
| M. aeruginosavorus |
B. exovorus |
B. bacteriovorus |
B. marinus |
||
|---|---|---|---|---|---|
|
ARL-13 |
EPB |
JSS |
HD100 |
SJ |
|
| α-proteobacteria | δ-proteobacteria—Bdellovibrionales | ||||
|
Predation mode (division mode) |
Epibiotic (binary) | Periplasmic (filamentous) | |||
| Genome length (Mbp) | 2.48 | 2.46 | 2.66 | 3.78 | 3.44 |
| GC % | 54.7 | 55.0 | 46.1 | 50.6 | 36.7 |
| Proteins | 2432 | 2460 | 2669 | 3586 | 3231 |
| Secreted proteins (%) | 438 (18.0) | 404 (16.4) | 615 (23.0) | 1225(34.2) | 706 (21.9) |
| HI variant | No | No | No | Yes | Yes |
| Hit locus | No | No | Putative | Yes | Yes |
| rRNA operons | 1 | 1 | 1 | 2 | 2 |
| tRNA genes | 40 | 43 | 33 | 36 | 36 |
| CRISPR | 0 | 0 | 1 | 0 | 0 |
| Antibiotic resistance | 23 | 26 | 19 | 35 | 27 |
| Efflux | 16 | 18 | 10 | 27 | 15 |
| Tetracycline | 2 | 2 | 4 | 3 | 3 |
| Lin/str/phe/mac | 3 | 3 | 3 | 2 | 4 |
| Rifampin | 1 | 1 | 1 | 1 | 1 |
| Glycopeptide | 0 | 0 | 1 | 1 | 1 |
| Macrolide | 1 | 1 | 0 | 0 | 2 |
| Mupirocin | 0 | 0 | 0 | 1 | 1 |
| MprF | 0 | 1 | 0 | 0 | 0 |
| OmpA | 8 | 8 | 5 | 9 | 3 |
| Peptidases (%a) | 93 (3.8) | 94 (3.8) | 113 (4.2) | 178 (5.0) | 151 (4.7) |
| Aspartic (%b) | 3 (3.2) | 3 (3.2) | 4 (3.5) | 3 (1.7) | 2 (1,.3) |
| Cysteine (%b) | 11 (11.8) | 10 (10.6) | 9 (7.9) | 18 (10) | 16 (10.6) |
| Metallo (%b) | 40 (43) | 40 (42.5) | 41 (36.3) | 53 (29.7) | 56 (37.1) |
| Serine (%b) | 36 (38.7) | 36 (38.2) | 56 (49.5) | 96 (53.9) | 69 (45.7) |
| Threonine (%b) | 0 | 0 | 0 | 1 (0.6) | 1 (0.6) |
| Unknown (%b) | 3 (3.2) | 5 (5.3) | 3 (2.6) | 7 (3.9) | 7 (4.6) |
| Peptidoglycan-modifying enzymes (PME) | 13 AmiA, AmiB, AmiC, DacA (x3), DacD (x3), SltY (x2), PbpG (x2) | 13 AmiA, AmiB, AmiC, DacA (x3), DacD (x3), SltY (x2), PbpG (x2) | 14 AmiA, AmiB, AmiC, MepA, SltY (x3), MltC (x2), MltD (x3), DacB, PbpE | 16 AmiC, MepA, SltY (x3), MltA, MltB, MltC, MltD, DacB (x3), PbpE (x2), DacA | 15 AmiA, AmiB (x2), AmiC, MepA, SltY (x3), MltA, MltC (x2), MltD (x3), DacB, PbpE |
| Predatory PMEc | 0 | 0 | Possibly 1 | 2 | 2 |
| Ribonuclease | 14 | 18 | 17 | 19 | 25 |
| Transcription factors (HTH) | 63 | 62 | 88 | 107 | 97 |
| Sigma factors | RpoNHD | RpoNHD | RpoNHDE Whig | RpoNHDE Whig | RpoNHDE Whig, FecI |
| Diguanylate cyclase | 7 | 6 | 1 | 5 | 5 |
| Integrase | 10 | 6 | 6 | 3 | 2 |
| Two-component systems | 7 Osmotic upshift (K+), pH, chemotaxis, low nitrogen availability, CckA/ChpT/CtrA, PleC-PleD | 6 Osmotic upshift (K+), low nitrogen availability, chemotaxis, cell density (quorum sensing), CckA/ChpT/CtrA, PleC-PleD | 7 Phosphate limitation, chemotaxis, low nitrogen availability, secretion stress misfolded proteins, low turgor pressure, aceto-acetate | 6 Phosphate limitation, chemotaxis, low turgor pressure, low nitrogen availability, aceto-acetate, red/far-red light | 2 Phosphate limitation, chemotaxis |
| Secretion | Sec-SRP, type I, type II | Sec-SRP, type I, type II | Sec-SRP, type II | Sec-SRP, type II | Sec-SRP, type-II |
| Hemolysin | 6 | 6 | 2 | 9 | 2 |
Figure 2.
Venn diagram of the proteins in the genomes of B. bacteriovorus HD100, B. marinus SJ, M. aeruginosavorus EPB and B. exovorus JSS. Proteins were clustered using BLAST with an E-value <10−4.
A specific comparison of the two M. aeruginosavorus strains ARL13 and EPB reveals that both have genomes of almost identical sizes, yet they differ in their putative proteomes, with 340 and 271 PCG found in one of the strains but absent from the other (Supplementary Table 5). Of these strain-specific PCG, 453 (74.1%) are hypothetical, a much larger fraction than in the total proteome (which is 40.9% in ARL13 and 38.6% in M. aeruginosavorus EPB). Of the non-hypothetical strain-specific PCG, a large fraction (30%) is characterized as phage-associated, membrane, or regulatory proteins.
(ii) Metabolism
A detailed metabolic annotation was performed on both the B. exovorus JSS and M. aeruginovorus EPB genomes (Table 1; Supplementary Table S2), showing that like other BALOs (Rendulic et al., 2004, Crossman et al., 2013) they possess full gene pathways for glycolysis and for the tricarboxylic acid cycle. Fatty acid elongation and the phosphogluconate pathway appear to be limited in B. exovorus JSS, but the full gene content is found in M. aeruginosavorus EPB. DNA metabolism is complete in periplasmic predators. However, it is restricted in both epibiotic predators by the lack of a complete purine biosynthesis pathway, that is, by the inability to make inosine de novo. The production of amino acids is also constrained with the full gene pathways for the seven essential amino acids phenylalanine, arginine, histidine, tryptophan, valine, isoleucine and tyrosine missing in the epibiotic predators, as they are in the periplasmic predators. Tryptophan, tyrosine and leucine are produced by Micavibrio but not by the other BALOs. Interestingly, the genomes of the Bdellovibrionales encode for ABC transporters for branched-chain amino acids whereas those of Micavibrio do not. Thus, the predators had similar profiles of missing amino-acid biosynthesis capabilities with differences unlinked to their type of predation. Complete gene pathways for riboflavin and vitamin B6 were found in the periplasmic but not in the epibiotic predators, while the nicotinate pathway is complete in the three Bdellovibrionales but not in Micavibrio. As shown earlier (Pasternak et al., 2013), B. exovorus JSS, like almost all bacterial predators, possess a mevalonate pathway for isoprenoid biosynthesis while Micavibrio encodes the DOXP pathway. None of the predators are able to synthesize the reserve compounds glycogen and polyhydroxyalkanoate; yet, the periplasmic predators but not the epibiotic predators possess a polyhydroxyalkanoate depolymerase. The siderophore aerobactin, a secondary metabolite that may be produced during GP (Karunker et al., 2013), is encoded only in periplasmic BALOs. The genomes of both epibiotic and periplasmic predators displayed a large complement of antibiotic-resistance (AR) genes, most encoding for efflux pumps. B. bacteriovorus JSS had the smallest number of AR genes while the soil periplasmic predator B. bacteriovorus HD100 encoded 30–80% more resistance genes than the other BALOs. Finally, a striking difference was found between the genomes of the two M. aeruginosavorus strains, EPB and ARL-13 (Wang et al., 2011), namely the presence in EPB (but not in ARL13) of a genomic island encoding for 46 proteins involved in anaerobic nitrate respiration, including a nitrate reductase complex, a nitrate/nitrite transporter, NnrS and NnrU proteins involved in response to NO, molybdopterin biosynthesis and the transcriptional regulator Crp/Fnr (Supplementary Figure S4-A). Another EPB-specific island contains 56 PCG, some of which encode RND transporters and may be involved in multidrug efflux (Supplementary Figure S4-B). ARL13 has several islands that do not appear in EPB (Supplementary Figure S4-C-E), one of them apparently involved in copper resistance and another in cation efflux.
(iii) Regulatory processes
DNA processing, that is, PCG for repair, transcription and translation machinery, is complete in all BALOs but differences were detected in the transcription factors required to operate it (Table 1). These differences appear to be more related to phylogeny than to phenotype: both Micavibrio strains possess the three sigma factors RpoD (‘general housekeeping'), RpoH (‘heat shock') and RpoN (‘nitrogen limitation') (Gruber and Gross, 2003). This complement was slightly expanded in the three Bdellovibrionales to include RpoE (‘extracytoplasmic/extreme heat stress') and WhiG (‘flagellar synthesis and motility') (Gruber and Gross, 2003). Additionally, transcription factors were much more numerous in the periplasmic Bdellovibrio than in the two epibiotic predators, with Micavibrio possessing a significantly lesser number (Supplementary Table S6). One specific group of transcription factors showing significantly higher gene abundance in periplasmic predators is MerR, which is thought to play a major role in pathogenesis and virulence of pathogenic bacteria (Kidd, 2011). Genes encoding for the synthesis of the second effector cyclic di-GMP, a regulator known to be involved in the AP/GP transition (Hobley et al., 2012), were strikingly more numerous in Micavibrio than in the periplasmic B. bacteriovorus and B. marinus, while only one representative was found in B. exovorus JSS. As for two-component systems, genes for five and six systems were found in the two Micavibrio, six in B. exovorus JSS and B. bacteriovorus HD100 but only two in B. marinus. The Micavibrio two-component systems were identical, with the EPB strain encoding for an additional cell density detection system not found in ARL-13. Genes for regulation of osmotic pressure, chemotaxis and nitrogen availability were common to all predators; regulation of phosphate limitation was found in the three Bdellovibrionales (that also produce polyphosphate) and that of acetoacetate in the two Bdellovibrio species.
(iv) Predatory arsenal
The known and inferred predatory arsenal of BALO predators includes attachment components that help the predatory cell latch onto the prey, and lytic enzymes used to degrade it. The genomes of both Micavibrio strains and JSS, like all BALOs, possess nearly full gene complements for type IVa pili (TFP) and type IVb pili (FLP), used to attach and possibly (in periplasmic BALOs) penetrate the prey cell (Evans et al., 2007; Mahmoud and Koval, 2010). The gene for the subunit forming the major filament in both the TFP and FLP in Micavibrio could not be found by sequence homology; we thus used structural analysis (Hansen and Forest, 2006) to detect the PilA (TFP pilin) and Flp1 (FLP pilus subunit) genes, thereby finally showing that in Micavibrio, as in the periplasmic predators, complete sets of both type IVa and type IVb pilus genes are present. A large complement of protease-coding genes was found in all the predators, with their total number as well as percentage out of total PCG increasing between the epibiotic and the periplasmic predators (Table 1). This set includes between 13 (M. aeruginosavorus) to 16 (B. bacteriovorus) genes for peptidoglycan-modifying enzymes, comprising transglycosylases, aminidases, DD-endopeptidases and DD-carboxypeptidases. Lerner et al. (2012) discovered two B. bacteriovorus peptidoglycan endopeptidases, namely Bd0816 and Bd3459, which have adapted into secreted predation-specific proteins that ‘sculpt' the prey cell peptidoglycan structures so as to prevent double invasion and create a stable intracellular niche for the predator, inside which it replicates and finally kills the prey. Genes for these two proteins were also found in the periplasmic B. marinus but not in either of the epibiotic M. aeruginosavorus strains. B. exovorus had a single possible homologue, BexJSS_2041, which is a carboxypeptidase but only bears a weak similarity to the two B. bacteriovorus predation-specific carboxypeptidases (BLAST e-value >10−3).
The largest class of protease-coding genes shifts from metallo-proteases in Micavibrio to serine proteases in the Bdellovibrionales, with a very large (80%) increase in B. bacteriovorus (Table 1). While none of the epibiotic-specific clusters (Supplementary Table S3) was a protease, the periplasmic-specific set (Supplementary Table S4) contained 21 clusters of genes encoding proteolytic enzymes, representing 9.6% (21/219) of the set, a twofold increase compared to their relative abundance in the total genomes (Table 1). Seventeen of these 21 clusters were specifically induced during the GP (Supplementary Table S4; Karunker et al., 2013). The relative activities of serine proteases in the two epibiotic predators were further examined using a casein degradation assay. Serine protease activity was significantly reduced in both M. aeruginosavorus EPB and B. exovorus JSS by the addition of a serine protease inhibitor (Supplementary Figure S5). Interestingly, older lytic cultures of EPB were more sensitive to such inhibition than fresh AP cells.
As a whole, the epibiotic and periplasmic predators share similar gene profiles of secretory pathways. All possess the Sec pathway but only the Bdellovibrionales complement it with a full (B. bacteriovorus) or almost full (B. exovorus, B. marinus SJ) Gsp proteins-based type II export system. The δ-proteobacterial predators B. exovorus, B. bacteriovorus and B. marinus lack the SecB translocase of the Sec pathway. Further examination showed that genes for this chaperone-like protein are missing from many δ-proteobacteria, irrespective of their predatory ability. B. bacteriovorus is also the only BALO with a full TAT secretion system pathway as epibiotic predators only possess a TatC homologue and B. marinus SJ lacks TatB. Thus, all BALOs have PCG involved in initial substrate recognition and binding (TatC; Alami et al., 2003) but most lack the components used for translocation (TatA; Gohlke et al., 2005). All BALOs, like many Gram-negative bacteria, possess TolC homologues that may form a part of antibacterial efflux systems (Koronakis et al., 2004); only M. aeruginosavorus possesses the HlyB component of a type I secretion pathway but it still lacks the HlyD membrane fusion PCG to form a full pathway.
Discussion
This study details how obligate epibiotic and periplasmic BALO predators differ in behavior, phenotype and genome content structure. It expands upon a previous study in which predatory bacteria were shown to possess significant and characteristic genomic signatures (Pasternak et al., 2013). It thus provides an additional approach for characterizing not only the predatory abilities of a bacterium but also the predatory strategy it might employ. The differences between epibiotic and periplasmic predation include: (i) the mode of predation itself, (ii) the method of cell reproduction, (iii) the ability to live in the absence of a host, (iv) the sizes of the genome and of the proteome and (v) the abundance of hydrolytic and nucleolytic enzymes. The largest difference found between the epibiotic and periplasmic predators, underlying many of the other differences, was in genome size. This difference was expressed quantitatively, as the number of protein-encoding genes with ca. 3400 vs ca. 2500 in the periplasmic and epibiotic predators, respectively; and qualitatively, as the significantly higher diversity of PCG clusters exclusively found in periplasmic predators (219) vs those found exclusively in epibiotic predators (15). Accordingly, the ‘periplasmic predatory' PCG spanned a large range of functions, including protein degradation enzymes (mainly proteases), transport and metabolism. This set, compared to the entire genome, had significantly more genes expressed during GP (95% vs 67% of genes; Karunker et al., 2013) and included more potentially secreted proteins (33.3% vs 28%). Many of these genes are genomically clustered into operons. As so many of the genes encode lytic and transport proteins it may be inferred that they constitute the core of the ‘predatome'. Noteworthy, the ‘epibiotic predatory' PCG set was quite different, containing no lytic enzymes and at least seven metabolism proteins as well as a HipA-N terminal domain-containing protein. HipA, which may act as a toxin, can also cause multidrug resistance (Lewis, 2010), potentially explaining resistance to some of the antibiotics tested. Interestingly, B. exovorus JSS was sensitive to fluoroquinolones while M. aeruginosavorus exhibited resistance. Examination of the Bdellovibrio genomes showed that they encode for topoisomerase VI, an ‘archaeal' and ‘plant' topoisomerase rarely found in Bacteria (Forterre et al., 2007). It has been proposed that this type II topoisomerase only weakly interacts with fluoroquinolones (Dridi et al., 2011). FtsN, another protein involved in cell multiplication found in the genomes of the epibiotic predators, is expressed during the predatory phase (Wang et al., 2011). In the divisome, this protein contributes to the progress of cell wall constriction by mediating synthesis of septal peptidoglycan (Möll and Thanbichler, 2009). It is present in many proteobacteria, including δ-proteobacteria and the predatory Myxococcus xanthus (Möll and Thanbichler, 2009), but absent from the periplasmic predators, suggesting that the machinery used to synchronously split the predatory filamentous cell (Fenton et al., 2010b) does not simply replicate the binary division apparatus at multiple sites. Other differences between the two types of predators, which are related to growth mechanisms, include the apparent inability to obtain host-independent (HI) mutants from epibiotic predators, in contrast to periplasmic predators (Stolp and Starr, 1963; Barel and Jurkevitch, 2001; Dashiff and Kadouri, 2009). This may be related to the absence of bd108 homologues (bd108 is a gene directly implicated in the HI phenotype); (Cotter and Thomashow, 1992; Wurtzel et al., 2010; Roschanski et al., 2011) in the genomes of B. exovorus JSS and M. aeruginosavorus. Yet, in a number of instances in late lytic cultures, epibiotic predators were observed dividing when unattached to prey cells (Figure 1e). Whether this was due to the breaking of attachment towards the end of the growth cycle or to the ability of unattached predators to grow and divide is yet to be resolved. Multiple attachment (Figure 1c), followed by penetration of more than one predatory cell into the prey, can also occur in B. bacteriovorus under high multiplicity of infection (Fenton et al., 2010a), as used in this study.
The examination of functions defined as important for predation showed that all BALO possess pilus genes that are expressed during the attack phase and during attachment to the prey (Dori-Bachash et al., 2008; Lambert et al., 2010; Wang et al., 2011). Interestingly, PilA, a protein that composes the pilus filament, was not detected by sequence homology in the epibiotic predators but only by structural comparisons. The presence of PilA in the B. exovorus JSS pilus was inferred using antibodies raised against the periplasmic predator B. bacteriovorus 109J's PilA, as the antisera inhibited attachment of B. exovorus JSS to prey cells (Mahmoud and Koval, 2010). Previous studies in Pseudomonas aeruginosa have shown that PilA antibodies were able to bind to a PilA C-terminal domain of variable sequence, suggesting that conformation and not sequence was important for antibody recognition (Campbell et al., 1997). Different predatory strategies may imply different anchoring mechanisms: in B. bacteriovorus, the predator needs to reach the cytoplasmic membrane in order to penetrate its prey (Abram et al., 1974), a process that may be mediated by type IV pili (Evans et al., 2007). Consequently, it has been proposed that B. bacteriovorus could use the retractile forces exerted by type IV pili to facilitate prey entry (Mahmoud and Koval, 2010). Further studies may be necessary to evaluate if sequence differences between the epibiotic and the periplasmic predators' pilin proteins underlie the different abilities of the various predatory phenotypes.
The differences between epibiotic and periplasmic predators are not phylogenetic as B. exovorus JSS, a strain closely related to the periplasmic predator B. bacteriovorus, and M. aeruginosavorus, an unrelated α-proteobacterium, both exhibit this phenotype. This, along with the examination of their genomic content and genome structure, suggests that epibiotic predation in BALO results from convergent evolution, and has evolved a number of times in unrelated taxa. Furthermore, M. aeruginosavorus represents a deep taxonomic branch in the α-proteobacteria with its closest relative within the SAR116 group, both forming a sister clade to the Rhodospirillales (Davidov et al., 2006; Wang et al., 2011). Because neither the SAR116 nor the Rhodospirillales contain any known predators, it is likely that M. aeruginosavorus evolved to be an epibiotic predator ‘de novo', from an unidentified non-predatory ancestor. B. exovorus JSS forms an inner group within the Bdellovibrionales, with sister clades of periplasmic predators (Koval et al., 2012). So, in contrast to M. aeruginosavorus, it is possible that B. exovorus JSS evolved as an epibiotic predator from an ancestral periplasmic predator: this process included the loss of the genes that enable prey invasion, like the specific DD-carboxypeptidases found in B. bacteriovorus and B. marinus while preserving nearly all the metabolic functions present in periplasmic BALOs. Indeed, the genomes of B. exovorus JSS and B. bacteriovorus HD100 are highly syntenious (Supplementary Figure S3), confirming that JSS is taxonomically firmly rooted within the Bdellovibrionales, all of which (to our knowledge) are periplasmic predators.
Another molecule that may be associated with life in the periplasm is the siderophore aerobactin, which is not found in either B. exovorus JSS or M. aeruginosavorus. Aerobactin is expressed during the GP (Karunker et al., 2013): as oxidizing conditions prevail in the periplasm (Depuydt et al., 2009), soluble ferrous iron (the redox state of iron in the cytoplasm; Saha et al., 2012) from the prey may become oxidized and thus unavailable to the predator. Aerobactin could then sequester ferric ion and deliver it (Saha et al., 2012) to the growing predatory cell. Although B. bacteriovorus HD100 lacks an identified aerobactin receptor, it possesses six TonB-dependent receptor genes, two of which are putative siderophore receptors expressed during GP. Interestingly, a siderophore transporter gene was found in B. exovorus JSS, suggesting it may scavenge iron secreted by other bacteria without producing its own, as has been found in other bacterial species (Jurkevitch et al., 1992). The genome of B. exovorus JSS may have been shaped by a second wave of genome streamlining, discarding the expanded degradative capabilities that are adaptive for periplasmic life in periplasmic predators. A first set of genome reduction events had previously shaped the obligate periplasmic ancestor to its dependence on prey-derived amino acids and vitamins, in a manner similar to that observed in parasites or obligate symbionts in which genome streamlining includes the loss of such metabolic abilities (Klasson and Andersson, 2004). What ecological pressures would explain the evolutionary path leading to such diversification in predatory bacteria is an interesting, albeit open question. However, no other signs of genome degradation, such as a decrease in GC content, are obvious in either epibiotic or periplasmic predators. This suggests that the attack phase of the predator, which is free-living and therefore requires a multitude of genes and regulatory pathways for nutrition, motion, and so on, prohibits the establishment of a Muller's ratchet as experienced by obligate symbionts that lose many control functions (Moran, 1996).
Acknowledgments
We thank Albina Afinogenova (previously at the Russian Academy of Sciences, Pushchino, Russia) for kindly providing M. aeruginosavorus ARL-13. This work was supported by grants of the Israel Science Foundation (Grants No. 1290/08 and 1583/12) and the Natural Sciences and Engineering Research Council of Canada. We also thank the anonymous reviewers for their helpful comments.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abram D, Melo CJ, Chou D. Penetration of Bdellovibrio bacteriovorus into host cells. J Bacteriol. 1974;118:663–680. doi: 10.1128/jb.118.2.663-680.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alami M, Lüke I, Deitermann S, Eisner G, Koch H-G, Brunner J, et al. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol Cell. 2003;12:937–946. doi: 10.1016/s1097-2765(03)00398-8. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves JMP, Buck GA. Automated system for gene annotation and metabolic pathway reconstruction using general sequence databases. Chem Biodiv. 2007;4:2593–2602. doi: 10.1002/cbdv.200790212. [DOI] [PubMed] [Google Scholar]
- Barel G, Jurkevitch E. Analysis of phenotypic diversity among host-independent mutants of Bdellovibrio bacteriovorus 109J. Arch Microbiol. 2001;176:211–216. doi: 10.1007/s002030100312. [DOI] [PubMed] [Google Scholar]
- Barel G, Sirota A, Volpin H, Jurkevitch E. Fate of predator and prey proteins during browth of Bdellovibrio bacteriovorus on Escherichia coli and Pseudomonas syringae prey. J Bacteriol. 2005;187:329–335. doi: 10.1128/JB.187.1.329-335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AP, Wong WY, Houston Jr M, Schweizer F, Cachia PJ, Irvin RT, et al. Interaction of the receptor binding domains of Pseudomonas aeruginosa pili strains PAK, PAO, KB7 and P1 to a cross-reactive antibody and receptor analog: implications for synthetic vaccine design. J Mol Biol. 1997;267:382–402. doi: 10.1006/jmbi.1996.0871. [DOI] [PubMed] [Google Scholar]
- Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, Parkhill J. ACT: the Artemis comparison tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Cotter T, Thomashow M. Identification of a Bdellovibrio bacteriovorus genetic locus, hit, associated with the host-independent phenotype. J Bacteriol. 1992;174:6018–6024. doi: 10.1128/jb.174.19.6018-6024.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossman LC, Chen H, Cerdeno-Tarraga A-M, Brooks K, Quail MA, Pineiro SA, et al. A small predatory core genome in the divergent marine Bacteriovorax marinus SJ and the terrestrial Bdellovibrio bacteriovorus. ISME J. 2013;7:148–160. doi: 10.1038/ismej.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashiff A, Kadouri D. A new method for isolating host-independent variants of Bdellovibrio bacteriovorus using E. coli auxotrophs. Op Microbiol J. 2009;3:87–91. doi: 10.2174/1874285800903010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidov Y, Huchon D, Koval SF, Jurkevitch E. A new α-proteobacterial clade of Bdellovibrio-like predators: implications for the mitochondrial endosymbiotic theory. Environ Microbiol. 2006;8:2179–2188. doi: 10.1111/j.1462-2920.2006.01101.x. [DOI] [PubMed] [Google Scholar]
- Depuydt M, Leonard SE, Vertommen D, Denoncin K, Morsomme P, Wahni K, et al. A periplasmic reducing system protects single cysteine residues from oxidation. Science. 2009;326:1109–1111. doi: 10.1126/science.1179557. [DOI] [PubMed] [Google Scholar]
- Dori-Bachash M, Dassa B, Pietrokovski S, Jurkevitch E. Proteome-based comparative analyses of growth stages reveal new cell cycle-dependent functions in the predatory bacterium Bdellovibrio bacteriovorus. Appl Environ Microbiol. 2008;74:7152–7162. doi: 10.1128/AEM.01736-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi B, Fardeau M-L, Ollivier B, Raoult D, Drancourt M. The antimicrobial resistance pattern of cultured human methanogens reflects the unique phylogenetic position of archaea. J Antimicrob Chemo. 2011;66:2038–2044. doi: 10.1093/jac/dkr251. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Evans KJ, Lambert C, Sockett RE. Predation by Bdellovibrio bacteriovorus HD100 requires Type IV pili. J Bacteriol. 2007;189:4850–4859. doi: 10.1128/JB.01942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton AK, Kanna M, Woods RD, Aizawa SI, Sockett RE. Shadowing the actions of a predator: backlit fluorescent microscopy reveals synchronous nonbinary septation of predatory Bdellovibrio inside prey and exit through discrete bdelloplast pores. J Bacteriol. 2010;192:6329–6335. doi: 10.1128/JB.00914-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton AK, Lambert C, Wagstaff PC, Sockett RE. Manipulating each MreB of Bdellovibrio bacteriovorus gives diverse morphological and predatory phenotypes. J Bacteriol. 2010;192:1299–1311. doi: 10.1128/JB.01157-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P, Gribaldo S, Gadelle D, Serre M-C. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Gohlke U, Pullan L, McDevitt CA, Porcelli I, de Leeuw E, Palmer T, et al. The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc Natl Acad USA. 2005;102:10482–10486. doi: 10.1073/pnas.0503558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. Multiple Sigma subunits and the partitioning of bacterial transcription space. Ann Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- Hansen JK, Forest KT. Type IV Pilin structures: insights on shared architecture, fiber assembly, receptor binding and Type II secretion. J Mol Microbiol Biotechnol. 2006;11:192–207. doi: 10.1159/000094054. [DOI] [PubMed] [Google Scholar]
- Hobley L, Fung R, Lambert C, Harris M, Dabhi J, King S, et al. Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog. 2012;8:e1002493. doi: 10.1371/journal.ppat.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, LoCascio P, Land M, Larimer F, Hauser L. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkevitch E. Isolation and Classification of Bdellovibrio and Like Organisms. Current Protocols in Microbiology. John Wiley & Sons, Inc; 2005. [DOI] [PubMed] [Google Scholar]
- Jurkevitch E, Davidov Y.2007Phylogenetic Diversity and Evolution of Predatory ProkaryotesIn: Jurkevitch E (ed)Predatory Prokaryotes – Biology, ecology, and evolution Springer-Verlag: Heidelberg [Google Scholar]
- Jurkevitch E, Hadar Y, Chen Y. Differential siderophore utilization and iron uptake by soil and rhizosphere bacteria. Appl Environ Microbiol. 1992;58:119–124. doi: 10.1128/aem.58.1.119-124.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunker I, Rotem O, Dori-Bachash M, Jurkevitch E, Sorek R. A global transcriptional switch between the attack and growth forms of Bdellovibrio bacteriovorus. PloS One. 2013;8:e61850. doi: 10.1371/journal.pone.0061850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessel M, Shilo M. Relationship of Bdellovibrio elongation and fission to host cell size. J Bacteriol. 1976;128:477–480. doi: 10.1128/jb.128.1.477-480.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd SP.2011Novel regulation in response to host-generated stresses: the MerR family of regulators in pathogenic bacteriaIn: Kidd SP (ed)Stress Response in Pathogenic Bacteria CABI: Cambridge, USA [Google Scholar]
- Klasson L, Andersson SGE. Evolution of minimal-gene-sets in host-dependent bacteria. Trends Microbiol. 2004;12:37–43. doi: 10.1016/j.tim.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Koronakis V, Eswaran J, Hughes C. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Ann Rev Biochem. 2004;73:467–489. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- Koval SF, Hynes SH, Flannagan RS, Pasternak Z, Davidov Y, Jurkevitch E. Bdellovibrio exovorus sp. nov., a novel predator of Caulobacter crescentus. Int J Syst Evol Microbiol. 2012;63:146–151. doi: 10.1099/ijs.0.039701-0. [DOI] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rodland E, Staerfeldt H, Rognes T, Ussery D. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert C, Chang CY, Capeness MJ, Sockett RE. The first bite—profiling the predatosome in the bacterial pathogen Bdellovibrio. PLoS One. 2010;5:e8599. doi: 10.1371/journal.pone.0008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, Brinkman FSL. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics. 2009;25:664–665. doi: 10.1093/bioinformatics/btp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner TR, Lovering AL, Bui NK, Uchida K, Aizawa S-I, Vollmer W, et al. Specialized peptidoglycan hydrolases sculpt the intra-bacterial niche of predatory Bdellovibrio and increase population fitness. PLoS Pathog. 2012;8:e1002524. doi: 10.1371/journal.ppat.1002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Persister cells. Ann Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics. 2008;24:863–865. doi: 10.1093/bioinformatics/btn043. [DOI] [PubMed] [Google Scholar]
- Lowe T, Eddy S. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud KK, Koval SF. Characterization of type IV pili in the life cycle of the predator bacterium Bdellovibrio. Microbiology. 2010;156:1040–1051. doi: 10.1099/mic.0.036137-0. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möll A, Thanbichler M. FtsN-like proteins are conserved components of the cell division machinery in proteobacteria. Mol Microbiol. 2009;72:1037–1053. doi: 10.1111/j.1365-2958.2009.06706.x. [DOI] [PubMed] [Google Scholar]
- Oliveros JC.2007. VENNY. An interactive tool for comparing lists with Venn Diagrams http://bioinfogp.cnb.csic.es/tools/venny/index.html .
- Pasternak Z, Pietrokovski S, Rotem O, Gophna U, Lurie-Weinberger MN, Jurkevitch E. By their genes ye shall know them: genomic signatures of predatory bacteria. ISME J. 2013;7:756–769. doi: 10.1038/ismej.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Meth. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, et al. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- Roschanski N, Klages S, Reinhardt R, Linscheid M, Strauch E. Identification of genes essential for prey-independent growth of Bdellovibrio bacteriovorus HD100. J Bacteriol. 2011;193:1745–1756. doi: 10.1128/JB.01343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R, Saha N, Donofrio RS, Bestervelt LL. Microbial siderophores: a mini review. J Basic Microbiol. 2012;53:303–317. doi: 10.1002/jobm.201100552. [DOI] [PubMed] [Google Scholar]
- Shilo M, Bruff B. Lysis of Gram negative bacteria by host-independent ectoparasitic Bdellovibrio bacteriovorus isolates. J Gen Microbiol. 1965;40:317–328. doi: 10.1099/00221287-40-3-317. [DOI] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol İ. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sockett RE. Predatory lifestyle of Bdellovibrio bacteriovorus. Ann Rev Microbiol. 2009;63:523–539. doi: 10.1146/annurev.micro.091208.073346. [DOI] [PubMed] [Google Scholar]
- Sommer MOA, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolp H, Starr M. Bdellovibrio bacteriovorus gen. et al sp. n., a predatory, ectoparasitic, and bacteriolytic microorganism. Antonie van Leeuwenhoek. 1963;29:217–248. doi: 10.1007/BF02046064. [DOI] [PubMed] [Google Scholar]
- Thomashow MF, Rittenberg SC. Penicillin-induced formation of osmotically stable spheroplasts in nongrowing Bdellovibrio bacteriovorus. J Bacteriol. 1978;133:1484–1491. doi: 10.1128/jb.133.3.1484-1491.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama I. MBGD: a platform for microbial comparative genomics based on the automated construction of orthologous groups. Nucleic Acids Res. 2007;35:D343–D346. doi: 10.1093/nar/gkl978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kadouri D, Wu M. Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13. BMC Genomics. 2011;12:453. doi: 10.1186/1471-2164-12-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Dori-Bachash M, Pietrokovski S, Jurkevitch E, Sorek R. Mutation detection with next-generation resequencing through a mediator genome. PLoS One. 2010;5:e15628. doi: 10.1371/journal.pone.0015628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.