Abstract
The monthly, seasonal and interannual variability of microbial eukaryote assemblages were examined at 5 m, the deep chlorophyll maximum, 150 m and 500 m at the San Pedro Ocean Time-series station (eastern North Pacific). The depths spanned transitions in temperature, light, nutrients and oxygen, and included a persistently hypoxic environment at 500 m. Terminal restriction fragment length polymorphism was used for the analysis of 237 samples that were collected between September 2000 and December 2010. Spatiotemporal variability patterns of microeukaryote assemblages indicated the presence of distinct shallow and deep communities at the SPOT station, presumably reflecting taxa that were specifically adapted for the conditions in those environments. Community similarity values between assemblages collected 1 month apart at each depth ranged between ∼20% and ∼84% (averages were ∼50–59%). The assemblage at 5 m was temporally more dynamic than deeper assemblages and also displayed substantial interannual variability during the first ∼3 years of the study. Evidence of seasonality was detected for the microbial eukaryote assemblage at 5 m between January 2008 and December 2010 and at 150 m between September 2000 and December 2003. Seasonality was not detected for assemblages at the deep chlorophyll a maximum, which varied in depth seasonally, or at 500 m. Microbial eukaryote assemblages exhibited cyclical patterns in at least 1 year at each depth, implying an annual resetting of communities. Substantial interannual variability was detected for assemblages at all depths and represented the largest source of temporal variability in this temperate coastal ecosystem.
Keywords: 18S rRNA gene, microbial eukaryotes, protists, temporal variability, time series
Introduction
Protistan abundance and community composition are governed by a multitude of environmental factors including prey availability, top-down grazing pressure (e.g., viruses, other microbial eukaryotes and metazoa) and abiotic factors (e.g., temperature, light, salinity and nutrients) (Sherr et al., 2007). In turn, protistan assemblages have substantial impacts on ecosystem processes and function. Improving our understanding of community assembly and reassembly, and the factors that control them, are essential for predicting ecosystem response to a changing environment (McGradySteed et al., 1997; Naeem and Li, 1997; Chapin et al., 2000).
Molecular snapshots of microbial eukaryote assemblages from varying geographic locations, depths and times have provided a better understanding of the overall diversity, community structure and distribution of microbial eukaryotes in different marine ecosystems (Massana and Pedros-Alio, 2008; Vaulot et al., 2008; Caron et al., 2012). Molecular approaches have also been applied to characterize short-term temporal dynamics (days to weeks) of protistan assemblages in natural ecosystems (Vigil et al., 2009) and bottle incubations (Countway et al., 2005; Kim et al., 2011; Weber et al., 2012). These studies have shown rapid shifts in community structure in response to changes in environmental conditions. Several studies have also compared seasonal differences of microbial eukaryote communities (Massana et al., 2004; Romari and Vaulot, 2004; Lepere et al., 2006; Medlin et al., 2006; Aguilera et al., 2007; Countway et al., 2010; Nolte et al., 2010; Piwosz and Pernthaler, 2010; Edgcomb et al., 2011; Orsi et al., 2011), although most of these reports spanned relatively short timescales (⩽1–2 years). Multiyear comparisons that capture interannual changes of microbial eukaryotes have focused largely on bloom-forming conspicuous phytoplankton taxa such as diatoms and dinoflagellates using traditional microscopical methods (Venrick, 1998; Kim et al., 2009; Hinder et al., 2012). Thus, significant gaps exist in our basic knowledge regarding the natural variability of microbial eukaryote assemblages in marine environments, especially below the euphotic zone.
Terminal restriction fragment length polymorphism (T-RFLP) is a common molecular fingerprinting approach that offers a relatively fast and inexpensive method to characterize natural assemblages of microbial eukaryotes. The method has proven to be highly reproducible and reliable for the characterization of dominant taxa within diverse assemblages of microbes (Schutte et al., 2008; Rossi et al., 2009; Balzano et al., 2012; Kim et al., 2012) since its introduction for the study of microbial ecology (Liu et al., 1997), although limitations of the method include the limited number of taxa that can be detected and the inability to definitively assign a species identity to fragment-based operational taxonomic units (OTUs) (Osborn et al., 2000; Schutte et al., 2008).
Here, we characterized the vertical structure as well as the temporal (monthly, seasonal and interannual) variability of microbial eukaryote assemblages at four depths (5 m, the deep chlorophyll maximum (DCM), 150 m and 500 m) that spanned transitions in temperature, light, nutrients and oxygen at the San Pedro Ocean Time-series (SPOT) station. Microbial eukaryote assemblages were characterized using 18S rRNA gene-based T-RFLP analysis on 237 samples collected at approximately monthly intervals on cruises between September 2000 and December 2010 from 5 m and the DCM (with the exception of a 3-year period between October 2006 and February 2008), and between September 2000 and December 2003 from 150 and 500 m. To our knowledge, this represents one of the largest fingerprinting-based studies of microbial eukaryote assemblages in a marine environment. A database of partial 18S rRNA gene sequences from 5 m and 500 m at the SPOT station was used to assign putative taxonomic identifications to the fragment-based OTUs detected (Kim et al., 2012). Patterns of spatiotemporal variability revealed distinct assemblages of microbial eukaryotes in the shallow and deep environments at the SPOT station. Monthly changes in community composition were generally small at all depths, but assemblages at 5 m were more dynamic compared with deeper assemblages. Evidence of seasonality was detected for microbial eukaryote assemblages at 5 m during the last 3 years of the study as well as at 150 m between September 2000 and December 2003. Microbial eukaryote assemblages followed a cyclical pattern in some years, indicating an annual resetting of communities at specific depths. Substantial interannual variability within assemblages of microbial eukaryotes was also observed at each depth, which often represented the largest source of temporal variability at the study site.
Materials and methods
Environmental data
Environmental parameters were measured as a part of the SPOT program (http://dornsife.usc.edu/spot/). In situ measurements of conductivity, temperature and depth (Sea-bird Electronics or SBE 911 plus CTD; Sea-Bird Electronics, Inc., Bellevue, WA, USA), chlorophyll fluorescence (Wet Labs WETStar fluorometer; WETLabs, Philomath, OR, USA) and dissolved oxygen (SBE 13 sensor; Sea-Bird Electronics, Inc.) were measured during the collection of each sample. Mixed-layer depths (MLDs) were estimated as the depths at which σ–θ (potential density) differed from surface (10 m) σ–θ by 0.125 kg m−3 (Levitus, 1982). Chlorophyll a concentrations were measured for samples collected from 5 m and the DCM using the standard fluorometric method, dissolved oxygen concentrations were measured using the Winkler titration method (Grasshoff et al., 2007) for samples collected at 100 m (Winkler titration data were not available for samples collected at 150 m) and nutrient concentrations were measured for samples collected every 10 m between the surface and 40 m using an Alpkem RFA Auto Analyzer (Alpkem Corporation, Clackamas, OR, USA; Gordon et al., 1993; Grasshoff et al., 2007) (0–40 m depth-integrated nitrate concentrations are reported in this study). Vertical profiles of environmental parameters at our study site have been documented previously (Beman et al., 2010; Countway et al., 2010; Hamersley et al., 2011; Schnetzer et al., 2011).
Daily averages of sea surface temperature (SST) from NASA's GOES Imager (Wu et al., 1999) and near-daily averages of SeaWiFS chlorophyll a concentrations from the GeoEye Orbview-2 satellite (Hooker and McClain, 2000) were obtained from the National Oceanic and Atmospheric Administration (NOAA) CoastWatch Browser for the West Coast Regional Node (http://coastwatch.pfeg.noaa.gov/coastwatch/CWBrowser.jsp).
Sample collection
Seawater samples were collected on board the R/V Seawatch or R/V Yellowfin from four depths (5 m, the DCM, 150 m and 500 m) at approximately monthly intervals between September 2000 and December 2003 at the SPOT station located in the eastern North Pacific (33°33′N, 118°24′W). The depth of the DCM was determined by in situ real-time fluorescence during each sample collection. Seawater samples were pre-screened through a 200 μm nitex mesh net using gravity filtration and in-line filters that were directly attached to the Niskin bottles. Additional seawater samples were collected from 5 m and the DCM between January 2004 and December 2010, with the exception of a period between October 2006 and February 2008. These samples were pre-screened through 200 and 80 μm nitex mesh nets, also using gravity filtration and in-line filters. Prefiltered seawater samples were vacuum filtered (<5 mm Hg) during each cruise onto 47-mm GF/F filters, flash frozen in liquid nitrogen and stored at −80 °C until further processing.
Molecular characterization of microbial eukaryote assemblages
A total of 237 samples were analyzed for this study (Table 1). A combination of mechanical and chemical methods was used to extract nucleic acids for all samples, and has been detailed in Countway et al. (2005). Terminal restriction fragment length polymorphism (T-RFLP) was used to characterize microbial eukaryote assemblages using the Euk-A (Medlin et al., 1988) (labeled with the Beckman D4 fluorochrome) and Euk-570R (Weekers et al., 1994) 18S rRNA gene primers and the HaeIII restriction enzyme, according to Kim et al. (2012). Briefly, ∼10 ng of DNA template was used for each PCR reaction. Triplicate PCR reactions were performed for each sample, and the products were pooled following visualization on a 1.2% SeaKem LE agarose gel. Pooled and purified PCR products were treated with Mung Bean nuclease to eliminate single-stranded products (Egert and Friedrich, 2003), and ∼300 ng of each sample was used for overnight digestion using the HaeIII enzyme. The selection of the restriction enzyme was based on the in silico analysis of sequences obtained from GenBank (Countway et al., 2005). The digested DNA products were precipitated and purified before being resuspended in a deionized formamide solution (Sample Loading Solution; Beckman Coulter, Inc., Brea, CA, USA). T-RFLP products were run on the Beckman CEQ8000 gel capillary electrophoresis platform, using the following settings: 2.9 kV at 60 °C for 70 s and injection times that ranged between 9 and 15 s. An analysis of replicate T-RFLP patterns showed high reproducibility with average similarities of 80–90%. The chromatogram resulting in the highest quality (based on relative fluorescence units, standards and positive controls) was used for each sample for further analysis. OTUs were defined by unique fragment lengths, and T-RFLP fragments were assigned to high-level taxonomic groups based on putative identifications of fragment lengths obtained from in silico digests of 1341 partial 18S rRNA gene sequences from a 5 and 500 m sample collected at the SPOT station (Kim et al., 2012).
Table 1. The total number of samples collected in each month from four depths (5 m, the depth of the DCM, 150 m and 500 m) at the SPOT station.
| Depth |
Month
|
Total | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Jan | Feb | Mar | Apr | May | Jun | Jul | Aug | Sept | Oct | Nov | Dec | ||
| 5 m | 6 | 6 | 8 | 7 | 8 | 8 | 9 | 9 | 9 | 8 | 5 | 7 | 90 |
| DCM | 6 | 6 | 8 | 7 | 8 | 7 | 9 | 9 | 7 | 8 | 5 | 6 | 86 |
| 150 m | 3 | 2 | 3 | 2 | 4 | 2 | 3 | 3 | 4 | 4 | 1 | 3 | 34 |
| 500 m | 3 | 2 | 2 | 2 | 3 | 1 | 3 | 2 | 3 | 3 | 1 | 2 | 27 |
| All depths combined | 18 | 16 | 21 | 18 | 23 | 18 | 24 | 23 | 23 | 23 | 12 | 18 | 237 |
Abbreviations: DCM, deep chlorophyll maximum; SPOT, San Pedro Ocean Time-series.
Samples were collected from 5 m and the DCM between September 2000 and December 2010, with the exception of a 3-year period between October 2006 and February 2008. Samples were collected from 150 and 500 m between September 2000 and December 2003.
Multivariate analyses
T-RFLP results were normalized to peak area (Kaplan and Kitts, 2004) and square-root transformed to downweight the contribution of highly dominant T-RFLP fragments in each sample. PRIMER (v.6) and PERMANOVA+β18 (PRIMER-E Ltd, Ivybridge, UK) was used to calculate Bray–Curtis similarity values, which were used for non-metric multidimensional scaling (MDS) and CLUSTER (including SIMPROF) analyses (Clarke, 1993). The parameters used for the SIMPROF test were: group average mode; permutations for mean profile: 1000; simulation permutations: 999; and significance level: 0.05. The parameters used for MDS analyses were: Kruskal stress formula: 1; minimum stress: 0.01. Stress values were calculated for MDS plots and reflect the level of distortion that results from representing similarity rankings between many samples in a two-dimensional space. Stress values of <0.2 generally indicate an accurate representation of similarity rankings (Clarke and Gorley, 2006). The RELATE test was used to calculate Spearman's correlations between each year and other years from the same depth as well as a cyclical model to test for an annual resetting of microbial eukaryote assemblages (Clarke and Gorley, 2006). Analysis of similarity is the multivariate analog of analysis of variance and was used to test the null hypothesis that there were no significant compositional differences between depths. The analysis of similarity test statistic (R) ranges between 0 and 1, where 1 indicates completely different assemblages. The Species Accumulation Plot function in the PRIMER software package was applied to determine the number of unique fragment-based OTUs at each depth (sample order was permutated, max permutations: 999). The BEST function was used for multivariate analyses to determine the combination of environmental variables that resulted in the strongest correlation with the patterns of community (dis)similarity at each depth. Environmental data were log transformed and normalized. There were two to four time-points from each depth that were missing one environmental variable, in which case the average of the variable from other time-points was used to fill in the missing data in order to retain the time-point for analysis. The number of permutations used to determine significance for the BEST analysis was 999.
Averages of all possible pairwise Bray–Curtis similarity values were used to compare the overall (dis)similarity of microbial eukaryote assemblages at each depth. Samples from each depth were also grouped by months apart (1–24 months lag), and averages of pairwise Bray–Curtis similarity values were calculated for each group to look for evidence of seasonality, as well as to compare monthly, seasonal and interannual community similarity values. Assemblages were determined to be seasonal if community similarity values were lowest between assemblages that were 6 and 18 months apart (approximately opposite seasons) and highest between assemblages that were 12 and 24 months apart (similar seasons), and resulted in a nonlinear regression of a sinusoidal pattern characteristic of seasonal plots (e.g., see Supplementary Figure S1A). Groups of samples that were more than 24 months apart reduced the power of the analysis because of the increasingly smaller number of samples with increased lag, and were excluded from the analyses.
Results
Environmental variables
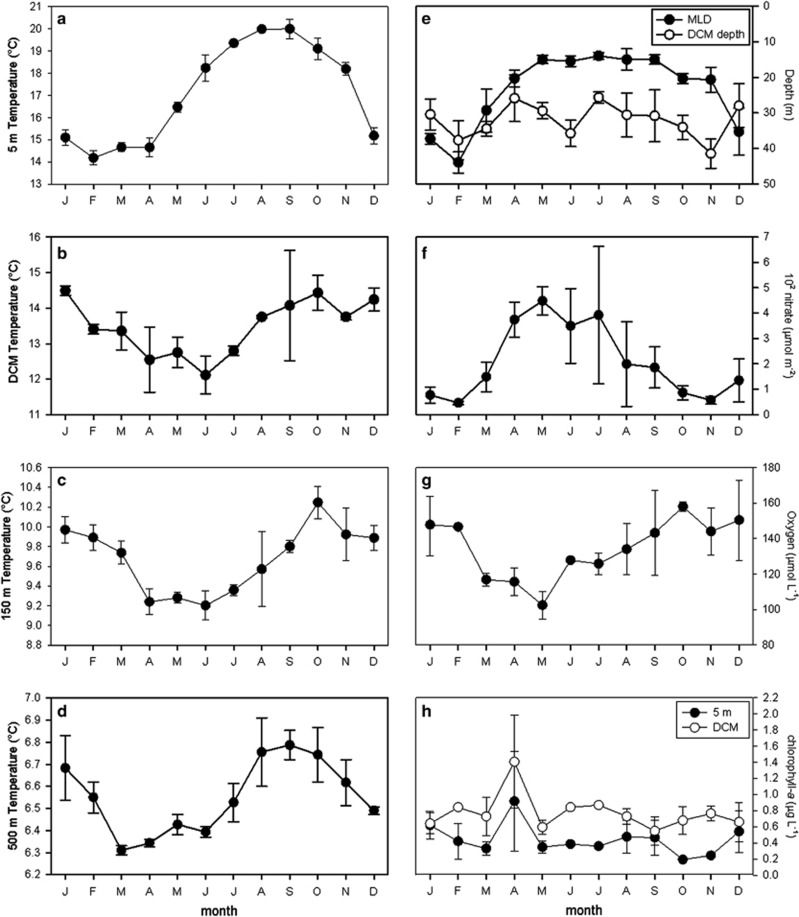
Annual SSTs at the SPOT station ranged between ∼13.5 °C and ∼21.5 °C (Figure 1a and Supplementary Figures S1A and D). Warmest temperatures occurred in August and September, and coolest temperatures occurred from December through April. Coolest temperatures at the DCM were observed between April and July, and warmest temperatures occurred between September and January (Figure 1b). Temperature ranges were more constrained at 150 m and especially at 500 m, but the temporal variability of temperatures in these deep environments resembled the patterns observed at the DCM (Figures 1b–d). Although vertical profiles of the photosynthetically active region at the SPOT station are not available for the current study, monthly measurements made in 2012 indicate that 1% photosynthetically active region levels never exceeded depths of 100 m, even during times of highest irradiance and lowest biomass.
Figure 1.
Monthly averages of physical and chemical environmental parameters measured from multiple depths at the SPOT station between September 2000 and December 2003. In situ temperature at 5 m (a), the DCM (b), 150 m (c) and 500 m (d); the depths of the DCM and mixed layer (e); depth-integrated (0–40 m) nitrate concentrations (f); oxygen concentrations (100 m) (g); and chlorophyll a concentrations at 5 m and the DCM (h). The error bars represent the standard error of the monthly means. Details of environmental variables are available in Supplementary Figure S1.
MLDs remained relatively shallow throughout each year, ranging between approximately 15 and 50 m (Figure 1e). MLDs correlated with SSTs (r2=0.65, P<0.01; Supplementary Figures S1D and F) and were deepest between December and February of each year before shoaling in March and April.
There was a persistent oxycline at approximately 50 m, and hypoxic conditions (<∼50 μM O2) were observed below ∼350 m throughout each year (see Countway et al., 2010 for seasonal profiles of oxygen concentrations in 2001). The monthly averages of oxygen concentrations at 100 m correlated with monthly averages of temperatures at 150 m (r2=0.84, P<0.001; Figures 1c and g). A DCM was detected throughout each year of this study, but the depth and shape of this biological feature varied throughout the year (see Countway et al., 2010 for seasonal vertical profiles of chlorophyll a). On average, the depth of the DCM was found above the MLD during winter months and below the mixed layer between March and November (Figure 1e). Nutrient concentrations at 5 m were low throughout each year (e.g., <0.3 μM nitrate at 5 m), reflecting the generally oligotrophic conditions at the SPOT station. Depth-integrated (0–40 m) nitrate concentrations were highest between April and July (Figure 1f and Supplementary Figure S1G). Highest chlorophyll a concentrations at 5 m and the DCM were observed during April, although chlorophyll a concentrations were generally low throughout each year (<∼2 μg l−1; Figure 1h and Supplementary Figures S1B and C). Interannual variability was observed for all environmental parameters, reflected by the standard error bars in Figure 1 and detailed in Supplementary Figure S1 (e.g., anomalously high chlorophyll a concentrations in September 2003 and April 2005; Supplementary Figure S1B).
Vertical distribution of microbial eukaryote assemblages
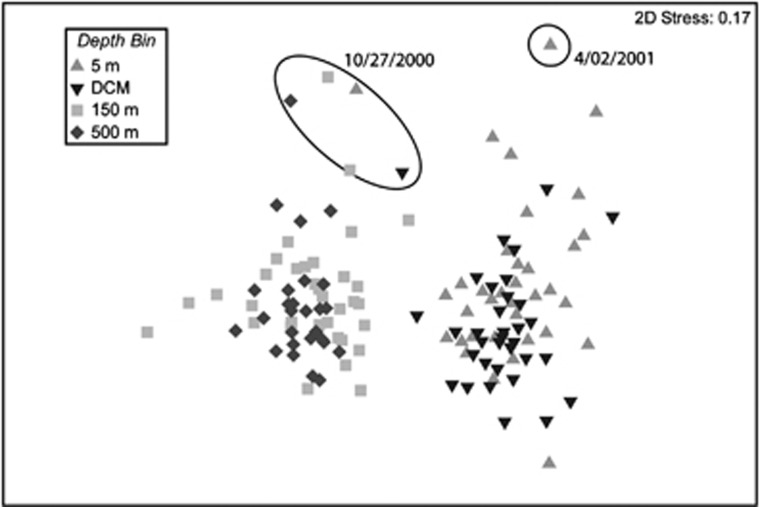
MDS and CLUSTER analyses of assemblages at four depths between September 2000 and December 2003 resolved two significantly different communities of microbial eukaryotes: a shallow community at 5 m and the DCM; and a deep community at 150 and 500 m (R=0.78, P=0.001) (Figure 2). Exceptions included assemblages collected on 27 October 2000 from all four depths and on 2 April 2001 from 5 m (circled in Figure 2). The assemblage of microbial eukaryotes at 5 m could not be differentiated from the assemblage at the DCM (R=0.08). However, pairwise comparisons of community similarity values between the two near-surface assemblages varied throughout each year and were, on average, highest between November and March, with the exception of January (∼60–64% Supplementary Figure S2). Community similarity values between assemblages at 5 m and the DCM were, on average, lowest in September and October (∼49%).
Figure 2.
MDS plot of T-RFLP results from seawater samples collected approximately every month between September 2000 and December 2003 from 4 depths (5 m, the DCM, 150 m and 500 m) at the SPOT station. The MDS plot is based on Bray–Curtis similarity values of square-root transformed T-RFLP data. The stress value is reported on the top right corner of the plot. Highly disparate samples that were observed on 27 October 2000 at all four depths and on 2 April 2001 at 5 m are circled.
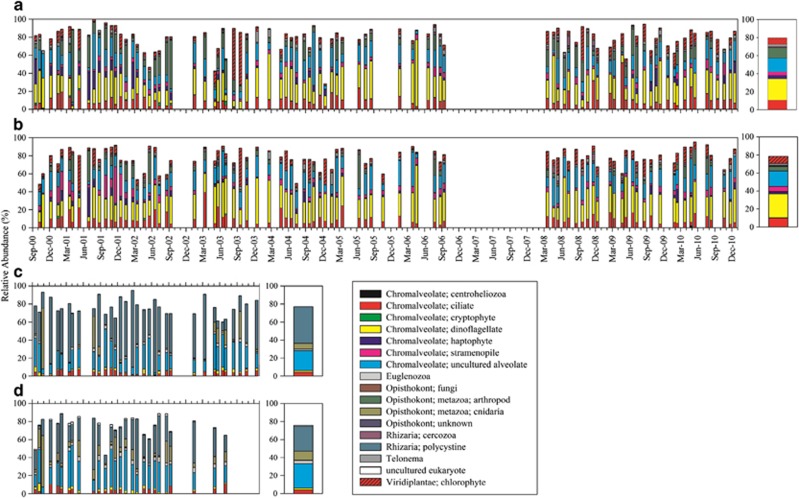
The taxonomic composition of assemblages at 5 m and the DCM were distinct from assemblages at 150 and 500 m (Figures 3a–d and Supplementary Figures S3A–D). Shallow communities were dominated by fragment lengths representative of arthropod, chlorophyte, ciliate, dinoflagellate, haptophyte, stramenopile, telonemid and uncultured alveolate (Groups I and II) taxa. Deep communities were largely comprised of cnidarian, euglenozoan, polycystine and uncultured alveolate (mostly Group II) taxonomic groups, as well as a smaller fraction of ciliate and dinoflagellate taxa. OTUs that could be assigned a high-level taxonomic identification accounted for an average of ∼75–80% of total abundance in each sample (right panels of Figures 3a–d). Only modest compositional differences were apparent between the monthly averages of relative abundances of high-level taxonomic groups at each depth (Supplementary Figures S3A–D), except for OTUs representative of polycystines, which dominated the 150 m assemblage during winter months (between December and March) (Supplementary Figure S3C).
Figure 3.
Relative abundances of high-level taxonomic groups based on T-RFLP data for approximately monthly samples collected from 5 m (a) and the DCM (b) between 2000 and 2010 (with the exception of months between October 2006 and February 2008); and from 150 m (c) and 500 m (d) between 2000 and 2003 at the SPOT station. Gaps represent missing data. The averages of relative abundances for each high-level taxonomic group are presented on the right panel for each depth. Taxonomic identifications for the fragments were assigned using a database of 1341 18S rRNA gene sequences with assigned taxonomies that were subjected to in silico restriction digestion to predict the length of terminally labeled fragments at the study site (obtained from Kim et al., 2012). Monthly averages of the relative abundances of high-level taxonomic group are summarized for each depth in Supplementary Figure S3.
Accumulation plots
The analysis of 237 samples resulted in an average of 35 OTUs per sample (there were no significant differences in the average number of OTUs between depths). However, accumulation plots showed that a larger number of unique taxa were detected in samples collected at 5 m and the DCM between September 2000 and December 2003 (169 and 168 unique fragment-based OTUs, respectively) compared with the number of unique taxa detected at 150 and 500 m (147 and 129 OTUs, respectively) (Supplementary Figure S4). Additional fragment analyses of samples collected from 5 m and the DCM between 2004 and 2010 resulted in 45 and 51 more OTUs, respectively.
Overall similarity of microbial eukaryote assemblages at specific depths
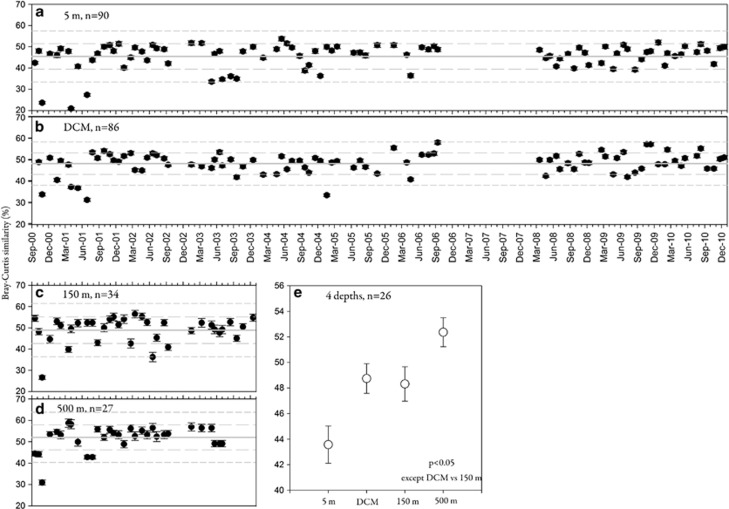
The assemblage at 5 m had a larger number of highly disparate samples compared with other depths between 2000 and 2003 (i.e., more samples that were >1 or 2 s.d.'s away from the average community similarity value; Figures 4a–d). There was also a larger number of highly disparate samples collected between 2000 and 2003 compared with samples collected between 2008 and 2010 from 5 m as well as the DCM (Figures 4a and b). The average of all possible pairwise community similarity values for samples collected between September 2000 and December 2003 at 5 m (∼43.6%) was significantly lower than the average obtained for all possible pairwise comparisons of the assemblages at deeper depths (P<0.05) (Figure 4e). Assemblages at 150 m and the DCM resulted in intermediate averages that were nearly identical (∼48%).
Figure 4.
The averages of all possible pairwise Bray–Curtis similarity values (plotted with standard error bars) for each sample collected from 5 m (a), the DCM (b), 150 m (c) and 500 m (d) at the SPOT station. Samples collected between 2000 and 2010 from 5 m and the DCM, and samples collected between 2000 and 2003 from 150 and 500 m, were included in the analysis. Sample sizes (n) are noted for each data set. The solid grey lines represent the means of all pairwise averages for each depth, and the dashed grey lines represent 1 and 2 s.d.s. The y-axes reflect Bray–Curtis similarity values (%) and are standardized to facilitate comparison. The averages of all pairwise comparisons for samples collected between September 2000 and December 2003 from each depth are shown in (e) (and only dates that included samples from each depth were included, n=26). Significant differences in the averages of pairwise comparisons were detected between all depths (P<0.05), except the comparison between the DCM and 150 m.
Monthly, seasonal and interannual variability of microbial eukaryote assemblages
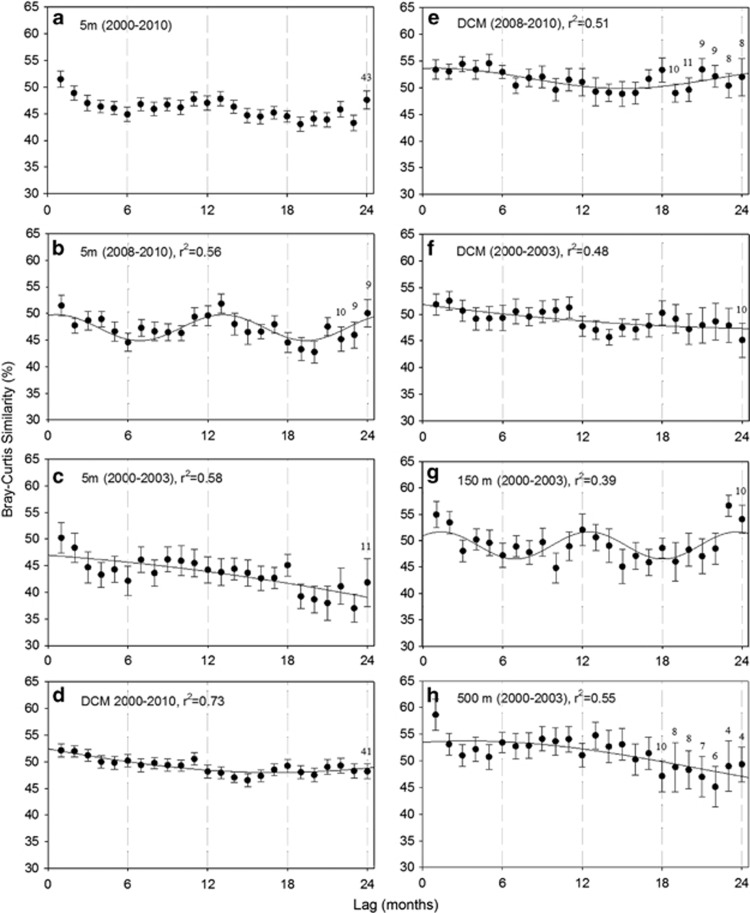
Examination of the entire time-series data for the assemblage at 5 m revealed a weak signal of seasonality that did not result in a good fit of a sinusoidal curve (Figure 5a). Samples that were collected 1 to 24 months apart displayed relatively similar average Bray–Curtis similarity values and variance. Similarity values between assemblages collected 1 month apart were as low as ∼20% and as high as ∼82% (with an average of ∼51%). The large number of highly disparate samples at 5 m between 2000 and 2003 (Figure 4a) prompted the analyses to test for seasonality in subsets of the 5 m data set.
Figure 5.
The averages of pairwise Bray–Curtis similarity values for all possible pairs of assemblages that occurred 1 to 24 months apart (plotted with s.e. bars). Results are shown for samples collected from 5 m between September 2000 and December 2010 (a), between January 2008 and December 2010 (b) and between September 2000 and December 2003 (c); from the DCM between September 2000 and December 2010 (d), between January 2008 and December 2010 (e) and between September 2000 and December 2003 (f); and for samples collected from 150 and 500 m between September 2000 and December 2003 (g and h, respectively). The smallest sample sizes are noted for each panel and are shown above the corresponding averages and s.e. bars. The x-axes represent the number of months lagged and the dotted lines mark approximately ‘opposite' (6 and 18 months) or ‘similar' (12 and 24 months) seasons. The y-axes represent average pairwise Bray–Curtis similarity values (%) and display the same range in each panel to facilitate comparison. Nonlinear regression resulted in a good fit of a sinusoidal pattern only for assemblages at 5 m between 2008 and 2010 (r2=0.56) (b) and for assemblages at 150 m between 2000 and 2003 (r2=0.39) (g).
Evidence of seasonality was detected for the assemblage at 5 m between January 2008 and December 2010 (r2=0.56; Figure 5b). Nonlinear regression resulted in the fit of a sinusoidal curve that resembled the seasonal SST plot in Supplementary Figure S1A. Lowest community similarity values were observed between assemblages that were 6 and 18 months apart (approximately opposite seasons), and high community similarity values returned annually (at 12- and 24-month intervals). Evidence of seasonality was not detected in the 5 m assemblage between 2000 and 2003, and the decreasing average community similarity values with increasing time apart indicate substantial interannual variability during the first ∼3 years of the study (Figure 5c).
Average community similarity values remained relatively consistent between assemblages that were collected 1 to 24 months apart at the DCM between 2000 and 2010, and did not result in evidence of seasonality (Figure 5d). Similar results were obtained for the DCM assemblage collected between January 2008 and December 2010 (Figure 5e), although the variance of average community similarity values increased with increasing time apart. Evidence of seasonality was also not detected for the DCM assemblage between September 2000 and December 2003 (Figure 5f). Averages of community similarity values for samples collected more than 12 months apart at the DCM were lower than the averages obtained for samples that were collected only 1–12 months apart (Figure 5f).
A pattern of seasonality was detected for the assemblage at 150 m between September 2000 and December 2003 (r2=0.39; Figure 5g). Evidence of seasonality was not detected for the assemblage at 500 m (Figure 5h). Assemblages that were collected only 1 month apart at 500 m resulted in the highest Bray–Curtis similarity average (∼59%), while samples that were collected more than 15 months apart resulted in the lowest community similarity averages in this hypoxic part of the water column.
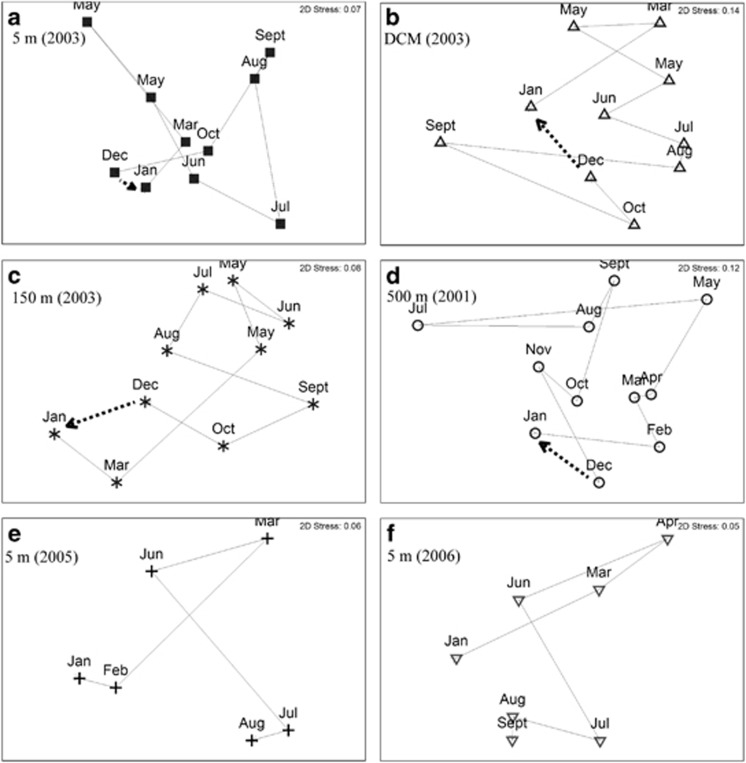
Microbial eukaryote assemblages exhibited cyclical annual patterns of similarity in at least 1 year at each depth (Table 2). The assemblage at 5 m displayed cyclicity in 5 out of the 9 years that were examined, and the assemblage at the DCM in 3 years. The cyclical pattern observed for assemblages at both 5 m and the DCM in 2003 were illustrated in MDS plots (Figures 6a and b, respectively). Assemblages at 150 and 500 m displayed cyclicity in 1 year between 2000 and 2003 (2003 and 2001, respectively) (Table 2 and Figures 6c and d). Cyclical patterns in all cases included an annual ‘resetting' of assemblages, as indicated by returning high similarity values between communities collected in December and January (a theoretical ‘resetting' of communities is indicated by dashed arrows in Figures 6a–d).
Table 2. Significant Spearman's correlations between each year at each depth and a cyclical model.
| Year |
Depth
|
|||
|---|---|---|---|---|
| 5 m | DCM | 150 m | 500 m | |
| 2001 | 0.232 (0.001) | — | — | 0.207 (0.018)* |
| 2002 | 0.3718 (0.008) | — | — | — |
| 2003 | 0.223 (0.017)* | 0.189 (0.025)* | 0.356 (0.012)* | |
| 2004 | — | 0.232 (0.024) | ||
| 2005 | — | — | ||
| 2006 | — | 0.443 (0.017) | ||
| 2008 | — | — | ||
| 2009 | 0.219 (0.035) | — | ||
| 2010 | 0.320 (0.001) | — | ||
Abbreviations: DCM, deep chlorophyll maximum; MDS, multidimensional scaling.
The cyclical model assumes a relatively equidistant distribution of assemblages around a center. Spearman's correlations are reported with P-values within parentheses. Em-dashes represent results that were not significant, and the empty cells represent no data. Years and depths that were depicted in MDS plots in Figures 6a–d are marked with asterisks.
Figure 6.
Two-dimensional MDS plots based on Bray–Curtis similarity values for assemblages of microbial eukaryotes that were collected approximately every month in 2003 from 5 m (a), the DCM (b), 150 m (c); in 2001 from 500 m (d); and in 2005 (e) and 2006 (f) from 5 m. Trajectories of relative changes in community composition from month to month are plotted on each MDS plot, and stress values are reported in the top right corner of each plot. Dashed arrows represent a theoretical resetting of assemblages for years and depths that tested significantly positive for cyclicity by the RELATE test (a–d, also see Table 2). Monthly similarities between assemblages in 2005 and 2006 at 5 m (e and f, respectively) were significantly correlated (also see Table 3).
Significant correlations (P<0.05) were detected between different years at the same depth for assemblages of microbial eukaryotes at 5 m as well as at the DCM (Table 3). Significant correlations between years were not detected for assemblages at 150 or 500 m. The strongest inter-year correlation (0.806) was observed for the assemblage at 5 m, between years 2005 and 2006 (Table 3 and Figures 6e and f). Figures 6e and f show that patterns of monthly community (dis)similarity in 2005 and 2006 at 5 m were correlated.
Table 3. Significant Spearman's correlations between microbial eukaryote assemblages from different years at the same depth were detected for assemblages at 5 m and at the DCM, but not for the assemblages at 150 or 500 m.
| Depth | Years compared | Spearman's correlation (P-value) |
|---|---|---|
| 5 m | 2002, 2006 | 0.368 (0.046) |
| 5 m | 2003, 2006 | 0.456 (0.020) |
| 5 m | 2005, 2006* | 0.806 (0.025) |
| 5 m | 2002, 2009 | 0.349 (0.047) |
| DCM | 2003, 2004 | 0.598 (0.014) |
| DCM | 2003, 2010 | 0.461 (0.020) |
Abbreviations: DCM, deep chlorophyll maximum; MDS, multidimensional scaling.
The table includes depth, years compared, Spearman's correlations and P-values within parentheses. The correlation between the monthly assemblage at 5 m in 2005 and the monthly assemblage at 5 m in 2006 (marked with an asterisk) is depicted in MDS plots (Figures 6e and f).
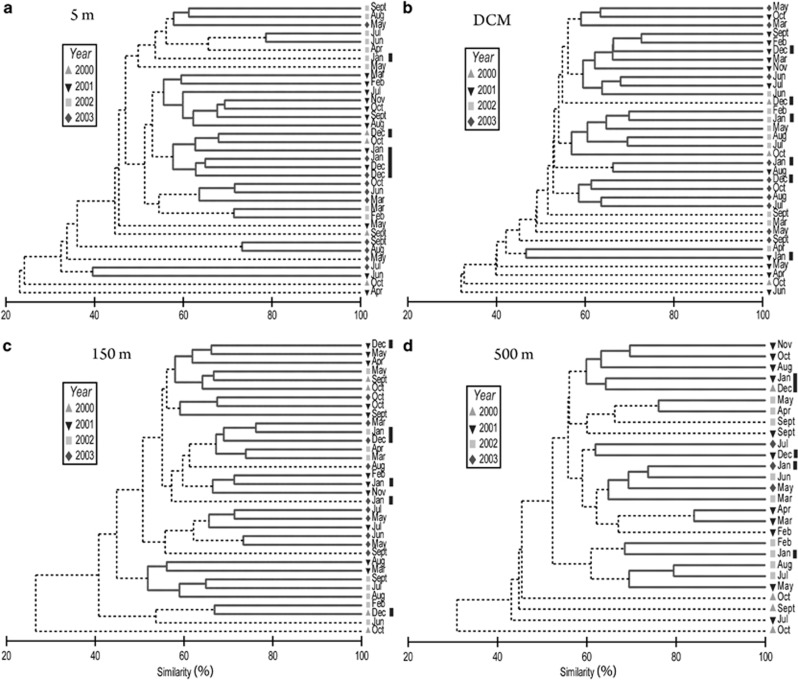
Assemblages at 5 m and the DCM between 2000 and 2003 largely clustered by year (Figures 7a and b), with the exception of assemblages in December and January at 5 m (Figure 7a, December and January samples were marked with bars to the right of the CLUSTER diagrams). Communities at 150 and 500 m (Figures 7c and d) also showed interannual variability, but the monthly assemblages did not cluster by year as tightly as observed for near-surface assemblages. The assemblage at 150 m in December and January between 2000 and 2003 shared high community similarity values (Figure 7c), however, similar to the assemblage at 5 m.
Figure 7.
CLUSTER results for assemblages collected approximately every month between September 2000 and December 2003 from 5 m (a), the DCM (b), 150 m (c) and 500 m (d), based on Bray–Curtis similarity values on square-root transformed T-RFLP data. Bray–Curtis similarity values (%) are on the x-axes and normalized. Solid lines denote differences that were not significant (P>0.05), and dotted lines denote significant differences (determined by SIMPROF). December and January samples are marked by bars to the right of each CLUSTER diagram.
Correlations between environmental variables and community dynamics
The monthly variability of microbial eukaryote assemblages at 5 m and the DCM between September 2000 and December 2003 significantly correlated with different combinations of environmental variables (Supplementary Table S1). Approximately 38% of the temporal variability of assemblages at 5 m correlated with nitrite and phosphate concentrations, salinity and temperature (P<0.01). The temporal variability of assemblages at the DCM correlated with dissolved oxygen concentrations and salinity (0.35, P<0.01). Assemblages at 150 m or 500 m did not result in significant correlations with any combination of the environmental parameters that were measured as a part of this study (Supplementary Table S1).
Discussion
The study site
The SPOT station provides an interesting coastal environment for comparison to long-term open-ocean monitoring sites such as the Hawaii Ocean Time-series (Dore et al., 2008) and Bermuda Atlantic Time-series (Steinberg et al., 2001) stations. The SPOT station is located approximately 16 km from one of the most densely populated regions of the United States as well as one of the busiest ports in the world (the port of Los Angeles, CA, USA) which contribute to a high flow of energy and carbon in the region. The San Pedro basin (maximum depth of ∼890 m) is bordered by Santa Catalina Island to the west and subsurface sills (∼740 m) to the north and south. The restricted circulation in deeper waters combined with terrigenous inputs and aerobic respiration contribute to persistent hypoxic conditions below ∼350 m, providing a unique environment for biological studies. Seasonal fluctuations in temperature during this study were relatively restricted, especially at depth (Figures 1a–d), which is not surprising for a region that experiences a Mediterranean-like climate. Despite the modest temporal fluctuations, seasonal patterns in temperature and dissolved oxygen concentration were still observed below the euphotic zone at 150 m (Figures 1c and g). Dissolved oxygen was not seasonally variable at 500 m (data not shown), and temperatures at that depth displayed limited seasonal fluctuations (∼0.5 °C; Figure 1d). The water column was stratified throughout each year, punctuated by winter mixing events to ∼50 m (Figure 1e), which is shallow compared to the oceanic time-series stations, HOT and BATS. The winter mixing events at the SPOT station entrained nutrients into surface waters that stimulated phytoplankton in the spring (Figures 1f and h). A DCM was also present throughout each year but varied seasonally in shape and depth, with summer and fall months characterized by a deeper DCM (Figure 1e).
Distinct shallow and deep assemblages of microbial eukaryotes
The spatiotemporal variability of microbial eukaryote assemblages in this study showed that the shallow (5 m and the DCM) and deeper environments (150 and 500 m) at the SPOT station were comprised of distinct communities throughout each year, presumably reflecting taxa that are specifically adapted for conditions in those environments (Figure 2). Previous studies by Countway et al. (2010) and Schnetzer et al. (2011) also showed distinct shallow and deep assemblages during four different seasons in 2001 and in June of 2001, respectively. The vertical gradients in environmental factors (e.g., light, temperature, nutrients and oxygen) observed at the SPOT station likely have important roles in shaping distinct shallow and deep microbial communities at the study site.
The relatively shallow mixed layer throughout each year (Figure 1e) limited mixing between shallow (<50 m) and deep taxa throughout the time-series study (Figure 2). The seasonal deepening of the mixed layer to ∼50 m (Figure 1e), however, did lead to higher community similarity values between assemblages at 5 m and the DCM between November and March, with the exception of January (Supplementary Figure S2). Following stratification and stabilization of the water column during periods of warming, assemblages at 5 m and the DCM became less similar and were most dissimilar when the DCM was found below the MLD (Figures 1a and e and Supplementary Figure S2). These results show how the microbial eukaryote assemblages at 5 m and the DCM at the SPOT station are seasonally coupled.
Assemblages in the euphotic zone at the SPOT site were generally dominated by arthropods, ciliates, chlorophytes, dinoflagellates, stramenopiles, telonemids and uncultured alveolates, common taxonomic groups that have been previously characterized using molecular approaches at the study site (Moorthi et al., 2006; Countway et al., 2010; Fitzpatrick et al., 2010; Schnetzer et al., 2011; Steele et al., 2011; Kim et al., 2012) (Figures 3a and b and Supplementary Figures S3A and B). Diatoms (stramenopiles) are common constituents of surface microbial communities at and near the SPOT station, and often dominate the eukaryotic biomass during spring (Venrick, 2002). In our study, however, DNA fragment lengths representative of diatoms (stramenopiles) did not show seasonal dominance. Other molecular-based studies have also reported a lack of seasonal dominance of diatoms, suggesting a potential bias or artifact in PCR-based approaches for detecting some diatom species (Potvin and Lovejoy, 2009). Dinoflagellates of the genera Ceratium and Lingulodinium are common microzooplankton grazers and phytoplankton in the region, respectively (Moorthi et al., 2006), and often dominated the T-RFLP profiles from 5 m and the DCM in the current study. Chlorophytes, including species of the genera Micromonas and Ostreococcus, were found year-round in surface waters at the SPOT station, and exhibited episodic dominance. Episodic dominance by Ostreococcus was also previously characterized at the study site using a quantitative PCR approach (Countway and Caron, 2006), which corresponded to the relative abundances of a DNA fragment length identified as Ostreococcus in the current T-RFLP data set (Kim et al., 2012). Picoeukaryotes have been detected in a wide range of geographical locations (Worden, 2006; Demir-Hilton et al., 2011) and presumably contribute significantly to biogeochemical cycles as primary producers and prey at our coastal ocean time-series station.
Deeper assemblages were largely comprised of Group II alveolates and polycystines, also consistent with previous studies at the SPOT station (Figures 3c and d and Supplementary Figures S3C and D) (Countway et al., 2010; Schnetzer et al., 2011). Group II alveolates were discovered by the application of culture-independent molecular approaches (Moreira and Lopez-Garcia, 2002). Although no cultured representative exists from this alveolate lineage, phylogenetic analyses of environmental sequences suggest that Group II alveolates are closely related to species of the genus Amoebophrya (a group consisting of parasitic alveolates) (Groisillier et al., 2006). Environmental sequences of Group II alveolates have been frequently uncovered from other deep-sea marine environments (Edgcomb et al., 2002; Stoeck and Epstein, 2003; Romari and Vaulot, 2004; Medlin et al., 2006). The results in this study show that this enigmatic group of alveolates represents a significant constituent of the microbial assemblage in the deep and hypoxic environment at the SPOT station year-round.
Polycystines are not commonly detected in deep-sea samples by microscopy, but sequences with high homology to sequences obtained from known polycystines have been detected throughout each year in the deep and hypoxic environments at the SPOT station in this study as well as in previous studies (Countway et al., 2010; Schnetzer et al., 2011). Molecular diversity surveys of deep-sea microbial eukaryotes from other locations have also reported 18S rRNA gene sequences with high sequence similarity to polycystines (Edgcomb et al., 2002; Countway et al., 2007; Not et al., 2007). The ecology of polycystines and other deep-sea dwelling microbial eukaryotes such as acantharea are not well known, but there is some evidence that some of these taxa are active members of the deep-sea community at the SPOT station (Gilg et al., 2010). The increasing evidence of a rich assemblage of deep-sea microbial eukaryotes and the potentially substantial impact they have on deep-sea biogeochemical cycles warrants further investigation.
A comparison of the overall temporal variability of microbial eukaryote assemblages at different depths
The microbial eukaryote community composition at 5 m was significantly more variable over time than assemblages at other depths (P<0.05; Figure 4e), presumably due to larger exposure to climatic fluctuations, the inherent patchiness of surface waters and the influence of mesoscale turbulence in the region (Hickey et al., 2003; Dong et al., 2009). The combinations of environmental parameters that significantly correlated with the variability of microbial eukaryote assemblages at 5 m and the DCM (Supplementary Table S1) were different, suggesting that different forcing factors may dominate in shaping the assemblages at these two near-surface environments. Abiotic and biotic factors that affect microbial eukaryotic community structure at the SPOT station continue to be investigated.
Assemblages at 500 m resulted in the highest average similarity value (Figure 4e), possibly due to temporally stable environmental conditions at that depth. Horizontal advection is restricted at depth, creating a relatively stable physical and chemical environment (Berelson, 1991; Hickey, 1991). Sampling from a consistent biological feature (the DCM) and at 150 m, where seasonal fluctuations in environmental conditions were dampened (Figures 1c and g), most likely contributed to the intermediate averages obtained for assemblages at the DCM and 150 m (Figure 4e).
Month-to-month variability and seasonality of microbial eukaryote assemblages
Month-to-month changes in the composition of microbial eukaryote assemblages at the SPOT station were generally modest compared with other studies that have characterized substantial shifts in community composition within time-frames of hours to weeks (1-month time lag in Figures 5a–h) (Countway et al., 2005; Vigil et al., 2009; Kim et al., 2011; Weber et al., 2012). Vigil et al. (2009) used T-RFLP to examine shifts in microbial eukaryote assemblages in estuarine ecosystems and showed that community similarity values ranged between ∼20 and <70% on timescales of days to weeks. Similarity values between assemblages from adjacent months in the current study were as high as ∼82%, which is not surprising, considering the small seasonal changes in environmental parameters observed at the station, especially at depth (Figures 1a–h). Molecular fingerprinting assays of the surface bacterial community at the SPOT station using automated ribosomal intergenic spacer analysis (ARISA) have also shown similarity values of over 80% between bacterial assemblages collected 1 month apart during some parts of the year (Chow et al., accepted).
Evidence of seasonality was detected for assemblages at 5 m in samples collected between 2008 and 2010 (Figure 5b), but not between 2000 and 2003 (Figure 5c). Countway et al. (2010) documented distinct assemblages of microbial eukaryotes at the study site during different seasons in 2001. The findings in this study show that the microbial eukaryote assemblages that were collected 1 to 6 months apart between 2000 and 2003 became increasingly dissimilar, reflecting a gradual seasonal change in composition. The increasingly lower averages in community similarity values with increasing time apart (>12 months) during the first ∼3 years of the study, however, also indicate that interannual differences were a large source of variability during that period. The assemblage at 5 m between 2008 and 2010 also became increasingly dissimilar with more time apart up to 6 months, but returned to a similar assemblage annually. An annual resetting of community composition at 5 m is mostly likely attributed to winter mixing and light-limiting conditions. Similar seasonal and annually reoccurring patterns were also reported for the surface bacterial community at the SPOT station (Fuhrman and Steele, 2008). Seasonality was not detected for the microbial eukaryote assemblage at the DCM in this study (Figures 5d–f), which was possibly a consequence of sampling from a persistent biological feature in the water column that varied in depth throughout each year (Figure 1e).
Assemblages of microbial eukaryotes below the euphotic zone at 150 m appeared to be seasonal, based on the analysis of samples that were collected between September 2000 and December 2003 (Figure 5g). To our knowledge, this is the first study to demonstrate seasonality for microbial eukaryote assemblages below the euphotic zone and within an oxycline. The seasonal pattern in community compositional changes is not surprising, considering the seasonal variability in temperature and oxygen concentration observed at that depth (Figures 1c and g). In addition, 150 m is located near the base of the euphotic zone and presumably experiences seasonal fluxes of dissolved and particulate materials from surface waters. Surprisingly, however, significant correlations between environmental parameters and community dynamics at 150 m were not observed (Supplementary Table S1). It is possible that biotic factors (e.g., ecological relationships between taxa) have a more significant role in shaping microbial eukaryote communities in the dark and relatively nutrient-rich environment at 150 m than the abiotic environmental variables that were measured in the current study. Assemblages at 500 m did not exhibit seasonality (Figure 5h), which may be a result of the physically and chemically stable environment at that depth. Assemblages of microbial eukaryotes were cyclical in only 1 year at both 150 and 500 m (Table 2), implying an annual resetting of communities at these depths, but it is not clear what environmental conditions contributed to the annual resetting of assemblages in these chronically dark and cold environments.
Interannual variability of microbial eukaryote assemblages
There were considerable interannual differences in the community composition of microbial eukaryotes at each depth, especially in surface waters (5 m) between September 2000 and December 2003 at the study site (Figures 7a–d). Assemblages at each depth exhibited an annual resetting of communities in some but not all years (Table 2), and correlations between years from the same depth were found in only a subset of our time-series data (Table 3). Sources of interannual variability in the region include influences by the colder and more nutrient-rich waters of the California Current as well as the warmer and relatively nutrient-poor countercurrent when trade winds are weakened in the summer (Hickey et al., 2003; Dong et al., 2009). The station is also surrounded by a complex seafloor topography and the Channel Islands that influence the circulation patterns in and around the station (Dong and McWilliams, 2007). Local and remote climatic events and wind stress also affect circulation patterns as well as mixing of the water column in this coastal ecosystem (Hayward, 2000) (although only relatively weak El Niño and La Niña events were detected during this study, data not shown). The SPOT station is also located approximately 16 km away from one of the largest urban centers and busiest seaports in the world, and can experience substantial terrestrial input following large storm events (Thunell et al., 1994).
Conclusion
Strong selective forces maintained distinct assemblages of microbial eukaryotes between shallow (<50 m) and deep environments. In general, month-to-month changes were small at all depths, but assemblages at 5 m displayed the most temporal variability overall. Evidence of seasonality was detected for microbial eukaryote assemblages at 5 m between January 2008 and December 2010, as well as at 150 m between January 2000 and December 2003. Assemblages at each depth showed cyclical patterns in at least 1 year throughout this study, suggesting an annual ‘resetting' of the communities at specific depths. Interannual differences were substantial for microbial eukaryote assemblages at all depths and represented the dominant source of temporal variability in this temperate ecosystem.
Acknowledgments
We thank the Wrigley Institute for Environmental Studies for providing ship time and the environmental data used in this study. Troy Gunderson was instrumental in sampling, CTD operations and processing of environmental samples. We also thank the captain and crew of the R/V Seawatch and R/V Yellowfin for their help on cruises. Jed Fuhrman and members of his lab provided helpful comments and discussions. Victoria Campbell, Jacob Cram, Alyssa Gellene, Ian Hewson, Alle Lie, David Needham, Alma Parada, Anand Patel and Joshua Steele helped with sample collection and processing of samples. This work was funded by NSF Grants MCB-0703159, MCB-0084231 and OCE-1136818.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aguilera A, Zettler E, Gomez F, Amaral-Zettler L, Rodriguez N, Amils R. Distribution and seasonal variability in the benthic eukaryotic community of Rio Tinto (SW, Spain), an acidic, high metal extreme environment. Syst Appl Microbiol. 2007;30:531–546. doi: 10.1016/j.syapm.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Balzano S, Marie D, Gourvil P, Vaulot D. Composition of the summer photosynthetic pico and nanoplankton communities in the Beaufort Sea assessed by T-RFLP and sequences of the 18S rRNA gene from flow cytometry sorted samples. ISME J. 2012;6:1480–1498. doi: 10.1038/ismej.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beman JM, Sachdeva R, Fuhrman JA. Population ecology of nitrifying Archaea and Bacteria in the Southern California Bight. Environ Microbiol. 2010;12:1282–1292. doi: 10.1111/j.1462-2920.2010.02172.x. [DOI] [PubMed] [Google Scholar]
- Berelson WM. The flushing of 2 deep-sea basins, Southern California borderland. Limnol Oceanogr. 1991;36:1150–1166. [Google Scholar]
- Caron DA, Countway PD, Jones AC, Kim DY, Schnetzer A. Marine protistan diversity. Annu Rev Mar Sci. 2012;4:467–493. doi: 10.1146/annurev-marine-120709-142802. [DOI] [PubMed] [Google Scholar]
- Chapin FS, III, Zavaleta ES, Eviner VT, Naylor RL, Vitousek PM, Reynolds HL, et al. Consequences of changing biodiversity. Nature. 2000;405:232–242. doi: 10.1038/35012241. [DOI] [PubMed] [Google Scholar]
- Chow CET, Kim DY, Sachdeva R, Caron DA, Fuhrman JA.Top-down controls on bacterial community structure: microbial network analysis of bacterial, viral and protistan communities ISME J (accepted). [DOI] [PMC free article] [PubMed]
- Clarke KR. Nonparametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–143. [Google Scholar]
- Clarke KR, Gorley RN. PRIMER-E. Plymouth; 2006. PRIMER v6: user manual/tutorial. [Google Scholar]
- Countway PD, Caron DA. Abundance and distribution of Ostreococcus sp. in the San Pedro Channel, California, as revealed by quantitative PCR. Appl Environ Microbiol. 2006;72:2496–2506. doi: 10.1128/AEM.72.4.2496-2506.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Countway PD, Gast RJ, Dennett MR, Savai P, Rose JM, Caron DA. Distinct protistan assemblages characerize the euphotic zone and deep sea (2500 m) of the western North Atlantic (Sargasso Sea and Gulf Stream) Environ Microbiol. 2007;9:1219–1232. doi: 10.1111/j.1462-2920.2007.01243.x. [DOI] [PubMed] [Google Scholar]
- Countway PD, Gast RJ, Savai P, Caron DA. Protistan diversity estimates based on 18S rDNA from seawater incubations in the western North Atlantic. J Eukaryot Microbiol. 2005;52:95–106. doi: 10.1111/j.1550-7408.2005.05202006.x. [DOI] [PubMed] [Google Scholar]
- Countway PD, Vigil PD, Schnetzer A, Moorthi SD, Caron DA. Seasonal analysis of protistan community structure and diversity at the USC Microbial Observatory (San Pedro Channel, North Pacific Ocean) Limnol Oceanogr. 2010;55:2381–2396. [Google Scholar]
- Demir-Hilton E, Sudek S, Cuvelier ML, Gentemann CL, Zehr JP, Worden AZ. Global distribution patterns of distinct clades of the photosynthetic picoeukaryote Ostreococcus. ISME J. 2011;5:1095–1107. doi: 10.1038/ismej.2010.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CM, Idica EY, McWilliams JC. Circulation and multiple-scale variability in the Southern California Bight. Prog Oceanogr. 2009;82:168–190. [Google Scholar]
- Dong CM, McWilliams JC. A numerical study of island wakes in the Southern California Bight. Cont Shelf Res. 2007;27:1233–1248. [Google Scholar]
- Dore JE, Letelier RM, Church MJ, Lukas R, Karl DM. Summer phytoplankton blooms in the oligotrophic North Pacific Subtropical Gyre: historical perspective and recent observations. Prog Oceanogr. 2008;76:2–38. [Google Scholar]
- Edgcomb V, Orsi W, Bunge J, Jeon S, Christen R, Leslin C, et al. Protistan microbial observatory in the Cariaco Basin, Caribbean. I. Pyrosequencing vs Sanger insights into species richness. ISME J. 2011;5:1344–1356. doi: 10.1038/ismej.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgcomb VP, Kysela DT, Teske A, Gomez AD, Sogin ML. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc Natl Acad Sci USA. 2002;99:7658–7662. doi: 10.1073/pnas.062186399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egert M, Friedrich MW. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl Environ Microbiol. 2003;69:2555–2562. doi: 10.1128/AEM.69.5.2555-2562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick E, Caron DA, Schnetzer A. Development and environmental application of a genus-specific quantitative PCR approach for Pseudo-nitzschia species. Mar Biol. 2010;157:1161–1169. [Google Scholar]
- Fuhrman JA, Steele JA. Community structure of marine bacterioplankton: patterns, networks, and relationships to function. Aquat Microb Ecol. 2008;53:69–81. [Google Scholar]
- Gilg IC, Amaral-Zettler LA, Countway PD, Moorthi S, Schnetzer A, Caron DA. Phylogenetic affiliations of Mesopelagic Acantharia and Acantharian-like environmental 18S rRNA genes off the Southern California Coast. Protist. 2010;161:197–211. doi: 10.1016/j.protis.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Gordon LI, Jennings JJC, Ross AA, Krest JM.1993. A Suggested Protocol for Continuous Flow Automated Analysis of Seawater Nutrients (Phosphate, Nitrate, Nitrite And Silicic Acid) in the WOCE Hydrographic Program and the Joint Global Ocean Fluxes Study, WHP Operations And Methods.WOCE Hydrographic Program Office,College of Oceanic and Atmospheric Sciences Oregon State University: Corvallis, Oregon p 52.
- Grasshoff K, Kremling K, Ehrhardt M.2007Methods of Seawater Anlaysis3 edn.Wiley-VCH GmbH: Weinheim Germany [Google Scholar]
- Groisillier A, Massana R, Valentin K, Vaulotl D, Guilloul L. Genetic diversity and habitats of two enigmatic marine alveolate lineages. Aquat Microb Ecol. 2006;42:277–291. [Google Scholar]
- Hamersley MR, Turk KA, Leinweber A, Gruber N, Zehr JP, Gunderson T, et al. Nitrogen fixation within the water column associated with two hypoxic basins in the Southern California Bight. Aquat Microb Ecol. 2011;63:193. [Google Scholar]
- Hayward TL. El Nino 1997-98 in the coastal waters of Southern California: a timeline of events. Cal Coop Ocean Fish. 2000;41:98–116. [Google Scholar]
- Hickey BM. Variability in 2 deep coastal basins (Santa Monica and San Pedro) off Southern California. J Geophys Res (Oceans) 1991;96:16689–16708. [Google Scholar]
- Hickey BM, Dobbins EL, Allen SE.2003Local and remote forcing of currents and temperature in the central Southern California Bight J Geophys Res (Oceans) 108doi:101029/2000JC000313. [Google Scholar]
- Hinder SL, Hays GC, Edwards M, Roberts EC, Walne AW, Gravenor MB. Changes in marine dinoflagellate and diatom abundance under climate change. Nat Clim Change. 2012;2:271–275. [Google Scholar]
- Hooker SB, McClain CR. The calibration and validation of SeaWiFS data. Prog Oceanogr. 2000;45:427–465. [Google Scholar]
- Kaplan CW, Kitts CL. Bacterial succession in a petroleum land treatment unit. Appl Environ Microbiol. 2004;70:1777–1786. doi: 10.1128/AEM.70.3.1777-1786.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Countway PD, Gast RJ, Caron DA. Rapid shifts in the structure and composition of a protistan assemblage during bottle incubations affect estimates of total protistan species richness. Microb Ecol. 2011;62:383–398. doi: 10.1007/s00248-011-9816-9. [DOI] [PubMed] [Google Scholar]
- Kim DY, Countway PD, Yamashito W, Caron DA. A combined sequence-based and fragment-based characterization of microbial eukaryote assemblages provides taxonomic context for the Terminal Restriction Fragment Length Polymorphism (T-RFLP) method. J Microbiol Meth. 2012;91:527–536. doi: 10.1016/j.mimet.2012.09.026. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Miller AJ, McGowan J, Carter ML. Coastal phytoplankton blooms in the Southern California Bight. Prog Oceanogr. 2009;82:137–147. [Google Scholar]
- Lepere C, Boucher D, Jardillier L, Domaizon I, Debroas D. Succession and regulation factors of small eukaryote community. composition in a lacustrine ecosystem (Lake pavin) Appl Environ Microbiol. 2006;72:2971–2981. doi: 10.1128/AEM.72.4.2971-2981.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitus S.1982Climatological Atlas of the World OceanNOAA Professional Paper 13U.S. Gov. Printing Office, Rockville, MD; pp 190. [Google Scholar]
- Liu WT, Marsh TL, Cheng H, Forney LJ. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, Balague V, Guillou L, Pedros-Alio C. Picoeukaryotic diversity in an oligotrophic coastal site studied by molecular and culturing approaches. FEMS Microbiol Ecol. 2004;50:231–243. doi: 10.1016/j.femsec.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Massana R, Pedros-Alio C. Unveiling new microbial eukaryotes in the surface ocean. Curr Opin Microbiol. 2008;11:213–218. doi: 10.1016/j.mib.2008.04.004. [DOI] [PubMed] [Google Scholar]
- McGradySteed J, Harris PM, Morin PJ. Biodiversity regulates ecosystem predictability. Nature. 1997;390:162–165. [Google Scholar]
- Medlin LK, Elwood HJ, Stickel S, Sogin ML. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Genetica. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- Medlin LK, Metfies K, Mehl H, Wiltshire K, Valentin K. Picoeukaryotic plankton diversity at the Helgoland time series site as assessed by three molecular methods. Microb Ecol. 2006;52:53–71. doi: 10.1007/s00248-005-0062-x. [DOI] [PubMed] [Google Scholar]
- Moorthi SD, Countway PD, Stauffer BA, Caron DA. Use of quantitative real-time PCR to investigate the dynamics of the red tide dinoflagellate Lingulodinium polyedrum. Microb Ecol. 2006;52:136–150. doi: 10.1007/s00248-006-9030-3. [DOI] [PubMed] [Google Scholar]
- Moreira D, Lopez-Garcia P. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 2002;10:31–38. doi: 10.1016/s0966-842x(01)02257-0. [DOI] [PubMed] [Google Scholar]
- Naeem S, Li SB. Biodiversity enhances ecosystem reliability. Nature. 1997;390:507–509. [Google Scholar]
- Nolte V, Pandey RV, Jost S, Medinger R, Ottenwalder B, Boenigk J, et al. Contrasting seasonal niche separation between rare and abundant taxa conceals the extent of protist diversity. Mol Ecol. 2010;19:2908–2915. doi: 10.1111/j.1365-294X.2010.04669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Not F, Gausling R, Azam F, Heidelberg JF, Worden AZ. Vertical distribution of picoeukaryotic diversity in the Sargasso Sea. Environ Microbiol. 2007;9:1233–1252. doi: 10.1111/j.1462-2920.2007.01247.x. [DOI] [PubMed] [Google Scholar]
- Orsi W, Edgcomb V, Jeon S, Leslin C, Bunge J, Taylor GT, et al. Protistan microbial observatory in the Cariaco Basin, Caribbean. II. Habitat specialization. ISME J. 2011;5:1357–1373. doi: 10.1038/ismej.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn AM, Moore ERB, Timmis KN. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ Microbiol. 2000;2:39–50. doi: 10.1046/j.1462-2920.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- Piwosz K, Pernthaler J. Seasonal population dynamics and trophic role of planktonic nanoflagellates in coastal surface waters of the Southern Baltic Sea. Environ Microbiol. 2010;12:364–377. doi: 10.1111/j.1462-2920.2009.02074.x. [DOI] [PubMed] [Google Scholar]
- Potvin M, Lovejoy C. PCR-based diversity estimates of artificial and environmental 18S rRNA gene libraries. J Eukaryot Microbiol. 2009;56:174–181. doi: 10.1111/j.1550-7408.2008.00386.x. [DOI] [PubMed] [Google Scholar]
- Romari K, Vaulot D. Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol Oceanogr. 2004;49:784–798. [Google Scholar]
- Rossi P, Gillet F, Rohrbach E, Diaby N, Holliger C. Statistical assessment of variability of terminal restriction fragment length polymorphism analysis applied to complex microbial communities. Appl Environ Microbiol. 2009;75:7268–7270. doi: 10.1128/AEM.00135-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetzer A, Moorthi SD, Countway PD, Gast RJ, Gilg IC, Caron DA. Depth matters: microbial eukaryote diversity and community structure in the eastern North Pacific revealed through environmental gene libraries. Deep Sea Res I. 2011;58:16–26. [Google Scholar]
- Schutte UME, Abdo Z, Bent SJ, Shyu C, Williams CJ, Pierson JD, et al. Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl Microbiol Biot. 2008;80:365–380. doi: 10.1007/s00253-008-1565-4. [DOI] [PubMed] [Google Scholar]
- Sherr BF, Sherr EB, Caron DA, Vaulot D, Worden AZ. Oceanic protists. Oceanography. 2007;20:130–134. [Google Scholar]
- Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY, et al. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 2011;5:1414–1425. doi: 10.1038/ismej.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg DK, Carlson CA, Bates NR, Johnson RJ, Michaels AF, Knap AH. Overview of the US JGOFS Bermuda Atlantic Time-series Study (BATS): a decade-scale look at ocean biology and biogeochemistry. Deep Sea Res II. 2001;48:1405–1447. [Google Scholar]
- Stoeck T, Epstein S. Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Appl Environ Microbiol. 2003;69:2657–2663. doi: 10.1128/AEM.69.5.2657-2663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thunell RC, Pilskaln CH, Tappa E, Sautter LR. Temporal variability in sediment fluxes in the San-Pedro Basin, Southern California Bight. Cont Shelf Res. 1994;14:333–352. [Google Scholar]
- Vaulot D, Eikrem W, Viprey M, Moreau H. The diversity of small eukaryotic phytoplankton (<=3 mu m) in marine ecosystems. FEMS Microbiol Rev. 2008;32:795–820. doi: 10.1111/j.1574-6976.2008.00121.x. [DOI] [PubMed] [Google Scholar]
- Venrick EL. Spring in the California current: the distribution of phytoplankton species, April 1993 and April 1995. Mar Ecol Prog Ser. 1998;167:73–88. [Google Scholar]
- Venrick EL. Floral patterns in the California current system off southern California: 1990–1996. J Mar Res. 2002;60:171–189. [Google Scholar]
- Vigil P, Countway PD, Rose J, Lonsdale DJ, Gobler CJ, Caron DA. Rapid shifts in dominant taxa among microbial eukaryotes in estuarine ecosystems. Aquat Microb Ecol. 2009;54:83–100. [Google Scholar]
- Weber F, Del Campo J, Wylezich C, Massana R, Jurgens K. Unveiling trophic functions of uncultured protist taxa by incubation experiments in the brackish Baltic Sea. PLoS One. 2012;7:e41970. doi: 10.1371/journal.pone.0041970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weekers PHH, Gast RJ, Fuerst PA, Byers TJ. Sequence variations in small-subunit ribosomal-RNAs of Hartmannella vermiformis and their phylogenetic implications. Mol Biol Evol. 1994;11:684–690. doi: 10.1093/oxfordjournals.molbev.a040147. [DOI] [PubMed] [Google Scholar]
- Worden AZ. Picoeukaryote diversity in coastal waters of the Pacific Ocean. Aquat Microb Ecol. 2006;43:165–175. [Google Scholar]
- Wu XQ, Menzel WP, Wade GS. Estimation of sea surface temperatures using GOES-8/9 radiance measurements. B Am Meteorol Soc. 1999;80:1127–1138. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.