Abstract
The concentrations of atmospheric carbon dioxide (CO2) and tropospheric ozone (O3) have been rising due to human activities. However, little is known about how such increases influence soil microbial communities. We hypothesized that elevated CO2 (eCO2) and elevated O3 (eO3) would significantly affect the functional composition, structure and metabolic potential of soil microbial communities, and that various functional groups would respond to such atmospheric changes differentially. To test these hypotheses, we analyzed 96 soil samples from a soybean free-air CO2 enrichment (SoyFACE) experimental site using a comprehensive functional gene microarray (GeoChip 3.0). The results showed the overall functional composition and structure of soil microbial communities shifted under eCO2, eO3 or eCO2+eO3. Key functional genes involved in carbon fixation and degradation, nitrogen fixation, denitrification and methane metabolism were stimulated under eCO2, whereas those involved in N fixation, denitrification and N mineralization were suppressed under eO3, resulting in the fact that the abundance of some eO3-supressed genes was promoted to ambient, or eCO2-induced levels by the interaction of eCO2+eO3. Such effects appeared distinct for each treatment and significantly correlated with soil properties and soybean yield. Overall, our analysis suggests possible mechanisms of microbial responses to global atmospheric change factors through the stimulation of C and N cycling by eCO2, the inhibition of N functional processes by eO3 and the interaction by eCO2 and eO3. This study provides new insights into our understanding of microbial functional processes in response to global atmospheric change in soybean agro-ecosystems.
Keywords: microbial responses/feedbacks, soil microbial community, elevated CO2, elevated O3, functional genes, soybean/SoyFACE, agro-ecosystem
Introduction
Because of fossil fuel combustion and land-use changes, the global atmospheric carbon dioxide (CO2) concentration has increased by more than 30% since the industrial revolution, and is projected to reach 700 p.p.m. by 2100 (IPCC, 2007). In addition, the average O3 concentration is expected to increase by 1–2% per year and reach 70 p.p.b. by 2100 (IPCC, 2007; Sitch et al., 2007). Such increases will be more rapid and have significant impacts on plant productivity, soil carbon and nitrogen dynamics and ecosystem functioning if anthropological activities continue unabated in the future (IPCC, 2007). Soybean, one of the largest food crops with an annual world production of >250 million metric tons in 2012 (http://www.soystats.com/2012/page_30.htm), is sensitive to O3 (Morgan et al., 2003). The productivity/yield of soybean largely depends on the ecosystem functional processes, such as nutrient cycling (for example, carbon and nitrogen fixation, nitrification, denitrification and methane cycling) and residue decomposition governed by microbial communities and their dynamics (Meriles et al., 2009). Therefore, toward sustainable agro-ecosystems, it is necessary to understand the effect of elevated CO2 (eCO2) and elevated O3 (eO3) on the functional diversity, composition, structure, metabolic potential and dynamics of soil microbial communities and their linkages with ecosystem functioning.
CO2 fertilization effects are well established by increased plant photosynthesis, growth, resource allocation and altered ecosystem functions (Reich et al., 2001; Ainsworth et al., 2002; Ainsworth and Long, 2005; Luo et al., 2006; Reich, 2009; Lindroth, 2010; Drake et al., 2011; van Groenigen et al., 2011; Zak et al., 2011; Biswas et al., 2013; Kumari et al., 2013; Twine et al., 2013), but the magnitude of eCO2 stimulation may be constrained by the nitrogen (N) supply (Zak et al., 2003; Reich et al., 2006) because of progressive N limitation under eCO2 (Luo et al., 2004; Reich et al., 2006). In contrast, O3 is a phytotoxic compound potentially suppressing crop yields and reducing above-ground plant growth, and more importantly, it may reduce below-ground functional processes, such as root growth, carbon (C) allocation and soil N status (Morgan et al., 2003; Feng and Kobayashi, 2009; Betzelberger et al., 2010; Lindroth, 2010; Zak et al., 2012; Biswas et al., 2013; Kumari et al., 2013). Under eCO2 and eO3, it has been suggested that CO2 might alleviate the negative effects of O3 on the above-ground processes by a decrease of O3 flux into leaves and by an increase of available photosynthates that may be used for detoxification processes (Allen, 1990; McKee et al., 1997; Volin et al., 1998). However, the impact of eCO2 and/or eO3 on the functional diversity, composition, structure and function of soil microbial communities is poorly understood. For example, no detectable effects of eCO2 on microbial community structure, microbial activity, potential soil N mineralization or nitrification were observed at a sweetgum free-air CO2 enrichment (FACE) experiment in TN, USA (Austin et al., 2009), whereas in a no-till wheat-soybean rotation agro-ecosystem, the community composition and structure significantly affected by eCO2 but not by eO3 or eCO2+eO3 (Cheng et al., 2011). Recently, more studies suggest that eCO2 and/or eO3 significantly alter microbial community composition, structure, functional potential/activity, interaction network and/or dynamics (Lesaulnier et al., 2008; Blagodatskaya et al., 2010; Drigo et al., 2010; Feng et al., 2010; Zhou et al., 2010, 2011; He et al., 2010b, 2012b; Deng et al., 2012; Drigo et al., 2013; Li et al., 2013). In addition, it has been shown that the response of soil microbial comunities to global change factors may be directly or indirectly mediated by plant genotypes/cultivars, the diversity of plant assemblages and/or other environmental factors (Talhelm et al., 2009; Singh et al., 2010; Drigo et al., 2013; Li et al., 2013). In addition, the effect of multiple global change factors (for example, eCO2, eO3, warming and precipitation) and their interactions on microbial communities might be highly uncertain (Castro et al., 2010; Gutknecht et al., 2012). Therefore, it is imporant to comprehensively examine the effect of eCO2 and eO3 on soil micorbial communities.
The advances of metagenomic technologies such as high-throughput sequencing (Margulies et al., 2005; Caporaso et al., 2012; Loman et al., 2012) and functional gene arrays (He et al., 2007, 2010a, 2012a) have revolutionized our analysis of microbial communities. For example, GeoChip 3.0, containing about 28 000 probes and covering about 57 000 genes in 292 functional groups, such as those involved in biogeochemical cycling of C, N, sulfur (S) and phosphorus (P) (He et al., 2010a), has been applied to analyze soil micorbial communities from various experimental sites (He et al., 2010b; Zhou et al., 2012; Li et al., 2013) and other habitats (He et al., 2012c). All results have demonstrated that GeoChip is a powerful tool to study the functional diversity, composition, structure and metabolic potential of microbial communities and link the microbial community structure to ecosystem functioning.
In this study, we hypothesize: (i) that the functional composition and structure of the soil microbial community would alter via changes in soil C and N inputs and soil chemistry (Dijkstra et al., 2005; Adair et al., 2009); and (ii) that various microbial functional groups (for example, C fixers, C degraders, N fixers and denitrifiers) would respond differentially due to changes in nutritional groups of microorganisms, leading to microbial utilization of more complex organic matter and C economy by plants and microorganisms (Elhottova et al., 1997) in response to eCO2 and/or eO3. To test these hypotheses, GeoChip 3.0 (He et al., 2010a) was used to detect functional genes and their associated populations. This study was conducted in a soybean FACE (SoyFACE) experimental site in Champaign, IL, USA. SoyFACE provides several advantages: (i) strong background knowledge about the effect of eCO2 and eO3 on soybean physiology, growth, yield and stress responses (Ainsworth et al., 2002; Morgan et al., 2003; Ainsworth and Long, 2005; Morgan et al., 2005; Rogers et al., 2009; Betzelberger et al., 2010); (ii) eliminating effects of plant diversity on soil microbial communities; and (iii) minimizing progressive N limitation at eCO2 with indeterminate and nodulating soybean. The results showed the functional composition, structure and metabolic potential of microbial communities significantly altered under eCO2, eO3 and eCO2+eO3, and such changes appeared distinct for each treatment and significantly correlated with soil properties and soybean yield. This study has important implications for microbial responses to global change in soybean agro-ecosystems.
Materials and methods
This is a summary description of experimental site, sampling and methods used in this study. More detailed information is provided in Supporting Information A.
Site description and sampling
This study was conducted at the SoyFACE experimental site in Champaign, IL, USA (40°02′N, 88°14′W) (http://www.soyface.uiuc.edu/index.htm) in 2008. The 32-h SoyFACE experiment was established on a farmland that had been cultivated with an annual rotation of soybean, Glycine max (L.) Merr. and corn, Zea mays L. for more than 25 years, and the soil at the site is a Drummer fine-silty, mixed, mesic Typic Endoaquoll, typical of wet, dark-colored ‘prairie soils' in northern and central Illinois (Pujol Pereira et al., 2011). More soil background properties, including soil pH, moisture, Bray P, K, Ca and Mg were previously documented (Peralta and Wander, 2008). SoyFACE aims to discover the effects of atmospheric change on the agronomy, productivity and ecology of Midwestern agro-ecosystems planted in a typical corn-soy rotation. The experiment was a randomized complete block design (n=4) with each block containing four treatments: (i) ambient (with ∼400 p.p.m. CO2 and ∼37.9 p.p.b. O3); (ii) eCO2 (∼550 p.p.m.); (iii) eO3 (∼ 61.3 p.p.b.); and (iv) eCO2+eO3 (∼550 p.p.m. CO2 and ∼61.3 p.p.b. O3). A total of 96 soil samples were collected in 2008 October from four soybean (Glycine max Merr.) plots under each of four treatments at both surface soil (0–5 cm) and subsoil (5–15 cm) layers with 48 samples for each soil layer, and 12 samples (three subsamples for each plot) for each treatment. All soil samples were immediately transferred to the laboratory and stored at −80 °C or 4 °C until DNA extraction or soil property analyses.
Crop yield and soil property analysis
Annual crop yield and soil property analyses were collected for each plot as previously described (Twine et al., 2013). To estimate the historical effects of eCO2 on seed yield production before the time of our sampling, soybean yield data from 2004 to 2006 were averaged. Soil NO3-N and NH4-N were extracted with 1.0 M KCl solution and quantified by a Flow Injection Autoanalyzer. Soil organic C and total N were determined using a LECO Truspec dry combustion carbon analyzer.
DNA extraction, amplification, target preparation and microarray hybridization
For each sample, soil microbial community DNA was extracted and purified as described previously (Zhou et al., 1996), and all DNA samples met the criteria: 260 nm/280 nm>1.70, and 260 nm/230 nm>1.8. Each purified DNA (50 ng) was first amplified using whole community genomic amplification (Wu et al., 2006), and 3.0 μg of amplified DNA was labeled and hybridized with GeoChip 3.0 on an HS4800 Pro Hybridization Station (Tecan US, Durham, NC, USA) at 42 °C for 16 h (He et al., 2010b).
GeoChip imaging, data processing and statistical analysis
Hybridized GeoChips were analyzed as previously described (He and Zhou, 2008; He et al., 2010b). A gene with a minimum of three positives out of 12 replicates for each treatment was considered positive and used for further statistical analyses. Permutational multivariate analysis of variance (PERMANOVA) was used to evaluate the contribution of various factors to microbial community variations with the Adonis function, and to partition sums of squares from a centroid based on a Bray-Curtis dissimilarity matrix implemented in R (R Development Core Team, 2011). Significance tests among four treatments were analyzed by multi-way analysis of variance (Geladi, 1989). Detrended correspondence analysis was used to determine the overall change in microbial community structures (He et al., 2010b).
Results
Effects of eCO2, eO3 and eCO2+eO3 on crop yield and soil properties
Historical soybean yields were significantly (P<0.05) greater by ∼14% and ∼12% under eCO2 and eCO2+eO3, respectively, whereas eO3 only caused an insignificant decrease (∼2.3%) in soybean yield (Table 1). Concentrations of soil nitrate (NO3-N), ammonium (NH4-N) and total nitrogen (TN) as well as soil total carbon (TC) and the TC/TN ratio (C/N) showed three different trends in both surface soil and subsoil samples. First, NO3-N was significantly (P<0.05) higher in the subsoil under eO3 and eCO2+eO3 compared with ambient or eCO2 samples but not significantly different between ambient and eCO2, or eO3 and eCO2+eO3. Second, NH4-N was significantly (P<0.05) lower in the surface soil under eO3 compared with ambient but not significantly different among eCO2, eO3 and eCO2+eO3. Third, NO3-N was significantly (P<0.05) different between two soil depths under ambient, eCO2 or eO3 despite no significant difference under eCO2+eO3. However, no significant changes were seen in TN, TC or C/N among different treatments or between two soil depths (Table 1). These results indicated that eCO2 and/or eO3 affected crop yield and soil N status.
Table 1. Effects of eCO2, eO3 and eCO2+eO3 on crop yield and soil properties at both surface soil and subsoil.
| Relative yield (%)a | Depth (cm) | NO3-N (lb/a) | NH4-N (lb/a) | TN (%) | TC (%) | C/N | |
|---|---|---|---|---|---|---|---|
| Ambient | 100.00b | 0–5 | 18.1±8.16A | 8.0±1.43A | 0.20±0.021A | 2.6±0.34A | 12.5±0.86A |
| 5–15 | 11.6±1.73b | 8.9±4.00a | 0.19±0.021a | 2.4±0.39a | 12.4±0.89a | ||
| P | 0.0131 | 0.4707 | 0.2559 | 0.1942 | 0.7822 | ||
| eCO2 | 113.97a | 0–5 | 17.2±4.73A | 7.4±1.84AB | 0.21±0.031A | 2.7±0.44A | 12.9±0.56A |
| 5–15 | 12.6±2.85b | 6.8±1.25a | 0.22±0.040a | 2.8±0.69a | 12.7±4.00a | ||
| P | 0.0086 | 0.3603 | 0.5008 | 0.6762 | 0.8654 | ||
| eO3 | 97.67b | 0–5 | 20.3±7.88A | 6.2±1.53B | 0.22±0.056A | 2.8±0.87A | 12.6±0.88A |
| 5–15 | 14.8±2.92a | 7.0±1.25a | 0.21±0.033a | 2.6±0.41a | 12.8±1.10a | ||
| P | 0.0336 | 0.1747 | 0.5994 | 0.4789 | 0.6277 | ||
| eCO2+eO3 | 112.03a | 0–5 | 17.1±5.26A | 7.5±1.47AB | 0.20±0.009A | 2.5±0.23A | 12.5±0.79A |
| 5–15 | 16.5±4.67a | 7.8±1.82a | 0.21±0.039a | 2.6±0.57a | 12.5±0.84a | ||
| P | 0.7704 | 0.6612 | 0.3961 | 0.5787 | 1.0000 |
Abbreviations: ANOVA, analysis of variance; eCO2, elevated CO2; eO3, elevated O3; TN, total nitrogen; TC, total carbon; C/N, TC/TN ratio.
Soil variables from each depth were analyzed separately and significances among four treatments (ambient, eCO2, eO3 and eCO2+eO3) were tested by ANOVA at the P<0.05 level. A and B indicate significant changes among treatments for surface soils, and a and b for subsoils and crop yield. Significances between two soil depths were performed by the Student t-test and P-values are given Bold face indicates significantly (P<0.05) changed correlations.
Crop yield data are the average of four plots in the previous two years (2004 and 2006) for each treatment.
Effects of block, depth, CO2 and eO3 on soil microbial communities
To assess whether block, soil depth, CO2 and O3 as well as their combinations affect soil microbial communities, Adonis analysis (Anderson, 2001) of all detected genes showed that these factors and their combinations significantly (P<0.01) impacted soil microbial communities with about 53% of the total variation explained by this model, suggesting that soil microbial community structure was shaped by all three treatments with eCO2 (3.5%) as the main factor, followed by eO3 (3.1%) and eCO2+eO3 (2.5%). Interestingly, a relatively strong block effect (5.2%) was observed, which explained even more total variance than those treatments: CO2, O3 or CO2+O3 (Supplementary Table S1). As depths significantly (P<0.01) affected microbial communities, further analyses were performed with two separate depths: surface soil (0–5 cm) and subsoil (5–15 cm).
Overall responses of soil microbial communities to eCO2, eO3 and eCO2+eO3
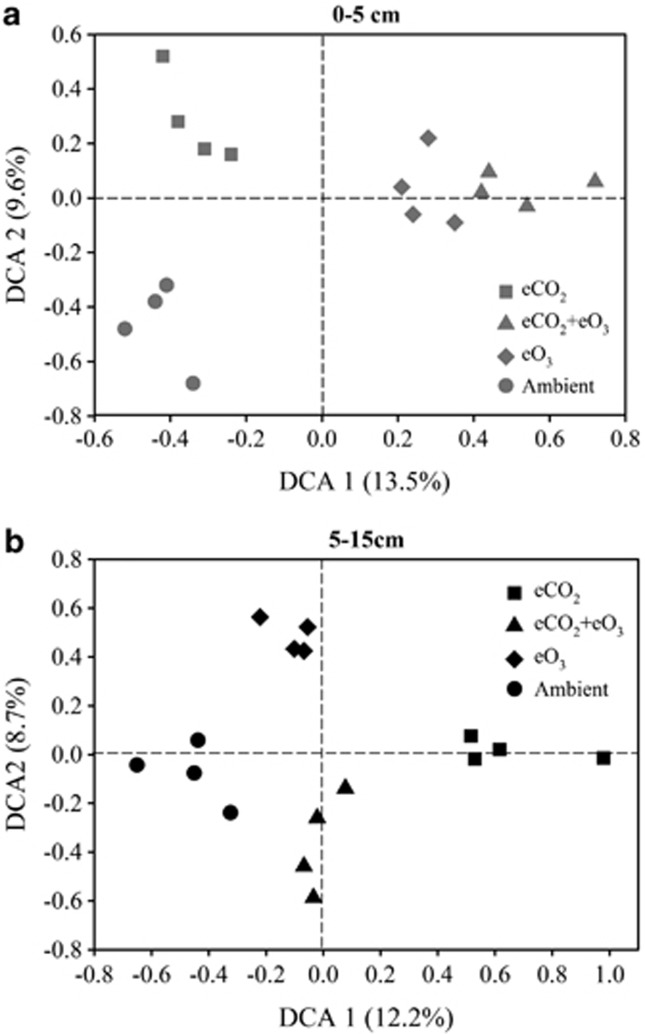
To examine the effect of eCO2 and eO3 on the functional diversity, composition and structure of soil microbial communities, 96 soil samples (48 from each soil depth) of four plots (each with three subsamples) from each of three treatments (eCO2, eO3 and eCO2+eO3) and the control (ambient) were analyzed by GeoChip 3.0. Detrended correspondence analysis of all detected functional genes showed that samples from all three treatments harbored gene assemblages distinct from ambient, and this was observed for both surface soil (Figure 1a) and subsoil (Figure 1b) samples. Samples from all four FACE treatments harbored distinct gene assemblages in the subsoil; however, the assemblages appeared less distinct under eO3 and under eCO2+eO3 in the surface soil (Figure 1). Similarly, such trends were also observed for comparisons between each treatment (eCO2, eO3 or eCO2+eO3) with ambient with 12 individual samples (Supplementary Figure S1), which were further confirmed by three statistical methods, ANOISM and adonis and Multi-Response Permutation Procedures (Supplementary Table S2). Generally, each community was significantly (P<0.05) different from others except eCO2+O3 vs ambient in the surface soil (P=0.066 for adonis, and P=0.052 for MRPP) and eO3 vs ambient and eCO2+O3 vs ambient (P>0.05) in the subsoil (Supplementary Table S2). The results suggest that the functional composition and structure of microbial communities significantly changed under eCO2, eO3 and eCO2+eO3, which is consistent with our PERMANOVA analysis above and that such effects appeared to be distinct for each treatment.
Figure 1.
Detrended corresponding analysis (DCA) of three treatments and control samples in the surface soil (a) and in the subsoil (b) with four plots for each condition (three subsamples from each plot were combined).
Effects of eCO2 and eO3 on key functional genes and processes
To further understand the effect of eCO2 and eO3 on specific functional processes of soil microbial communities, key genes involved in C, N, S and P cycling were further examined below.
C fixation genes
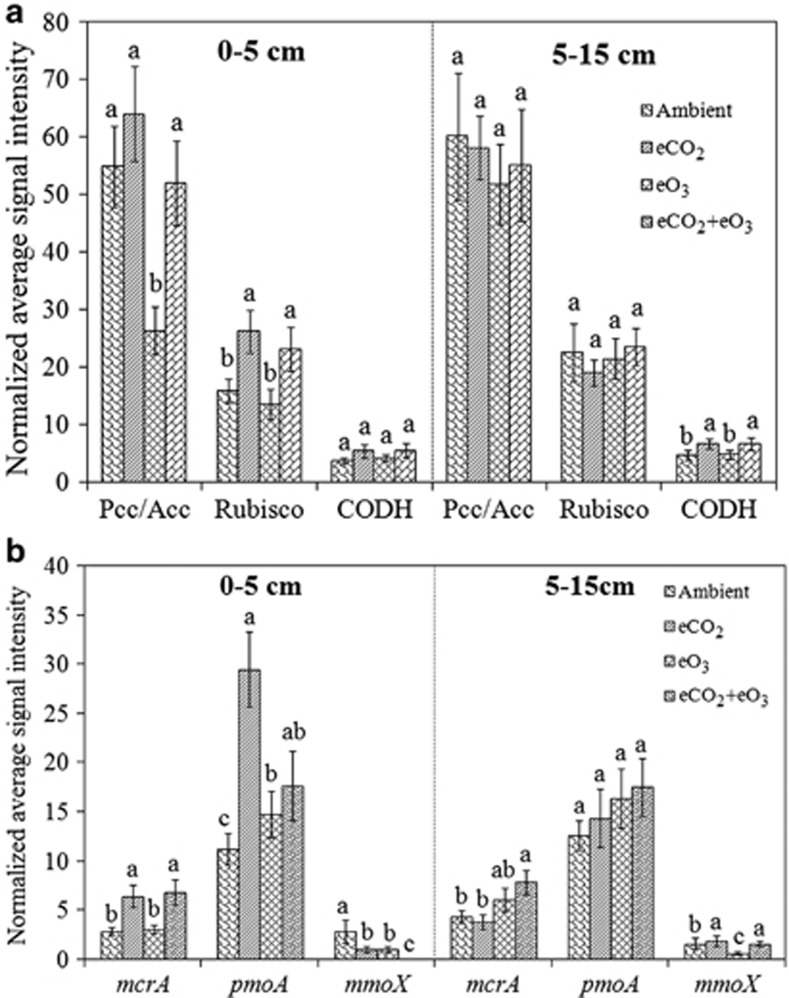
A total of 365, 450, 391 and 420 probes had positive signals under ambient, eCO2, eO3 and eCO2+eO3, respectively, in the surface soil, and 453, 432, 426 and 411 probes, respectively, in the subsoil, which are involved in C fixation, methane metabolism and C degradation (Supplementary Table S3). For C fixation, genes involved in CODH (carbon monoxide dehydrogenase), Pcc/Acc (propionyl-CoA/acetyl-CoA carboxylase) and Rubisco (Ribulose-1,5-bisphosphate carboxylase oxygenase) pathways were detected. Under eCO2 and eCO2+eO3, the signal intensities of Rubisco genes in the surface soil and CODH genes in the subsoil significantly (P<0.05) increased (Figure 2a), suggesting a potentially increased in microbial C fixation. Under eO3, the signal intensity of Pcc/Acc genes significantly (P<0.05) decreased in the surface soil despite no significant differences for Rubisco or CODH genes or all three genes in the subsoil (Figure 2a). Further analysis of all detected Rubisco gene sequences showed that all four forms of Rubisco genes were detected, but most of them belonged to Form I, a major form for CO2 fixation (Supplementary Figure S2). In the surface soil, a total of 52 sequences were detected with 35 clustered into Form I, and examples include gi22415761 from Synechocystis trididemni, gi91690340 from Burkholderia xenovorans LB400, gi148254105 from Bradyrhizobium sp. and gi91802339 from Nitrobacter hamburgensis X14 (Supplementary Figure S2A). In addition, 59 Rubisco gene sequences were detected in the subsoil samples, and similar results were observed (Supplementary Figure S2B). The results indicate that eCO2 and eCO2+eO3 may potentially lead to an increased C fixation, and that eO3 may cause a decreased C fixation in SoyFACE ecosystems.
Figure 2.
Normalized signal intensities of detected genes involved in C fixation pathways (a) and methane metabolism (b) in both surface soil (0–5 cm) and subsoil (5–15 cm) samples. Rubisco: ribulose-1,5-bisphosphate carboxylase/oxygenase; CODH: carbon monoxide dehydrogenase; Pcc/Acc: propionyl-CoA/acetyl-CoA carboxylase; mcrA: the alpha-subunit of methyl coenzyme M reductase for methane production; pmoA: particulate methane monooxygenase for methane oxidation; mmoX: methane monooxygenase for methane oxidation. All data are presented as mean±s.e. Significance among the treatments was analyzed by multi-way analysis of variance (ANOVA). The significance of a, b and c is at P<0.05 level.
C degradation genes
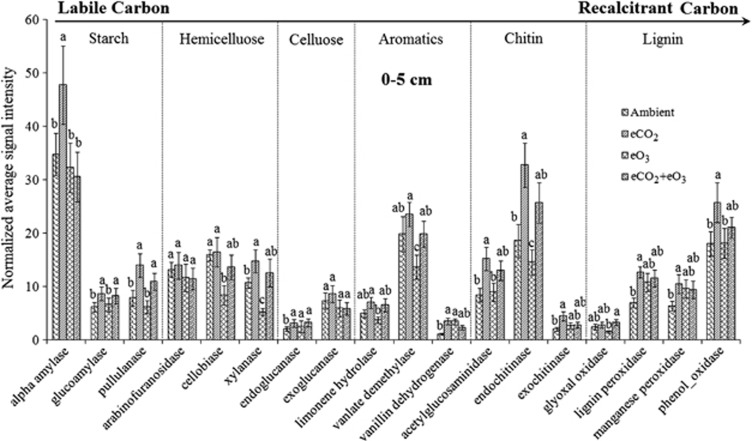
The pattern of signal intensities for functional genes involved in C degradation changed in response to all three treatments. In the surface soil, eCO2 either increased or had no effects on the abundance of detected C degradation genes. For example, the abundance of most C degradation genes significantly (P<0.05) increased under eCO2, including those encoding amylase, glucoamylase, pullulanase, fungal arabinofuranosidase, xylanase, endoglucanase, acetylglucosaminidase and exochitinase for labile C degradation, and those encoding lignin peroxidase, manganese peroxidase and phenol oxidase for recalcitrant C degradation (Figure 3). Under eO3, the abundance of most C degradation genes, especially those for recalcitrant C remained unchanged except with significant (P<0.05) increases for fungal arabinofuranosidase and endoglucanase, and significant decreases for xylanase, cellobiase and exochitinase (Figure 3). Under eCO2+eO3, only genes for glucoamylase, pullulanase and fungal arabinofuranosidase significantly (P<0.05) increased, and there were no significant changes for other C degradation genes detected (Figure 3). For the subsoil samples, very few significant changes were observed for C degradation genes (Supplementary Figure S3). The results indicate that degradation of both labile and recalcitrant C might increase under eCO2 with fewer changes under eCO2+eO3, whereas the abundance of C degradation genes largely remained unchanged under eO3.
Figure 3.
Effects of eCO2, eO3 and eCO2+eO3 on carbon degradation genes detected in the surface soil (0–5 cm). The complexity of carbon is presented in the order from labile to recalcitrant. All data are presented as mean±s.e. Significance among the treatments was analyzed by multi-way ANOVA. The significance of a and b is at P<0.05 level.
N cycling genes
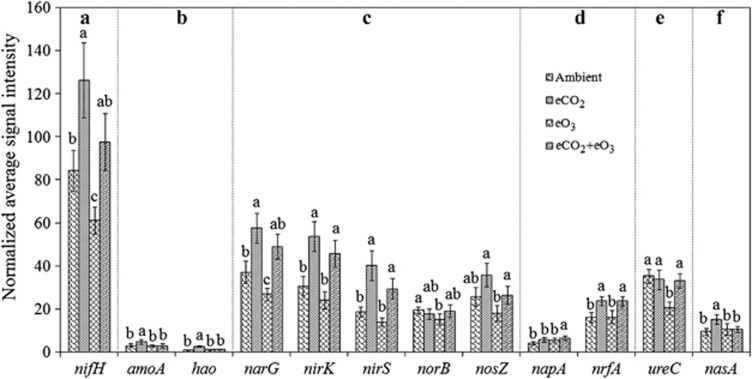
A total of 434, 531, 436 and 441 genes involved in N fixation, nitrification, ammonification, denitrification, dissimilatory N reduction and assimilatory N reduction were detected under ambient, eCO2, eO3 and eCO2+eO3, respectively, in the surface soil, and 536, 483, 503 and 480, respectively, in the subsoil (Supplementary Table S3). In the surface soil, the abundance of 186 detected nifH genes was significantly higher (P<0.05) under eCO2, lower (P<0.05) under eO3 and unchanged under eCO2+eO3 compared with ambient (Figure 4, Supplementary Figure S4A). Among four defined nifH clusters, only Cluster II and III genes were significantly (P<0.05) greater under eCO2, most of the detected Cluster II and III genes were closely related to known microorganisms, such as Rhizobium, Azospirillum, Methanococcus, Desulfovibrio, Methanosarcina and Bradyrhizobium species (Supplementary Figure S4A). In the subsoil, although similar clusters were formed with 217 nifH sequences detected, no significant differences were observed among three treatments and ambient in each cluster (Supplementary Figure S4B).
Figure 4.
Normalized signal intensities of key N cycling genes under eCO2, eO3 and eCO2+eO3 and control conditions in the surface soil (0–5 cm). (a) N2 fixation, nifH encoding nitrogenase; (b) Nitrification, amoA encoding ammonia monooxygenase; (c) Denitrification, including narG for nitrate reductase, nirS and nirK for nitrite reductase, norB for nitric oxide reductase and nosZ for nitrous oxide reductase; (d) Dissimilatory N reduction to ammonium, including napA for nitrate reductase and nrfA for c-type cytochrome nitrite reductase; (e). Ammonification, including gdh for glutamate dehydrogenase and ureC encoding urease; (f) Assimilatory N reduction, nasA encoding nitrate reductase. All data are presented as mean±s.e. Significance among the treatments was analyzed by multi-way ANOVA. The significance of a, b and c is at P<0.05 level.
eCO2 significantly (P<0.05) increased the abundance of narG, nirS and nirK related to denitrification, and the abundances of nirS and nirK also significantly (P<0.05) increased under eCO2+eO3. Although the abundance of norB and nosZ genes remained unaffected under both eCO2 and eCO2+eO3, the abundance of narG and norB genes were significantly (P<0.05) decreased under eO3. In addition, the abundance of amoA, nrfA and nasA increased under eCO2, and ureC decreased under eO3 (Figure 4). In addition, fewer significant changes in the abundance of N cycling genes were observed in the subsoil (Supplementary Figure S5) than in the surface soil. These results indicated that eCO2 might stimulate N cycling by a general increase in the abundance of N cycling genes, and that eO3 might inhibit N cycling by a general decrease in the abundance of N cycling genes, especially in the surface soil.
Greenhouse gas emission genes
The emission of greenhouse gases (for example, CH4 and N2O) was reported to be increased under eCO2 (van Groenigen et al., 2011). GeoChip 3.0 mainly targets mcrA for CH4 generation, pmoA and mmoX for CH4 oxidation (Figure 2b), norB for N2O production and nosZ for N2O reduction (Figure 4c). In the surface soil, the mcrA abundance was significantly (P<0.05) higher under eCO2 and eCO2+eO3, whereas eO3 did not substantially affect the mcrA abundance compared with ambient (Figure 2b). In the subsoil, a significant (P<0.05) increase in the mcrA abundance was observed only under eCO2+eO3 without significant differences detected under eCO2 or eO3 (Figure 2b). In addition, the norB abundance significantly (P<0.05) decreased under eO3, but no significant changes were seen under eCO2 or eCO2+eO3 in the surface soil (Figure 4c) or in the subsoil (Supplementary Figure S5C). The results indicated that eCO2 and/or eCO2+eO3 had the potential to stimulate CH4 emission and that eO3 could potentially decrease N2O emission.
P and S cycling genes
Under eCO2, the abundance of exopolyphosphatase (Ppx) and polyphosphate kinase (Ppk) genes significantly (P<0.05) increased in the surface soil but not in the subsoil. Under eCO2+eO3, only the Ppx gene abundance significantly (P<0.05) increased, and there were no significant changes under eO3 (Supplementary Figure S6A). Similarly, for S cycling, an increase in the signal intensity of dsrA and sox was only observed at eCO2 in the surface soil, and there were no significant changes under eO3 or eCO2+eO3 (Supplementary Figure S6B). These results suggest that eCO2 may enhance both P and S cycling in the surface soil, but eO3 and eCO2+eO3 had little impact, especially in the subsoil.
Relationships between the community structure and soil properties or crop yield
To link the microbial community structure and soil properties and crop yield, correlation analyses were performed by the Mantel test. The signal intensity of all detected genes did not show significant (P>0.05) correlations with soil properties (for example, NO3-N, NH4-N, TC, TN and C/N) or crop yield in the surface soil (Table 2), or subsoil (Supplementary Table S4) samples. However, further analysis of relationships between individual functional genes detected in the surface soil and soil properties and crop yield indicated that 36 functional gene families had significant correlations with soil properties (26 families) or crop yield (16 families), including those involved in C degradation, N cycling and bioremediation of aromatics, herbicides and pesticides (Table 2). For example, genes involved in C degradation (fungal arabinofuranosidase, cellobiase and glucoamylase genes) and denitrification (norB and nosZ) were significantly (P<0.05) correlated with crop yield. In addition, genes involved in recalcitrant C degradation (mnp), N reduction (nasA) and aromatics degradation (bclA, dfbA and tphA) were significantly (P<0.05) correlated with both soil properties and crop yield. In addition, there were significant correlations between genes involved in N reduction (nrfA) or biodegradation of aromatics (for example, amiE, arhA, benD, bphA, cymA and pheA) and soil variables (Table 2). It is noted that most of the aromatic compounds are the products of lignin degradation, which are also related to C degradation. Similarly, 30 functional gene families showed significant correlations with soil properties (25 families) or crop yield (7 families) in the subsoil (Supplementary Table S4). These results suggested that the microbial community functional structure was significantly correlated with soil C and N dynamics, and crop yield.
Table 2. Correlations (P-values) between soil properties or plant yield and signal intensities of functional genes by Mantel analysis of surface (0–5 cm) soil samples.
| Gene/enzyme | Functional process | NO3-N | NH4-N | TN | TC | C/N | Yield |
|---|---|---|---|---|---|---|---|
| All detected | 0.091 | 0.272 | 0.293 | 0.431 | 0.275 | 0.126 | |
| ara (fungi) | C degradation | 0.201 | 0.619 | 0.064 | 0.063 | 0.700 | 0.018 |
| Cellobiase | C degradation | 0.543 | 0.077 | 0.780 | 0.753 | 0.781 | 0.026 |
| Endochitinase | C degradation | 0.603 | 0.605 | 0.863 | 0.539 | 0.014 | 0.421 |
| Glucoamylase | C degradation | 0.648 | 0.935 | 0.301 | 0.193 | 0.295 | 0.023 |
| mnp | C degradation | 0.503 | 0.637 | 0.031 | 0.064 | 0.538 | 0.025 |
| nasA | N reduction | 0.372 | 0.662 | 0.047 | 0.032 | 0.252 | 0.000 |
| norB | Denitrification | 0.278 | 0.687 | 0.189 | 0.076 | 0.129 | 0.028 |
| nosZ | Denitrification | 0.643 | 0.302 | 0.090 | 0.108 | 0.065 | 0.044 |
| nrfA | N reduction | 0.595 | 0.619 | 0.606 | 0.359 | 0.028 | 0.358 |
| cnrC | Cobalt and nickel | 0.611 | 0.354 | 0.491 | 0.519 | 0.625 | 0.017 |
| cueO | Copper | 0.481 | 0.727 | 0.051 | 0.022 | 0.183 | 0.062 |
| metC | Mercury | 0.008 | 0.049 | 0.885 | 0.942 | 0.370 | 0.771 |
| terD | Tellurium | 0.901 | 0.579 | 0.104 | 0.039 | 0.337 | 0.169 |
| amiE | Aromatics | 0.241 | 0.045 | 0.884 | 0.824 | 0.311 | 0.911 |
| arhA | Aromatics | 0.003 | 0.060 | 0.483 | 0.516 | 0.592 | 0.380 |
| bclA | Aromatics | 0.203 | 0.365 | 0.049 | 0.072 | 0.811 | 0.021 |
| benD | Aromatics | 0.015 | 0.073 | 0.988 | 0.975 | 0.463 | 0.983 |
| bphA | Aromatics | 0.382 | 0.042 | 0.916 | 0.827 | 0.541 | 0.414 |
| cymA | Aromatics | 0.550 | 0.617 | 0.685 | 0.240 | 0.025 | 0.626 |
| dfbA | Aromatics | 0.244 | 0.655 | 0.068 | 0.027 | 0.054 | 0.025 |
| hcaACD | Aromatics | 0.164 | 0.047 | 0.798 | 0.756 | 0.861 | 0.969 |
| hdnO | Aromatics | 0.005 | 0.129 | 0.559 | 0.684 | 0.574 | 0.692 |
| nagG | Aromatics | 0.464 | 0.585 | 0.093 | 0.073 | 0.164 | 0.027 |
| nbaC | Aromatics | 0.327 | 0.312 | 0.053 | 0.059 | 0.155 | 0.042 |
| phdCI | Aromatics | 0.920 | 0.580 | 0.954 | 0.828 | 0.026 | 0.156 |
| pheA | Aromatics | 0.474 | 0.112 | 0.022 | 0.108 | 0.597 | 0.267 |
| tmoABE | Aromatics | 0.787 | 0.923 | 0.666 | 0.372 | 0.029 | 0.261 |
| tphA | Aromatics | 0.444 | 0.688 | 0.283 | 0.186 | 0.016 | 0.003 |
| tutFDG | Aromatics | 0.021 | 0.019 | 0.428 | 0.676 | 0.784 | 0.876 |
| xylC | Aromatics | 0.548 | 0.968 | 0.094 | 0.156 | 0.851 | 0.046 |
| mauAB | Herbicides | 0.262 | 0.390 | 0.066 | 0.054 | 0.630 | 0.044 |
| pcpB | Herbicides | 0.163 | 0.033 | 0.991 | 0.998 | 0.760 | 0.792 |
| chnE | Hydrocarbons | 0.587 | 0.986 | 0.086 | 0.034 | 0.213 | 0.276 |
| cpnA | Hydrocarbons | 0.046 | 0.246 | 0.488 | 0.269 | 0.011 | 0.963 |
| xamO | Hydrocarbons | 0.902 | 0.663 | 0.193 | 0.135 | 0.360 | 0.016 |
| adpB | Pesticides | 0.229 | 0.005 | 0.810 | 0.928 | 0.529 | 0.424 |
Abbreviations: TN, total nitrogen; TC, total carbon; C/N, TC/TN ratio.
Bold face indicates significantly (P<0.05) changed correlations.
Discussion
Understanding the response of soil microbial communities of terrestrial ecosystems to atmospheric changes is necessary to predict future global change. In this study, we comprehensively examined functional responses of soil microbial communities to eCO2, eO3 and eCO2+eCO3 using GeoChip technology. The results showed that the functional composition and structure of soil microbial communities shifted, and accordingly, the abundance of key functional genes for C fixation and degradation, N fixation, denitrification, greenhouse gas emission and N mineralization significantly changed under eCO2, eO3 and/or eCO2+eCO3. Such changes were distinct for each treatment and significantly correlated with soil properties and soybean yield. Overall, our analyses suggest possible mechanisms of microbial responses to global atmospheric changes through the stimulation of C and N cycling by eCO2, the inhibition of N functional processes by eO3 and their interactive effects by eCO2 and eO3. This study has important implications for microbial responses and feedbacks to global change and their impacts on crop productivity and ecosystem functioning in soybean agro-ecosystems.
Our core hypothesis is that eCO2, eO3 and eCO2+eO3 would significantly affect the functional composition and structure of soil microbial communities largely by indirect effects, such as altered C and N inputs into soil and soil microenvironments (Ainsworth et al., 2002; Dijkstra et al., 2005; Adair et al., 2009; Feng et al., 2010). Several previous studies showed that the microbial community composition and structure was affected by eCO2 (Feng et al., 2010; He et al., 2010b, 2012b; Deng et al., 2012; Hayden et al., 2012), eO3 and/or eCO2+eO3 (Phillips et al., 2002; Kasurinen et al., 2005; Kanerva et al., 2008). For example, a previous study at the BioCON site showed that eCO2 increased plant and microbial biomass, soil pH and moisture and significantly shifted the functional and phylogentic/taxonmoic composition, structure and network interactions of soil micorbial communities (Zhou et al., 2010; He et al., 2010b, 2012b; Zhou et al., 2011; Deng et al., 2012). In this study, the results support this hypothesis demonstrated by adonis and Detrended correspondence analysis analyses of all detected functional genes. As soybean yield increased under eCO2 and eCO2+eO3, it is expected that C and N input into soil increase and this along with available C and N and other factors (for example, soil moisture, pH and food-web interactions) may affect microbial responses either singly or in combination, leading to changes in the microbial community structure (Zak et al., 2000). However, two recent studies showed that eO3 did not alter the overall microbial community structure (Cheng et al., 2011; Li et al., 2013), which are not consistent with our observations in this study. Such differences may be due to different O3 levels, different replicates used, different exposure time and/or different soil chemistry in those sites. Indeed, in this study, we observed that NH4-N decreases in the surface soil and NO3-N increases in the subsoil despite no significant changes in total soil N. In addition, other eO3-induced changes in plants like root exudate may shift the soil microbial community structure (Phillips et al., 2011), although they were not measured in this study. It should be noted that a randomized complete block design is used in the SoyFACE experiment and a relatively strong block effect was observed. The spatial variations in many environmental variables that may affect soil microbial communities have the potential to be responsible for the block effect, such as soil pH, sunshine angle and plant growth. Most importantly, the treatment effects by CO2 and O3 were still significant via analysis of variance analysis, implying consistent impacts of CO2 and O3 on the soil microbial community across all environmental heterogeneous conditions tested in this study. The results also appeared to be generally consistent with a previous study at the Duke Forest FACE site, showing that spatial factor could explain 20% of the variation in the microbial community structure and CO2 or N treatment for less than 3% of the variation (Ge et al., 2010).
With the change in the microbial community structure under global changes factors, one of most important questions is whether the change in the soil microbial community structure affects microbial functional processes and ecosystem services, such as C and N dynamics. In this study, we found three distinct patterns for microbial responses and feedbacks to eCO2 and eO3: (i) the stimulation of C and N cycling genes by eCO2, (ii) the inhibition of N fixation, denitrification and N mineralization by eO3 and (iii) an interactive effects by eCO2+eO3. The results generally support one of our hypotheses that various microbial functional groups (for example, C fixers, C degraders, N2 fixers and denitrifiers) would differentially respond to eCO2 and eO3.
The first scenario is how microbial communities modify their functional processes by eCO2. Previous studies showed inconsistent responses of soil C and/or N to eCO2 with positive (Carney et al., 2007; Heimann and Reichstein, 2008), negative (Jastrow et al., 2005) or no significant effects (Hungate et al., 2009). A previous study at the BioCON, a grassland ecosystem showed that key genes involved in C fixation and labile C degradation were stimulated under eCO2 but there were no signifcant changes for recalcitrant C degradation genes (He et al., 2010b). This is generally consistent with the current study using the same GeoChip technology (GeoChip 3.0) except that the abundances of lignin degradation genes encoding lignin peroxidase, manganese peroxidase and phenol oxidase were also significantly increased under eCO2. Such differences may be ecosystem-dependent. A previous study showed that as soybean yield and root biomass increased under eCO2, soil organic matter turnover was accelerated but soil moisture and nutrients were not limiting factors at the SoyFACE (Peralta and Wander, 2008). Compared with grassland ecosystems, the soybean agro-ecosystem is N-rich and has competive advantages over non-legumious species at eCO2 by capitializing on eCO2 benefits (for example, increaseing N fixation) and limiting deleterious eCO2 effects (Rogers et al., 2009). Therefore, with more labile C inputs into soil via litter and root exudation, N cycling may be enhanced in soybean agrosystems. Indeed, the present study showed that the abundances of most N cycling genes (for example, nifH, amoA, narG, nirS, nirK, nrfA and nasA) signifcantly increased at eCO2. On the other hand, a meta-analysis suggests that soil N may modulate soil C cycling at eCO2, and that eCO2-induced soil C inputs are generally offset by increased heterotrophic respiration, resulting in no significant changes in soil C content (Dieleman et al., 2010), which appeared to be the case for this study. The results point toward a possible positive microbial feedback to eCO2, although further investigation is necessary.
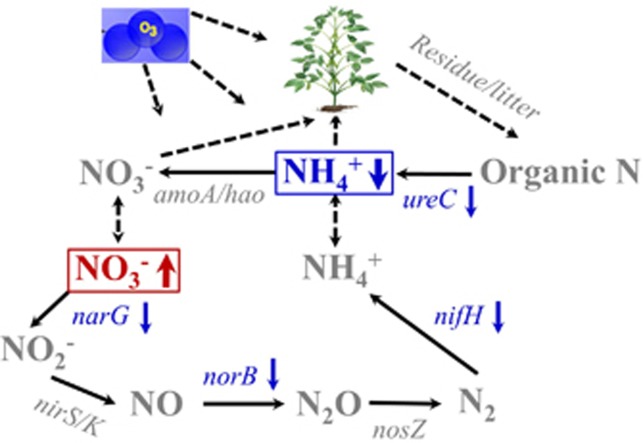
The second scenario is microbial responses to eO3. An early study showed that N concentration in soybean plants was not affected by eO3, but N fixation decreased due to a reduced photosynthate translocation to nodules (Pausch et al., 1996), whereas a recent study at the SoyFACE indicated that eO3 could lead to an increase in soil N availability in both bulk and rhizosphere soils by reducing the mineralization rates of plant-derived residues (Pujol Pereira et al., 2011). In addition, a previous study with soil enzyme activity analysis showed that 1,4-β-glucosidase activity was suppressed, but there were no significant changes for 1,4-β-N-acetylglucosaminidase and other C degradation enzymes under O3 (Larson et al., 2002), and another study of wheat rhizosphere microbial communities at an O3-FACE site identified a decreased abundance of fhs for acetogenesis, but most C and N cycling genes remained unchanged under eO3 (Li et al., 2013). The present study showed that under eO3, the abundances of almost all key C and N cycling genes remained unchanged, or significantly decreased (for example, Pcc/Acc, xylanase, cellobiase, endochitinase, nifH, narG, norB and ureC), and accordingly, we found signicantly higher NO3-N in the subsoil and signifcantly lower NH4-N in the surface soil compared with ambient. Therefore, based on our results and current knowledge, a simple conceptual model is constructed to summarize microbial N cycling and N dynamics in response to eO3 (Figure 5). Under eO3, plant residue was input into the surface soil and mineralized (for example, ureC) to NH4+, which also came from microbial N fixation (for example, nifH). NH4+ could moblize in soil and/or be used by plants and microorganisms. The abundances of both nifH and ureC significantly decreased, resulting in a decrease of NH4-N in the surface soil. NH4+ was transformed to NO3- by nitrification (for example,, amoA and hao), which could mobilize in soil or/and be used by plants and microorganisms. NO3- was then transformed to NO2−, NO, N2O and N2 by denitrification (for example, narG, nirS/K, norB and nosZ). As the narG and norB abundances signficantly decreased at eO3, denitrification, especially the first step (NO3- to NO2− by narG) was probably inhibited, leading to an increased NO3- in the subsoil. This conceptual model may suggest a possible negative microbial feedback to eO3 in this soybean agro-ecosystem.
Figure 5.
A conceptual model for microbial N cycling in response to eO3. Red color indicates an increase in NO3− in the subsoil; blue color indicates decreases in NH4+ in the surface soil or abundances of N cycling genes; gray indicates those parameters were not measured or were not changed under eO3.
The third scenario is how eCO2 and eO3 interact to affect microbial functional processes and ecosystem functioning. It has been proposed that eO3-induced negative effects on soil microbial communities may be ameliorated by eCO2, and a couple of mechanisms are considered. First, under eCO2, the uptake of O3 by plants may be reduced (Allen, 1990; McKee et al., 2000) due to reduced stomatal conductance (Wittig et al., 2009), which may reduce the loss of plant productivity. Second, an increase of available C, especially labile C and root exudation under eCO2 may help plants and microorganisms detoxify and activate cell repair processes (Allen, 1990; McKee et al., 1997). Although there were no significant interactions between eCO2 and eO3 in plant responses to eCO2 and eO3 (Valkama et al., 2007), previous studies showed that eO3 eliminated eCO2-induced effects, such as increased C inputs and microbial enzyme activity in soil (Larson et al., 2002; Phillips et al., 2002), leading to a negative feedback on soil N availability under CO2+O3 (Holmes et al., 2006). In this study, under eCO2 and eO3, we found that eCO2 promoted abundances of some eO3-suppressed genes to ambient, or even to eCO2-induced levels. For example, the abundances of four eO3-inhibited C cycling genes (Pcc/Acc, xylanase, cellobiase and endochitinase genes) exhibited ambient/eCO2 levels under eCO2+eO3. In addition, the abundances of four eO3-suppressed N cycling genes (nifH, narG, norB and ureC) were returned to ambient or promoted to eCO2 levels under eCO2+eO3. In addition, a few C and N cycling genes (for example, pulA, napA, nirK and nirS) showing no signifcant differences under eO3 were significantly increased under eCO2+eO3. The results suggest that the intereactive effects by eCO2+eO3 may be highly uncertain, largely depending on their concentrations, exposure time, soil properties, and ecosystems.
The results from all three scenarios showed that the change in soil microbial community structure affected microbial functional processes and ecosystem services, and such effects appeared distinct for each treatment. Some key functional genes involved in C and N cycling were identified, but their attributions to soil C and N dynamics remain unknown, waranting further investigation by high-throughput sequencing and other metagenomics approaches, such as stable isotope probing, metatranscriptomics and meteproteomics. Also, we analyzed microbial communities from bulk soil in this study, and it will be interesting to analyze the response of rhizosphere micorbial communities including arbuscular mycorrhizal fungi to eCO2 and eO3. In addition, we only examined one time point (late vegetation) in this study, and a future study may focus on the temporal dynamics of soil microbial communities in response to elevated greenhouse gases. Finally, it is necessary to understand the interaction of C and N cycling in soil microbial communities under different global change factors in the future.
Acknowledgments
Assistance with sample collection was provided by Ariane L Peralta, Yu-rui Chang, Sara F Paver, Diana N Flanagan and Anthony C Yannarell. We thank Lisa Ainsworth and Andrew Leakey for helpful comments on this manuscript. This work is supported by the US Department of Agriculture (Project 2007-35319-18305) through the NSF-USDA Microbial Observatories Program, by the US Department of Energy, Biological Systems Research on the Role of Microbial Communities in Carbon Cycling Program (DE-SC0004601). The GeoChips and associated computational pipelines used in this study were supported by ENIGMA-Ecosystems and Networks Integrated with Genes and Molecular Assemblies through the Office of Science, Office of Biological and Environmental Research, the US Department of Energy under Contract No. DE-AC02-05CH11231.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Adair EC, Reich P, Hobbie S, Knops J. Interactive effects of time, CO2, N, and diversity on total belowground carbon allocation and ecosystem carbon storage in a grassland community. Ecosystems. 2009;12:1037–1052. [Google Scholar]
- Ainsworth EA, Davey PA, Bernacchi CJ, Dermody OC, Heaton EA, Moore DJ, et al. A meta-analysis of elevated [CO2] effects on soybean (Glycine max) physiology, growth and yield. Glob Change Biol. 2002;8:695–709. [Google Scholar]
- Ainsworth EA, Long SP. What have we learned from 15 years of free-air CO2 enrichment (FACE)? A meta-analytic review of the responses of photosynthesis, canopy properties and plant production to rising CO2. New Phytol. 2005;165:351–372. doi: 10.1111/j.1469-8137.2004.01224.x. [DOI] [PubMed] [Google Scholar]
- Allen LH. Plant responses to rising carbon dioxide and potential interactions with air pollutants. J Environ Quality. 1990;19:15–34. [Google Scholar]
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- Austin EE, Castro HF, Sides KE, Schadt CW, Classen AT. Assessment of 10 years of CO2 fumigation on soil microbial communities and function in a sweetgum plantation. Soil Biol Biochem. 2009;41:514–520. [Google Scholar]
- Betzelberger AM, Gillespie KM, McGrath JM, Koester RP, Nelson RL, Ainsworth EA. Effects of chronic elevated ozone concentration on antioxidant capacity, photosynthesis and seed yield of 10 soybean cultivars. Plant Cell Environ. 2010;33:1569–1581. doi: 10.1111/j.1365-3040.2010.02165.x. [DOI] [PubMed] [Google Scholar]
- Biswas DK, Xu H, Li YG, Ma BL, Jiang GM. Modification of photosynthesis and growth responses to elevated CO2 by ozone in two cultivars of winter wheat with different years of release. J Exp Bot. 2013;64:1485–1496. doi: 10.1093/jxb/ert005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagodatskaya E, Blagodatsky S, Dorodnikov M, Kuzyakov Y. Elevated atmospheric CO2 increases microbial growth rates in soil: results of three CO2 enrichment experiments. Glob Change Biol. 2010;16:836–848. [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney MC, Hungate BA, Drake BG, Megonigal JP. Altered soil microbial community at elevated CO2 leads to loss of soil carbon. Proc Natl Acad Sci USA. 2007;104:4990–4995. doi: 10.1073/pnas.0610045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro HF, Classen AT, Austin EE, Norby RJ, Schadt CW. Soil microbial community responses to multiple experimental climate change drivers. Appl Environ Microbiol. 2010;76:999–1007. doi: 10.1128/AEM.02874-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Booker FL, Burkey KO, Tu C, Shew HD, Rufty TW, et al. Soil microbial responses to elevated CO2 and O3 in a nitrogen-aggrading agroecosystem. PLoS One. 2011;6:e21377. doi: 10.1371/journal.pone.0021377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, He Z, Xu M, Qin Y, Van Nostrand JD, Wu L, et al. Elevated carbon dioxide alters the structure of soil microbial communities. Appl Environ Microbiol. 2012;78:2991–2995. doi: 10.1128/AEM.06924-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieleman WIJ, Luyssaert S, Rey A, De Angelis P, Barton CVM, Broadmeadow MSJ, et al. Soil [N] modulates soil C cycling in CO2-fumigated tree stands: a meta-analysis. Plant Cell Environ. 2010;33:2001–2011. doi: 10.1111/j.1365-3040.2010.02201.x. [DOI] [PubMed] [Google Scholar]
- Dijkstra FA, Hobbie SE, Reich PB, Knops JMH. Divergent effects of elevated CO2, N fertilization, and plant diversity on soil C and N dynamics in a grassland field experiment. Plant Soil. 2005;272:41–52. [Google Scholar]
- Drake JE, Gallet-Budynek A, Hofmockel KS, Bernhardt ES, Billings SA, Jackson RB, et al. Increases in the flux of carbon belowground stimulate nitrogen uptake and sustain the long-term enhancement of forest productivity under elevated CO2. Ecol Lett. 2011;14:349–357. doi: 10.1111/j.1461-0248.2011.01593.x. [DOI] [PubMed] [Google Scholar]
- Drigo B, Pijl AS, Duyts H, Kielak AM, Gamper HA, Houtekamer MJ, et al. Shifting carbon flow from roots into associated microbial communities in response to elevated atmospheric CO2. Proc Natl Acad Sci USA. 2010;107:10938–10942. doi: 10.1073/pnas.0912421107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drigo B, Kowalchuk GA, Knapp BA, Pijl AS, Boschker HTS, van Veen JA. Impacts of 3 years of elevated atmospheric CO2 on rhizosphere carbon flow and microbial community dynamics. Glob Change Biol. 2013;19:621–636. doi: 10.1111/gcb.12045. [DOI] [PubMed] [Google Scholar]
- Elhottova D, Triska J, Santruckova H, Kveton J, Santrucek J, Simkova M. Rhizosphere microflora of winter wheat plants cultivated under elevated CO2. Plant Soil. 1997;197:251–259. [Google Scholar]
- Feng X, Simpson AJ, Schlesinger WH, Simpson MJ. Altered microbial community structure and organic matter composition under elevated CO2 and N fertilization in the duke forest. Glob Change Biol. 2010;16:2104–2116. [Google Scholar]
- Feng Z, Kobayashi K. Assessing the impacts of current and future concentrations of surface ozone on crop yield with meta-analysis. Atmos Environ. 2009;43:1510–1519. [Google Scholar]
- Ge Y, Chen C, Xu Z, Oren R, He J-Z. The spatial factor, rather than elevated CO2, controls the soil bacterial community in a temperate forest ecosystem. Appl Environ Microbiol. 2010;76:7429–7436. doi: 10.1128/AEM.00831-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geladi P. Analysis of multi-way (multimode) data. Chemo Intell Lab Sys. 1989;7:11–30. [Google Scholar]
- Gutknecht JLM, Field CB, Balser TC. Microbial communities and their responses to simulated global change fluctuate greatly over multiple years. Glob Change Biol. 2012;18:2256–2269. [Google Scholar]
- Hayden HL, Mele PM, Bougoure DS, Allan CY, Norng S, Piceno YM, et al. Changes in the microbial community structure of bacteria, archaea and fungi in response to elevated CO2 and warming in an Australian native grassland soil. Environ Microbiol. 2012;14:3081–3096. doi: 10.1111/j.1462-2920.2012.02855.x. [DOI] [PubMed] [Google Scholar]
- He Z, Gentry TJ, Schadt CW, Wu L, Liebich J, Chong SC, et al. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 2007;1:67–77. doi: 10.1038/ismej.2007.2. [DOI] [PubMed] [Google Scholar]
- He Z, Zhou J. Empirical evaluation of a new method for calculating signal-to-noise ratio for microarray data analysis. Appl Environ Microbiol. 2008;74:2957–2966. doi: 10.1128/AEM.02536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Deng Y, Van Nostrand JD, Tu Q, Xu M, Hemme CL, et al. GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 2010;4:1167–1179. doi: 10.1038/ismej.2010.46. [DOI] [PubMed] [Google Scholar]
- He Z, Xu M, Deng Y, Kang S, Kellogg L, Wu L, et al. Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol Lett. 2010;13:564–575. doi: 10.1111/j.1461-0248.2010.01453.x. [DOI] [PubMed] [Google Scholar]
- He Z, Deng Y, Zhou J. Development of functional gene microarrays for microbial community analysis. Curr Opin Biotechnol. 2012;23:49–55. doi: 10.1016/j.copbio.2011.11.001. [DOI] [PubMed] [Google Scholar]
- He Z, Piceno Y, Deng Y, Xu M, Lu Z, DeSantis T, et al. The phylogenetic composition and structure of soil microbial communities shifts in response to elevated carbon dioxide. ISME J. 2012;6:259–272. doi: 10.1038/ismej.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Van Nostrand JD, Zhou J. Applications of functional gene microarrays for profiling microbial communities. Curr Opin Biotechnol. 2012;23:460–466. doi: 10.1016/j.copbio.2011.12.021. [DOI] [PubMed] [Google Scholar]
- Heimann M, Reichstein M. Terrestrial ecosystem carbon dynamics and climate feedbacks. Nature. 2008;451:289–292. doi: 10.1038/nature06591. [DOI] [PubMed] [Google Scholar]
- Holmes WE, Zak DR, Pregitzer KS, King JS. Elevated CO2 and O3 alter soil nitrogen transformations beneath trembling aspen, paper birch, and sugar maple. Ecosystems. 2006;9:1354–1363. [Google Scholar]
- Hungate BA, Van Groenigen K-J, Six J, Jastrow JD, Luo Y, De Graaff M-A, et al. Assessing the effect of elevated carbon dioxide on soil carbon: a comparison of four meta-analyses. Glob Change Biol. 2009;15:2020–2034. [Google Scholar]
- IPCC (Intergovernmental Panel on Climate Change) Climate change 2007: The physical science basis: fourth assessment report of the intergovernmental panel on climate. change. Cambridge Univ. Press; 2007. [Google Scholar]
- Jastrow JD, Miller RM, Matamala R, Norby RJ, Boutton TW. Elevated atmospheric carbon dioxide increases soil carbon. Glob Change Biol. 2005;11:2057–2064. doi: 10.1111/j.1365-2486.2005.01077.x. [DOI] [PubMed] [Google Scholar]
- Kanerva T, Palojärvi A, Rämö K, Manninen S. Changes in soil microbial community structure under elevated tropospheric O3 and CO2. Soil Biol Biochem. 2008;40:2502–2510. [Google Scholar]
- Kasurinen A, Keinänen MM, Kaipainen S, Nilsson L-O, Vapaavuori E, Kontro MH, et al. Below-ground responses of silver birch trees exposed to elevated CO2 and O3 levels during three growing seasons. Glob Change Biol. 2005;11:1167–1179. [Google Scholar]
- Kumari S, Agrawal M, Tiwari S. Impact of elevated CO2 and elevated O3 on Beta vulgaris L: pigments, metabolites, antioxidants, growth and yield. Environ Pollu. 2013;174:279–288. doi: 10.1016/j.envpol.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Larson J, Zak D, Sinsabaugh R. Extracellular enzyme activity beneath temperate trees growing under elevated carbon dioxide and ozone. Soil Sci Soc Am J. 2002;66:1848–1856. [Google Scholar]
- Lesaulnier C, Papamichail D, McCorkle S, Ollivier B, Skiena S, Taghavi S, et al. Elevated atmospheric CO2 affects soil microbial diversity associated with trembling aspen. Environ Microbiol. 2008;10:926–941. doi: 10.1111/j.1462-2920.2007.01512.x. [DOI] [PubMed] [Google Scholar]
- Li X, Deng Y, Li Q, Lu C, Wang J, Zhang H, et al. Shifts of functional gene representation in wheat rhizosphere microbial communities under elevated ozone. ISME J. 2013;7:660–671. doi: 10.1038/ismej.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth R. Impacts of elevated atmospheric CO2 and O3 on forests: phytochemistry, trophic interactions, and ecosystem dynamics. J Chem Ecol. 2010;36:2–21. doi: 10.1007/s10886-009-9731-4. [DOI] [PubMed] [Google Scholar]
- Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotech. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- Luo Y, Su B, Currie WS, Dukes JS, Finzi A, Hartwig U, et al. Progressive nitrogen limitation of ecosystem responses to rising atmospheric carbon dioxide. Bioscience. 2004;54:731–739. [Google Scholar]
- Luo Y, Hui D, Zhang D. Elevated CO2 stimulates net accumulations of carbon and nitrogen in land ecosystems: a meta-analysis. Ecology. 2006;87:53–63. doi: 10.1890/04-1724. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee IF, Eiblmeier M, Polle A. Enhanced ozone-tolerance in wheat grown at an elevated CO2 concentration: ozone exclusion and detoxification. New Phytol. 1997;137:275–284. doi: 10.1046/j.1469-8137.1997.00801.x. [DOI] [PubMed] [Google Scholar]
- McKee IF, Mulholland BJ, Craigon J, Black CR, Long SP. Elevated concentrations of atmospheric CO2 protect against and compensate for O3 damage to photosynthetic tissues of field-grown wheat. New Phytol. 2000;146:427–435. [Google Scholar]
- Meriles JM, Vargas Gil S, Conforto C, Figoni G, Lovera E, March GJ, et al. Soil microbial communities under different soybean cropping systems: characterization of microbial population dynamics, soil microbial activity, microbial biomass, and fatty acid profiles. Soil Tillage Res. 2009;103:271–281. [Google Scholar]
- Morgan PB, Ainsworth EA, Long SP. How does elevated ozone impact soybean? A meta-analysis of photosynthesis, growth and yield. Plant Cell Environ. 2003;26:1317–1328. [Google Scholar]
- Morgan PB, Bollero GA, Nelson RL, Dohleman FG, Long SP. Smaller than predicted increase in aboveground net primary production and yield of field-grown soybean under fully open-air [CO2] elevation. Glob Change Biol. 2005;11:1856–1865. [Google Scholar]
- Pausch RC, Mulchi CL, Lee EH, Meisinger JJ. Use of 13C and 15N isotopes to investigate O3 effects on C and N metabolism in soybeans. Part II. Nitrogen uptake, fixation, and partitioning. Agr Ecosyst Environ. 1996;60:61–69. [Google Scholar]
- Peralta A, Wander M. Soil organic matter dynamics under soybean exposed to elevated [CO2] Plant Soil. 2008;303:69–81. [Google Scholar]
- Phillips RL, Zak DR, Holmes WE, White DC. Microbial community composition and function beneath temperate trees exposed to elevated atmospheric carbon dioxide and ozone. Oecologia. 2002;131:236–244. doi: 10.1007/s00442-002-0868-x. [DOI] [PubMed] [Google Scholar]
- Phillips RP, Finzi AC, Bernhardt ES. Enhanced root exudation induces microbial feedbacks to N cycling in a pine forest under long-term CO2 fumigation. Ecol Lett. 2011;14:187–194. doi: 10.1111/j.1461-0248.2010.01570.x. [DOI] [PubMed] [Google Scholar]
- Pujol Pereira EI, Chung H, Scow K, Sadowsky MJ, van Kessel C, Six J. Soil nitrogen transformations under elevated atmospheric CO2 and O3 during the soybean growing season. Environ Pollut. 2011;159:401–407. doi: 10.1016/j.envpol.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria; 2011. [Google Scholar]
- Reich PB, Knops J, Tilman D, Craine J, Ellsworth D, Tjoelker M, et al. Plant diversity enhances ecosystem responses to elevated CO2 and nitrogen deposition. Nature. 2001;410:809–812. doi: 10.1038/35071062. [DOI] [PubMed] [Google Scholar]
- Reich PB, Hobbie SE, Lee T, Ellsworth DS, West JB, Tilman D, et al. Nitrogen limitation constrains sustainability of ecosystem response to CO2. Nature. 2006;440:922–925. doi: 10.1038/nature04486. [DOI] [PubMed] [Google Scholar]
- Reich PB. Elevated CO2 reduces losses of plant diversity caused by nitrogen deposition. Science. 2009;326:1399–1402. doi: 10.1126/science.1178820. [DOI] [PubMed] [Google Scholar]
- Rogers A, Ainsworth EA, Leakey ADB. Will elevated carbon dioxide concentration amplify the benefits of nitrogen fixation in legumes. Plant Physiol. 2009;151:1009–1016. doi: 10.1104/pp.109.144113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BK, Bardgett RD, Smith P, Reay DS. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Micro. 2010;8:779–790. doi: 10.1038/nrmicro2439. [DOI] [PubMed] [Google Scholar]
- Sitch S, Cox PM, Collins WJ, Huntingford C. Indirect radiative forcing of climate change through ozone effects on the land-carbon sink. Nature. 2007;448:791–794. doi: 10.1038/nature06059. [DOI] [PubMed] [Google Scholar]
- Talhelm AF, Pregitzer KS, Zak DR. Species-specific responses to atmospheric carbon dioxide and tropospheric ozone mediate changes in soil carbon. Ecol Lett. 2009;12:1219–1228. doi: 10.1111/j.1461-0248.2009.01380.x. [DOI] [PubMed] [Google Scholar]
- Twine TE, Bryant JJ, Richter K, Bernacchi CJ, McConnaughay KD, Morris SJ, et al. Impacts of elevated CO2 concentration on the productivity and surface energy budget of the soybean and maize agroecosystem in the Midwest US. Glob Change Biol. 2013;19:2838–2852. doi: 10.1111/gcb.12270. [DOI] [PubMed] [Google Scholar]
- Valkama E, Koricheva J, Oksanen E. Effects of elevated O3, alone and in combination with elevated CO2, on tree leaf chemistry and insect herbivore performance: a meta-analysis. Glob Change Biol. 2007;13:184–201. [Google Scholar]
- van Groenigen KJ, Osenberg CW, Hungate BA. Increased soil emissions of potent greenhouse gases under increased atmospheric CO2. Nature. 2011;475:214–216. doi: 10.1038/nature10176. [DOI] [PubMed] [Google Scholar]
- Volin JC, Reich PB, Givnish TJ. Elevated carbon dioxide ameliorates the effects of ozone on photosynthesis and growth: species respond similarly regardless of photosynthetic pathway or plant functional group. New Phytol. 1998;138:315–325. doi: 10.1046/j.1469-8137.1998.00100.x. [DOI] [PubMed] [Google Scholar]
- Wittig VE, Ainsworth EA, Naidu SL, Karnosky DF, Long SP. Quantifying the impact of current and future tropospheric ozone on tree biomass, growth, physiology and biochemistry: a quantitative meta-analysis. Glob Change Biol. 2009;15:396–424. [Google Scholar]
- Wu L, Liu X, Schadt CW, Zhou J. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl Environ Microbiol. 2006;72:4931–4941. doi: 10.1128/AEM.02738-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak DR, Holmes WE, Finzi AC, Norby RJ, Schlesinger WH. Soil nitrogen cycling under elevated CO2: a sysnthesis of forest FACE experiemts. Ecol Appl. 2003;13:1508–1514. [Google Scholar]
- Zak DR, Kubiske ME, Pregitzer KS, Burton AJ. Atmospheric CO2 and O3 alter competition for soil nitrogen in developing forests. Glob Change Biol. 2012;18:1480–1488. [Google Scholar]
- Zak DR, Pregitzer KS, King JS, Holmes WE. Elevated atmospheric CO2, fine roots and the response of soil microorganisms: a review and hypothesis. New Phytol. 2000;147:201–222. [Google Scholar]
- Zak DR, Pregitzer KS, Kubiske ME, Burton AJ. Forest productivity under elevated CO2 and O3: positive feedbacks to soil N cycling sustain decade-long net primary productivity enhancement by CO2. Ecol Lett. 2011;14:1220–1226. doi: 10.1111/j.1461-0248.2011.01692.x. [DOI] [PubMed] [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Deng Y, Luo F, He Z, Tu Q, Zhi X. Functional molecular ecological networks. MBio. 2010;1:e00169–10. doi: 10.1128/mBio.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Deng Y, Luo F, He Z, Yang Y. Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. MBio. 2011;2:e00122–11. doi: 10.1128/mBio.00122-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Xue K, Xie J, Deng Y, Wu L, Cheng X, et al. Microbial mediation of carbon-cycle feedbacks to climate warming. Nature Clim Change. 2012;2:106–110. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.