Abstract
Interspecific competition is an important driver of community assembly in plants and animals, but phylogenetic evidence for interspecific competition in bacterial communities has been elusive. This could indicate that other processes such as habitat filtering or neutral processes are more important in bacterial community assembly. Alternatively, this could be a consequence of the lack of a consistent and meaningful species definition in bacteria. We hypothesize that competition in bacterial community assembly has gone undetected at least partly because overly broad measures of bacterial diversity units were used in previous studies. First, we tested our hypothesis in a simulation where we showed that how species are defined can dramatically affect whether phylogenetic overdispersion (a signal consistent with competitive exclusion) will be detected. Second, we demonstrated that using finer-scale Operational Taxonomic Units (OTUs) (with more stringent 16S rRNA sequence identity cutoffs or based on fast-evolving protein coding genes) in natural populations revealed previously undetected overdispersion. Finally, we argue that bacterial ecotypes, diversity units incorporating ecological and evolutionary theory, are superior to OTUs for the purpose of studying community assembly.
Keywords: bacterial species, community assembly, ecotype, interspecific competition, OTU
Introduction
Interspecific competition is one of the central pillars upon which evolutionary and ecological theory rests. Competition between species is fundamental to many pivotal ecological questions. Specifically, why do species exist in some habitats, but not in others? What processes determine the complement of species in any particular habitat? While plant and animal ecologists have made great progress in understanding how competition affects the composition of species in a community (Webb et al., 2002; Purvis et al., 2008; Cavender-Bares et al., 2009), the role of interspecific competition in building bacterial communities is still unclear (Horner-Devine and Bohannan, 2006).
Phylogenetic evidence has been used by plant and animal ecologists to detect the influence of competition on community assembly. The more closely related two species are, the greater their ecological similarity, and the more intense the competition between them is expected to be (Darwin, 1859; Cooper et al., 2008; Cavender-Bares et al., 2009; Wiens et al., 2010). As a result, species frequently find it more difficult to invade habitats occupied by their sister species (Fargione et al., 2003; Tilman, 2004). Competitive exclusion among close relatives can reveal itself via a specific phylogenetic signature called phylogenetic overdispersion, in which species found in the same habitat are more distantly related than expected by chance (Elton, 1946; MacArthur and Levins, 1967). Competition is only one of the many processes known to play a role in community assembly. Another is habitat filtering, where closely-related species sharing a trait or suite of traits persist in a given habitat; this can stem from difficulty in adapting to the abiotic conditions of another habitat. The expected phylogenetic signature of habitat filtering is the exact opposite of that for competitive exclusion. That is, co-occurring species are typically more closely related than expected (phylogenetic clustering).
Habitat filtering and competition can operate simultaneously in real communities, but their influence varies at different spatial and taxonomic scales (Weiher and Keddy, 1995; Webb et al., 2002; Cavender-Bares et al., 2004, 2006; Horner-Devine and Bohannan, 2006; Silvertown et al., 2006; Emerson and Gillespie, 2008; Purvis et al., 2008; Vamosi et al., 2009). When communities are studied at broad spatial and taxonomic scales, habitat filtering is expected to be dominant, because taxa and habitats are more heterogeneous. Inversely, competitive exclusion is expected to be more intense and influential in communities of smaller spatial and taxonomic scales. Accordingly, ecologists have found ample evidence of competition and habitat filtering in animal and plant communities, but the strength of the interactions varies at different scales. While studies have shown evidence of habitat filtering in natural bacterial communities, surprisingly little evidence has been uncovered to suggest that competition also plays an important role (Horner-Devine and Bohannan, 2006; Newton et al., 2007; Bryant et al., 2008; Pontarp et al., 2012; Stegen et al., 2012; Wang et al., 2012). Resource competition has been shown to shape the assembly of bacterial microcosm communities in laboratory experiments (Kurihara et al., 1990; Gerrish and Lenski, 1998; Rainey and Travisano, 1998; Hibbing et al., 2010), but there are very few documented instances of phylogenetic overdispersion in natural bacterial communities.
One explanation for the lack of phylogenetic evidence for competition is that habitat filtering or neutral processes (Tilman, 2004) is predominant in bacterial community assembly and that competitive exclusion plays only a limited role. Another possibility is that competition, although significant, does not always lead to phylogenetic overdispersion. This could be due to the lack of niche conservatism (Losos et al., 2003; Rice et al., 2003; Knouft et al., 2006; Losos, 2008), endemic adaptive radiation (Wiedenbeck and Cohan, 2011) or because competitive ability differences between species exceed their niche differences (Mayfield and Levine, 2010). Here we test an alternative hypothesis that phylogenetic analyses used to look for phylogenetic overdispersion in bacterial communities have been done at the wrong taxonomic scale, such that overdispersion cannot be readily detected, even if present (Horner-Devine and Bohannan, 2006).
The phylogenetic methods used to detect competition in plant and animal communities are challenged when applied to bacteria by the lack of a clear species concept (Cohan, 2002). In particular, if current molecular approaches for characterizing bacterial diversity result in taxa that are too broadly inclusive, this would hinder the detection of interspecific competition using phylogenetic methods. For example, in the case of plants and animals, we would not expect to detect competitive exclusion as a major factor in community assembly if the family or order is used as the diversity unit in the analysis of phylogenetic community structure (Vamosi et al., 2009). But currently accepted definitions of bacterial species result in taxa that are more analogous to families and orders among plants and animals than to their species (Staley, 2006)! The most commonly used approximations of bacterial species are Operational Taxonomic Units (OTUs), which are based solely on gene sequence similarity, most often the 16S rRNA gene. OTUs based on the commonly used 97% or 99% identity cutoffs at the 16S rRNA locus are known to encompass large swaths of genomic (Staley, 2006; Goris et al., 2007) and ecological (Ward et al., 2006) diversity within them (Wiedenbeck and Cohan, 2011), and therefore have the potential to bias bacterial diversity datasets against the detection of phylogenetic overdispersion.
We predicted that a finer scale of species delineation based on a narrower identity threshold, or less conserved markers (e.g., fast evolving protein-coding genes) would increase our power to detect phylogenetic overdispersion. We tested this hypothesis by analyzing the phylogenetic relatedness of several bacterial datasets using 16S rRNA and protein-coding genes and a range of identity thresholds to define the species boundary. In addition, since recently diverged bacterial ecotypes (ecologically homogeneous populations) may represent the units of bacterial diversity that are most closely equivalent to plant and animal species (Cohan and Perry, 2007; Wiedenbeck and Cohan, 2011), we also tested the effect of using bacterial ecotypes as the species unit for phylogenetic analyses. Our results suggest that phylogenetic overdispersion is more prevalent in bacterial communities than has previously been appreciated.
Materials and methods
Sequence datasets
Four sequence datasets from a wide range of environments were used in this study (Table 1). Marine Pelagibacter sequences were obtained by BLASTN searching the Global Ocean Survey (GOS) All ORFs database (Sun et al., 2010) with 31 Candidatus Pelagibacter ubique HTCC1062 protein-coding marker genes (Wu and Eisen, 2008) (e-value <=1e-10). The marine Vibrio dataset consisted of 1025 hsp60 sequences of the genus Vibrio, 541 bp in length, sampled in the spring and fall from particles of different sizes in a coastal marine environment (Hunt et al., 2008). The skin microbiome dataset (Grice et al., 2009) included skin bacteria from 10 healthy volunteers, each of which was sampled at 21 different skin sites, including moist, dry and sebaceous skin. This set contained 116391 near full-length 16S rRNA Sanger sequences. The gut microbiome dataset (Ley et al., 2006) contained gut bacteria sampled from 12 obese individuals over a time course of 52 weeks during which the obese subjects undertook one of two weight-loss regimens. The dataset contained 18052 near full-length 16S rRNA Sanger sequences. Pelagibacter sequences were retrieved from CAMERA (http://camera.calit2.net). All other sequences were retrieved from Genbank.
Table 1. Datasets used for Phylocom analysis.
| Dataset | Habitats | Gene | Reference |
|---|---|---|---|
| Marine Pelagibacter | GOS sampling sites | 16S rRNA and 10 protein-coding genes | (Rusch et al., 2007) |
| Marine Vibrio | Habitats based on particle sizes/sampling seasons | hsp60 | (Hunt et al., 2008) |
| Skin microbiome | Human subjects | 16S rRNA | (Grice et al., 2009) |
| Gut microbiome | Human subjects | 16S rRNA | (Ley et al., 2006) |
OTU generation
For GOS Pelagibacter sequences, translated protein-coding sequences were aligned by HMMer3 (Eddy, 2011) using profile Hidden Markov Models of known Pelagibacter marker genes. The protein alignments were then converted back to DNA alignments using in-house scripts. Given the fragmentary nature of the GOS ORFs, a sliding window approach (width: 200 bp, increment: 20 bp) was used to select alignment regions for further analysis. To be selected, an alignment region must contain at least 500 sequences and no sequence could have more than 10 gaps in the alignment region. 10 of the 31 Pelagibacter marker genes with enough sequences passed these criteria and were used in the subsequent Phylocom analysis (Table 2). Vibrio hsp60 sequences were aligned by their amino acid sequences using MUSCLE (Edgar, 2004), and then converted back to a DNA alignment. The 16S rRNA sequences were aligned using the PyNAST algorithm in QIIME (Caporaso et al., 2010) and were classified to the genus level using RDP Classifier, version 2 (Wang et al., 2007) using the default settings. OTUs were generated with MOTHUR (Schloss et al., 2009) using complete-linkage (furthest neighbor) clustering. Sequence with the minimum average distance to the other sequences of the same OTU was chosen as the representative sequence for the OTU.
Table 2. Summary of Pelagibacter overdispersion trends based on 10 protein-coding marker genes.
|
NRI |
NTI |
|||||||
|---|---|---|---|---|---|---|---|---|
| Gene | R2 | P | slope | Sig. Pos. | R2 | P | slope | Sig. Pos. |
| dnaG | 0.85 | 0.03 | + | ✓ | 0.9 | 0.001 | + | ✓ |
| infC | 0.64 | 0.03 | + | ✓ | 0.78 | 0.008 | + | ✓ |
| nusA | 0.88 | 0.002 | + | ✓ | 0.74 | 0.01 | + | ✓ |
| pyrG | 0.86 | 0.003 | + | ✓ | 0.69 | 0.02 | + | ✓ |
| rplB | 0.40 | 0.13 | + | 0.63 | 0.03 | + | ✓ | |
| rplK | 0.85 | 0.003 | + | ✓ | 0.83 | 0.004 | + | ✓ |
| rplS | 0.69 | 0.02 | + | ✓ | 0.58 | 0.05 | + | ✓ |
| rplT | 0.39 | 0.13 | + | 0.42 | 0.11 | + | ||
| rpoB | 0.87 | 0.002 | + | ✓ | 0.90 | 0.001 | + | ✓ |
| rpsC | 0.78 | 0.009 | + | ✓ | 0.56 | 0.05 | + | ✓ |
Each row represents the results of one marker gene analyzed in the GOS Pelagibacter dataset. The R2 and P values of the correlation between the identity cut-off and the fraction of communities called significantly overdispersed by Phylocom are listed. A positive slope indicates that more communities were overdispersed at more stringent species cutoffs. Check marks in the Sig. Pos. column denote significant positive slopes, similar to the pattern displayed in Figure 3. Results are shown for both the NTI and NRI metrics.
Simulating the effects of the granularity of species units
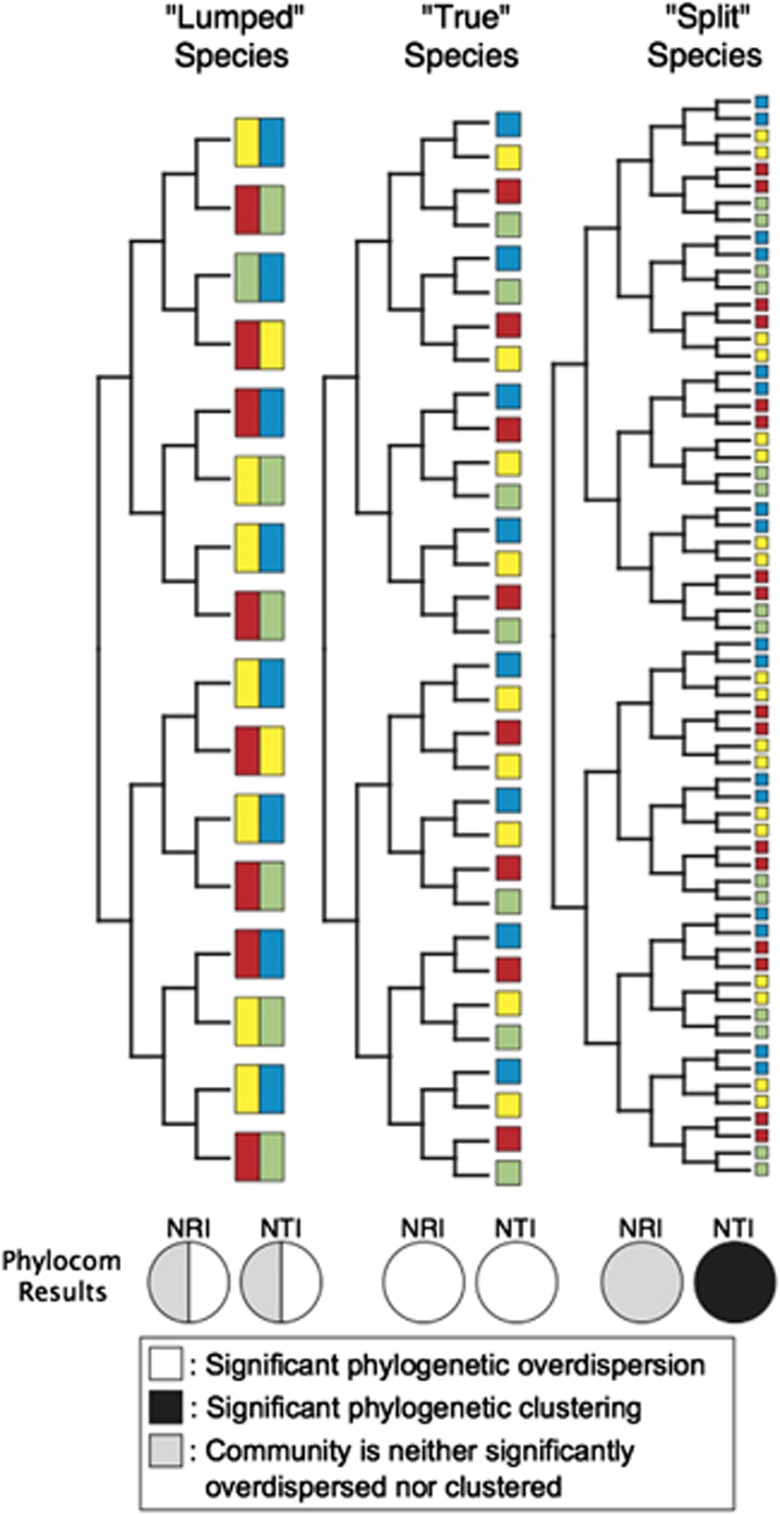
In order to determine whether species definition might theoretically affect the results of the Phylocom analysis in the absence of other factors, we used Phylocom to analyze a small simulated dataset (Figure 1). We generated a dataset in which 32 hypothetical ‘true' species were assigned to four communities, such that the ‘true' condition for all four communities was phylogenetic overdispersion. We then modified the species unit by splitting each ‘true' species into two, creating 64 ‘split' species. We also lumped each sister-species pair together into a single species resulting in 16 ‘lumped' species. Phylocom analysis was then run on all three datasets to determine whether changing how species are defined would affect the outcome.
Figure 1.
The effects of lumping and splitting of species on the phylogenetic analysis of community structure. This figure displays the setup and results of the simulation experiment. Each of the 32 leaves in the ‘true' species tree represents a ‘true' species. Colored squares at the leaves of the phylogeny represent the community within which the species is found. There are a total of four different communities in this simulation (green, blue, red and yellow). Solid colored squares indicate the species is found only in that community. When two sister species are lumped together, the ‘lumped' species will then be perceived to be present in two communities instead of one, so the communities for the lumped species are displayed as divided squares. The pie charts at the bottom of each tree indicate the percentages of communities that are significantly clustered, overdispersed or not significant according to the Phylocom analysis. For example, among the lumped species, two of the communities were called as significantly overdispersed by phylocom, while the other two were neither significantly clustered, nor significantly overdispersed.
Phylocom analysis
We used Phylocom (Webb et al., 2008) version 4.1 to compute the Net Relatedness Index (NRI) and Nearest Taxon Index (NTI). Both measure the phylogenetic relatedness of species within a community. The key difference is that NRI measures the average phylogenetic distance between all co-occurring species, while NTI considers only the average distance between co-occurring closest phylogenetic relatives. The statistical significance of observed NRI and NTI values was estimated by constructing 9999 simulated phylogenies in which the species were shuffled randomly between communities (the phylogeny shuffle model). The rank order position of the NTI and NRI values for the observed data relative to those of the simulations was used to calculate the statistical significance. All analyses were run both with and without taking into account taxa abundance, but the results were nearly identical in all cases. All results displayed in this manuscript are abundance-weighted. Samples used in Phylocom analyses were listed in Table 1. Community was defined as a group of bacterial species that were found in the same sample. For the GOS dataset, only samples with at least 20 Pelagibacter sequences were included for Phylocom analysis. A sample was compared to both the global pool and its local pool. For example, an Indian Ocean sample was compared to all GOS samples and also only to other Indian Ocean samples. For skin microbiome data, each of the 21 skin sites of one human subject was treated as a separate sample and was compared to the same skin site of the other human subjects. As species in the same genus are expected to be more likely to compete than those in different genera (Darwin, 1859; Cooper et al., 2008; Cavender-Bares et al., 2009; Wiens et al., 2010), we only used sequences that belong to the same genus in the Phylocom analysis to increase our ability to detect overdispersion.
Demarcation of ecotypes using ecotype simulation and AdaptML
We used Ecotype Simulation (ES) (Koeppel et al., 2008) version 0.6 and AdaptML (Hunt et al., 2008) to demarcate ecotypes for the Vibrio hsp60 data. The current version of ES is only capable of analyzing around 300 sequences at once within a reasonable time frame. Since the Vibrio dataset contained many more sequences, we employed a divide-and-conquer approach. Using a guiding tree, we subdivided the sequences into clades of<200 sequences and ran ES separately on each clade. We then demarcated ecotypes on the entire tree by finding the most inclusive clades that are each consistent with being a single ecotype (Koeppel et al., 2008). AdaptML for the Vibrio dataset was run with the particle size and season as environmental parameters following Hunt et al. (Hunt et al., 2008). Our AdaptML analysis returned habitats virtually identical to those of Hunt et al. (2008), with the exception that we had seven habitats instead of six. This is likely due to slight variations in tree topology resulting from using different tree-building algorithms. AdaptML was also used to demarcate ecotypes for the Pelagibacter rplK sequences from the GOS dataset using the temperature, salinity, chlorophyll density and water depth as environmental parameters. AdaptML was run using the default settings.
Results and discussion
Broad species units can obscure phylogenetic overdispersion
Before testing our hypothesis in natural systems, we first ran a simple simulation with the aim of demonstrating that changing how species are defined could alter the outcome of this type of phylogenetic analysis, in the absence of other factors. We simulated a set of hypothetical ‘true' species distributed across habitats such that all communities were phylogenetically overdispersed (Figure 1). We then modified the simulated communities in two ways, once by ‘lumping' two sister species into one unit and once by ‘splitting', in which the ‘true' species were split into two co-occurring sister units. We used Phylocom to quantify the degree of phylogenetic clustering and overdispersion with two indices: NRI and NTI (see Materials and Methods for details). Positive values of either index for a community indicate that the species within that community are phylogenetically clustered, while negative values indicate phylogenetic overdispersion. We found that the breadth of the species unit can dramatically alter how much phylogenetic overdispersion is detected (Figure 1). After lumping the ‘true' species into broader units, we failed to detect overdispersion in half of the communities. Consistent with our expectations, the effect of splitting species produced different results depending on the metric. NRI did not show any phylogenetic structures in the communities. In contrast, because species splitting always resulted in a nearest neighbor from the same community, NTI actually returned a false result indicating significant phylogenetic clustering. Actual phylogenies and habitat distributions of species in nature are of course unlikely to be so simplistic, but this simulation demonstrates that species definition can dramatically affect whether or not the signature of interspecific competition is detectable by phylogenetic analyses. Overdispersion is most apparent when the proper species unit is applied. Lumping or splitting will obscure the signature of competition and reduce the sensitivity of the phylogenetic methods. We went on to test whether this finding was also supported by results from natural bacterial communities.
Narrower species units reveal phylogenetic overdispersion in Pelagibacter
Because ‘lumping' species obscured the phylogenetic overdispersion in our simulated example, we predicted that finer-scale bacterial diversity units based on a more stringent identity threshold, or less conserved markers (e.g., fast evolving protein-coding genes) should then be more likely to reveal it. We tested this hypothesis by carrying out Phylocom analyses of several bacterial datasets using 16S rRNA and protein-coding genes and a range of identity thresholds to define the species boundary. We focused our analyses on sequences of the same genus because species in the same genus are expected to be more likely to compete than those in different genera (Darwin, 1859; Cooper et al., 2008; Cavender-Bares et al., 2009; Wiens et al., 2010).
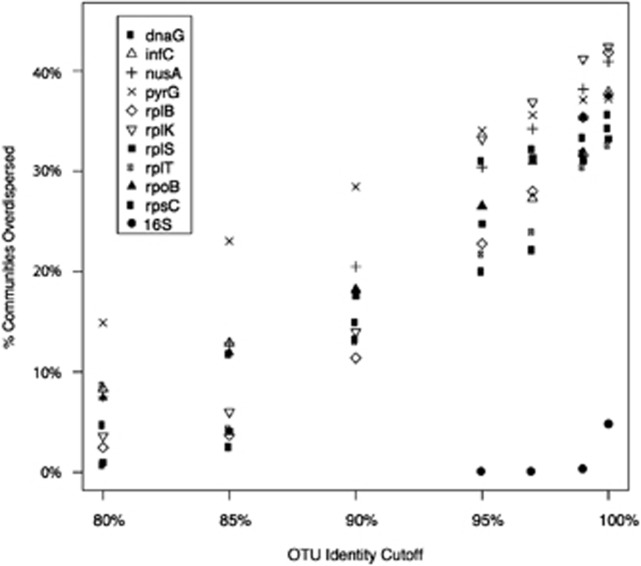
We first assessed the phylogenetic relatedness of Pelagibacter at the sampling sites of the GOS expedition (Rusch et al., 2007). Pelagibacter is the most abundant bacterium in the ocean surface water and is also widely dispersed (Morris et al., 2002). OTUs generated at a variety of sequence identity cutoffs were used as approximations of species. Our analyses of 10 protein-coding genes revealed a pattern consistent with the trend predicted by our simulation. When analyzed using all GOS samples, the fraction of communities that showed negative NRI values (overdispersion) increased in all marker genes as narrower diversity units were applied (Figure 2). The number of statistically significant overdispersed communities also increased with narrower species definition (Figure 3, Table 2). The increase in overdispersion was accompanied by a decrease in phylogenetic clustering. This effect was pronounced and prevalent across all markers (Supplementary Table S1). We noted that while overdispersion tended to increase, and clustering decrease as species definitions were narrowed, in most cases the number of clustered communities still exceeded the number of overdispersed communities. We also carried out Phylocom analysis of a regional pool by comparing samples from Indian Oceans only. The results were similar to the findings described above.
Figure 2.
Phylogenetic overdispersion increases when species are more narrowly defined. This figure displays the Phylocom results for 16S rRNA and 10 protein-coding genes from the GOS Pelagibacter sequences. For the protein-coding genes, the OTU identity cutoff correlates positively with the fraction of overdispersed communities. For 16S rRNA, no or little overdispersion was detected when 95%, 97%, 99% or 100% identity cutoffs were used.
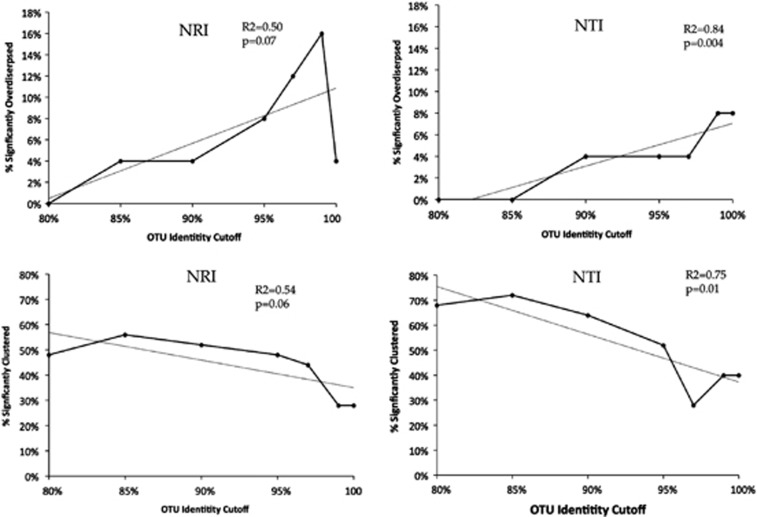
Figure 3.
Statistically significant phylogenetic overdispersion increases when species are more narrowly defined. This figure displays the Phylocom results for the rplK gene of the GOS Pelagibacter. The OTU identity cutoff correlates positively with the fraction of significantly overdispersed communities and negatively with the fraction of significantly-clustered communities.
Interestingly, for all protein markers the maximum number of significantly overdispersed communities was detected using identity cutoffs greater than 97% (data not shown). This is much narrower than the 97% or 99% 16S rRNA OTUs typically used to approximate bacterial species. In Pelagibacter, for example, two species with 99% identical 16S rRNA gene share only 80% DNA sequence identity at the rplK gene. Therefore, 99% 16S rRNA OTUs are roughly equivalent to 80% rplK OTUs. Accordingly, we did not detect any statistically significant overdispersion when we analyzed the GOS data using the 16S rRNA gene at 97%, 99% or 100% identity cutoffs. This result suggests that 16S rRNA OTUs, the widely used bacterial diversity unit, might be too broad to detect phylogenetic overdispersion in Pelagibacter communities, as seen in other bacterial communities (Horner-Devine and Bohannan, 2006; Pontarp et al., 2012).
These results demonstrate that broad definitions of bacterial species (e.g., 16S rRNA OTUs) tend to indicate habitat filtering as the dominant driver of community assembly, while narrower definitions (e.g., OTUs of protein-coding genes) suggest the possibility of a stronger role for interspecific competition. That we were able to observe this trend with both the NRI and NTI metrics is especially striking given previously observed effects of tree size (the number of taxa) on NRI and NTI. Prior studies have shown that NTI underpredicts overdispersion as the number of terminal taxa increases (Swenson, 2009). The fact that we observed an increased number of overdispersed communities according to the NTI metric as we narrowed the species cutoff (and therefore increased the number of terminal taxa) suggests that the effect may be even more pronounced than our results indicate.
Phylogenetic overdispersion is present in other bacterial communities
We next tested whether the pattern we observed in the GOS Pelagibacter data was also present in other bacterial communities. We analyzed several microbial datasets representing various contrasting habitat types (marine (Hunt et al., 2008) and human body sites (Ley et al., 2006; Grice et al., 2009)), with different genetic markers (16S rRNA and a protein-coding gene) (Table 1).
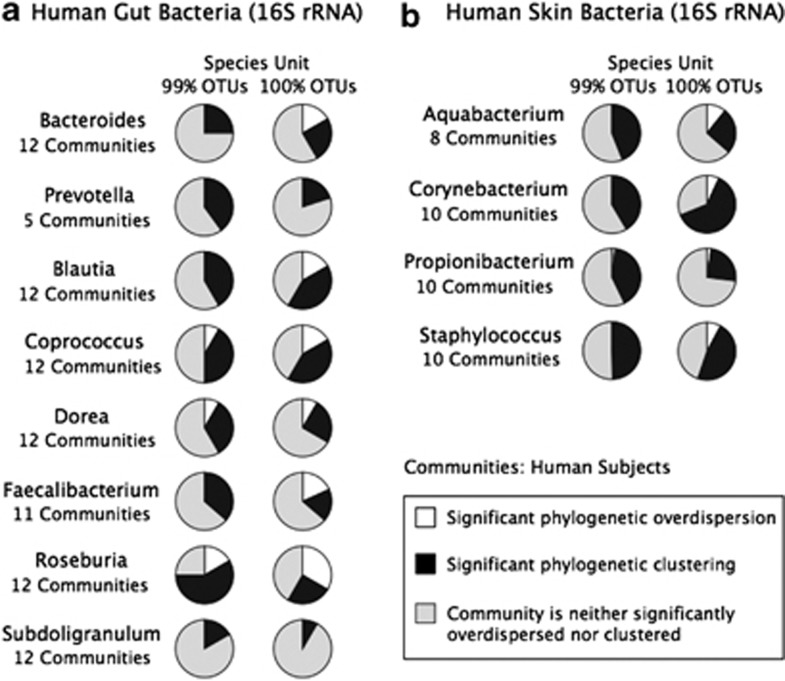
We discovered that for the human microbiome 16S rRNA datasets, there was very little evidence of phylogenetic overdispersion across habitats at a broader species definition (99% sequence identity) (Figure 4). The number of phylogenetically clustered communities was much greater than the number of overdispersed communities in every case. Taken on its own, this finding would appear to indicate that bacterial communities are predominantly assembled via habitat filtering rather than by competition-driven dispersion, as observed previously (Horner-Devine and Bohannan, 2006; Pontarp et al., 2012). However, when we narrowed the definition of species to a 100% sequence identity cutoff, the proportion of communities showing overdispersion increased substantially. A similar trend was observed in the Vibrio hsp60 dataset using the NRI metric (Figure 5a). No overdispersion was detected when OTU was defined using 97% identity cutoff. In comparison, one third of the communities were overdispersed when 99% and 100% identity cutoffs were used. These results further support the hypothesis that finer-scale species delineations are necessary for phylogenetic overdispersion to be readily detectable.
Figure 4.
Phylocom analysis of additional datasets. Phylocom results for all genera analyzed from the 16S rRNA sequences of the gut microbiome dataset (a), the skin microbiome dataset (b). Only NRI results are displayed. For simplicity, the skin microbiome pie charts displayed the Phylocom results averaged over all 21 skin sites.
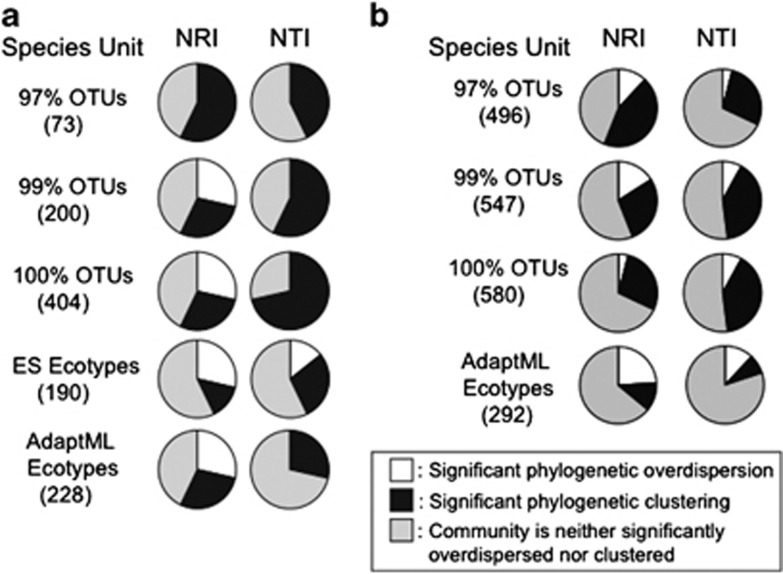
Figure 5.
Phylocom analyses using OTUs and ecotypes. This figure displays the Phylocom results for the gene hsp60 from the marine Vibrio dataset (a), and for the rplK gene in the GOS Pelagibacter dataset (b). As in previous figures, the pie charts show the fraction of communities that were significantly overdispersed, clustered or not significantly different from the null model. Species units were either OTUs (at 97%, 99%, and 100% identity cutoffs) or ecotypes (estimated using the AdaptML or ES algorithms). The numbers in parentheses indicate the number of species units.
Ecotypes reveal phylogenetic overdispersion in Vibrio and Pelagibacter
Recent models of bacterial speciation have suggested that very recently diverged bacterial ecotypes, whose discovery requires the resolution of rapidly evolving sequences, are better approximation of bacterial species (Cohan and Perry, 2007; Wiedenbeck and Cohan, 2011). We generated ecotypes in a marine Vibrio dataset using the ES (Koeppel et al., 2008) and AdaptML (Hunt et al., 2008) algorithms. ES identifies ecotypes by comparing the observed pattern of sequence diversity in a bacterial community to those of simulated communities ‘evolved' based on the stable ecotype model (Cohan and Perry, 2007; Cohan and Koeppel, 2008). AdaptML, by contrast, demarcates ecotypes by inferring the evolutionary history of habitat transitions. It identifies an ecotype as the largest clade whose members share an inferred habitat. Unlike OTU, neither ES nor AdaptML requires an arbitrary identity cutoff to demarcate ecotypes. Since the ecotypes defined by these algorithms are expected to be more evolutionarily and ecologically meaningful than OTUs, our expectation was that phylogenetic overdispersion would be easier to detect if ecotypes were used to approximate species.
The Vibrio hsp60 dataset of Hunt et al. (2008) represents an ideal dataset to test our hypothesis that ecotypes might be better species units for detecting phylogenetic overdispersion. It has the categorical ecological data necessary for analysis with AdaptML, and the pattern of sequence diversity fits the assumptions of the stable ecotype model, as required by ES (Supplementary Figure S1). We generated ecotypes using both the ES and AdaptML algorithms and then used ecotypes as the input species units for Phylocom analysis. Overall, the ecotypes generated by both algorithms showed more overdispersion and less phylogenetic clustering than OTUs. Most strikingly, the only case in which significant phylogenetic overdispersion was detected using the NTI metric was when ES ecotypes were used as the species unit (Figure 5a).
We also used AdaptML to generate ecotypes based on the rplK gene sequences from the GOS Pelagibacter dataset. Phlyocom analysis was performed using these ecotypes as species units, and the results were compared against results from the identical sequence set using OTUs as species. Strikingly, the analysis of the ecotypes indicated that a greater fraction of the communities were significantly overdispersed (and fewer significantly clustered) than had been shown by OTUs at any cutoff (Figure 5b).
Although OTUs provide a ‘quick and dirty' approach to characterizing bacterial diversity and can be useful in many circumstances, they is no substitute for coherent and meaningful units of bacterial ecology and evolution (Gevers et al., 2005; Cohan and Perry, 2007; Ward et al., 2008; Koeppel and Wu, 2013). Our findings suggest that there is no one right OTU identity cutoff that works well for detecting overdispersion. When using the NTI metric, we were able to detect phylogenetic overdispersion of ecotypes in communities that showed no overdispersion of OTUs (Figure 5a). Even very narrow species approximations may have difficulty in detecting phylogenetic overdispersion if they are based solely on sequence identity. This was true even when OTUs were clustered based on 100% identity at a protein-coding locus, a substantially narrower unit of diversity than the typically used 97% or 99% 16S rRNA OTUs (Stackebrandt and Goebel, 1994; Schloss and Handelsman, 2006; Stackebrandt and Ebers, 2006). While it has been well established that a consistent definition of species is necessary for many different types of ecological analysis (Hughes et al., 2001), our results starkly demonstrate the extent to which conclusions about bacterial ecology and evolution can be affected when different species units are employed in phylogenetic analyses.
Implications for future research
Our results highlight the advantages of using protein-coding genes as markers for studying microbial community assembly. We demonstrated that the narrower the species definition, the more phylogenetic overdispersion could be detected. Since the nucleotide sequences of protein-coding genes evolve faster than the 16S rRNA gene, using protein-coding genes should produce finer-scale bacterial species that are ecologically more meaningful. Therefore, the recent advance of metagenomics to examine genomes of different lineages from environmental samples will increase the power of phylogenetic methods for bacterial assembly analysis.
Our results also underscore the need for deep sequencing in microbial ecology studies. The more deeply sequenced a community is, the finer the taxonomic scales at which it can be examined and analyzed. There is a general expectation that community assembly at broader taxonomic scales will be predominated by habitat filtering (Cavender-Bares et al., 2006; Horner-Devine and Bohannan, 2006). However, due to limited sequencing depth in previous studies, bacterial community assembly were analyzed mostly at very broad taxonomic scales (either using the entire taxonomic breadth of bacteria in a community or at the phylum level) (Horner-Devine and Bohannan, 2006; Silvertown et al., 2006; Bryant et al., 2008; Pontarp et al., 2012; Wang et al., 2012). Without deep sequencing data, our analyses of community assembly at the genus level would not have been possible because there would not have been enough sequences of the same genus for Phylocom analysis. The depth of sequencing can also affect the way we demarcate species. For example, although GOS was a large-scale sampling expedition, our rarefaction analysis of the marker sequence data indicated that each individual sampling site was still undersampled (data not shown). The insufficient sampling prevented us from analyzing the GOS data using ES ecotypes because the sequence data did not adequately capture the microdiversity that was necessary for ES analysis.
If interspecific competition does play a significant role in bacterial community assembly in general, then it is possible to use its phylogenetic signature to evaluate the effectiveness of bacterial species definitions. Our simulated example indicates that phylogenetic overdispersion is at maximum when the proper species unit is used in the Phylocom analysis. Splitting or lumping species all produce lowered estimates of phylogenetic overdispersion. Therefore, the degree of phylogenetic overdispersion can be used as an objective function to benchmark species units. Using this criterion, our Phylocom analyses of Vibrio and Pelagibacter datasets indicated that ecotypes are better approximation of bacterial species than OTUs because for the same set of sequence data, overdispersion estimated using ecotype as species was greater than those estimated with OTUs.
Challenge of linking phylogenetic patterns to assembly processes
Phylogenetic structures have been successfully used to infer the underlying assembly processes. However, linking phylogenetic patterns to processes is not always straightforward because of their many-to-many relationships. For example, overdispersion does not always indicate competition. Overdispersion can result from habitat filtering when distant relatives share convergent traits. Overdispersion can also indicate facilitation between distantly related species (Cavender-Bares et al., 2004; Verdú et al., 2009). Both are unlikely to be the case in our study because we focused on closely-related species. One advantage of working with closely related taxa is that phylogenetic patterns are more likely to be indicative of the assembly processes, as suggested previously (Kraft et al., 2007; Fine and Kembel, 2011; Stegen et al., 2012). This is because the assumption of phylogenetic niche conservatism is more likely to be valid between closely-related taxa. Nevertheless, the possibility of phage predation causing phylogenetic overdispersion in bacterial communities (Sullivan et al., 2003; Acinas et al., 2004; Thompson et al., 2005; Holmfeldt et al., 2007; Lennon et al., 2007) cannot be excluded in our study.
Conversely, competition does not always drive phylogenetic overdispersion. The core assumption of the competition-relatedness hypothesis—that closely related species compete more intensely than distantly-related species has been challenged recently (Mayfield and Levine, 2010). According to the modern coexistence theory, species coexistence is driven by the interaction of two types of species differences: niche differences and competitive ability differences. When species differ primarily in their niche preference, closely-related species will have similar niches, and therefore are less likely to coexist, resulting in phylogenetic overdispersion. On the other hand, when species differ primarily in their competitive ability, closely-related species will have similar competitive ability and thus are more likely to coexist, resulting in phylogenetic clustering. Under the Mayfield and Levine model, although competition can lead to either overdispersion or clustering, overdispersion can still only result from competition. In other words, overdispersion would indicate competition when alternative explanations are exhausted, as discussed above.
Conclusions
Our results demonstrate that the definition of species matters a great deal to phylogenetic analyses of community assembly. Using many genes, numerous lineages and a wide range of habitats, we have shown that the use of finer-scale species units such as ecotypes can reveal phylogenetic overdispersion in communities where it was not apparent with broader units. Although habitat filtering could well be the dominant force, our results suggest the possibility of a more prominent role for interspecific competition in bacterial community assembly than had previously been recognized. Our findings therefore illustrate the need for careful consideration of how to delineate bacterial species in bacterial evolution and ecology studies.
Acknowledgments
We would like to thank Frederick M Cohan for valuable discussions.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, et al. Fine-scale phylogenetic architecture of a complex bacterial community. Nature. 2004;430:551–554. doi: 10.1038/nature02649. [DOI] [PubMed] [Google Scholar]
- Bryant JA, Lamanna C, Morlon H, Kerkhoff AJ, Enquist BJ, Green JL. Colloquium paper: microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc Natl Acad Sci USA. 2008;105:11505–11511. doi: 10.1073/pnas.0801920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavender-Bares J, Ackerly DD, Baum DA, Bazzaz FA. Phylogenetic overdispersion in Floridian Oak communities. Am Nat. 2004;163:823–843. doi: 10.1086/386375. [DOI] [PubMed] [Google Scholar]
- Cavender-Bares J, Kozak KH, Fine PVA, Kembel SW. The merging of community ecology and phylogenetic biology. Ecol Lett. 2009;12:693–715. doi: 10.1111/j.1461-0248.2009.01314.x. [DOI] [PubMed] [Google Scholar]
- Cavender-Bares J, Keen A, Miles B. Phylogenetic structure of Floridian plant communities depends on taxonomic and spatial scale. Ecology. 2006;87:109–122. doi: 10.1890/0012-9658(2006)87[109:psofpc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Cohan FM, Koeppel AF. The origins of ecological diversity in prokaryotes. Curr Biol. 2008;18:R1024–R1034. doi: 10.1016/j.cub.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Cohan FM, Perry EB. A systematics for discovering the fundamental units of bacterial diversity. Curr Biol. 2007;17:R373–R386. doi: 10.1016/j.cub.2007.03.032. [DOI] [PubMed] [Google Scholar]
- Cohan FM. What are bacterial species. Annu Rev Microbiol. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- Cooper N, Rodriguez J, Purvis A. A common tendency for phylogenetic overdispersion in mammalian assemblages. Proc Biol Sci. 2008;275:2031–2037. doi: 10.1098/rspb.2008.0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. On the Origin of Species by Means of Natural Selection. John Murray: London; 1859. [Google Scholar]
- Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton CS. Competition and the structure of ecological communities. J Anim Ecol. 1946;15:54–68. [Google Scholar]
- Emerson BC, Gillespie RG. Phylogenetic analysis of community assembly and structure over space and time. Trends in ecol evol. 2008;23:619–630. doi: 10.1016/j.tree.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Fargione J, Brown CS, Tilman D. Community assembly and invasion: an experimental test of neutral versus niche processes. Proc Natl Acad Sci USA. 2003;100:8916–8920. doi: 10.1073/pnas.1033107100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine PVA, Kembel SW. Phylogenetic community structure and phylogenetic turnover across space and edaphic gradients in western Amazonian tree communities. Ecography. 2011;34:552–565. [Google Scholar]
- Gerrish PJ, Lenski RE. The fate of competing beneficial mutations in an asexual population. Genetica. 1998;102-103:127–144. [PubMed] [Google Scholar]
- Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, et al. Opinion: Re-evaluating prokaryotic species. Nature Rev Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- Goris J, Konstantinidis K, Klappenbach J, Coenye T, Vandamme P, Tiedje J. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nature Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt K, Middleboe M, Nybroe O, Rieman L. Large variabilities in host strain susceptibility and phage host range govern interactions between lytic marine phages and their flavobacterium hosts. Appl Environ Microbiol. 2007;73:216730–216739. doi: 10.1128/AEM.01399-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner-Devine MC, Bohannan BJM. Phylogenetic clustering and overdispersion in bacterial communities. Ecology. 2006;87:100–108. doi: 10.1890/0012-9658(2006)87[100:pcaoib]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJM. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl Environ Microbiol. 2001;67:4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- Knouft JH, Losos JB, Glor RE, Kolbe JJ. Phylogenetic analysis of the evolution of the niche in lizard of the Anolis sagrei group. Ecology. 2006;87:S29–S38. doi: 10.1890/0012-9658(2006)87[29:paoteo]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Koeppel A, Perry EB, Sikorski J, Krizanc D, Warner A, Ward DM, et al. Identifying the fundamental units of bacterial diversity: a paradigm shift to incorporate ecology into bacterial systematics. Proc Natl Acad Sci USA. 2008;105:2504–2509. doi: 10.1073/pnas.0712205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppel AF, Wu M. Surprisingly extensive mixed phylogenetic and ecological signals among bacterial Operational Taxonomic Units. Nucleic Acids Res. 2013;41:5175–5188. doi: 10.1093/nar/gkt241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft NJB, Cornwell WK, Webb CO, Ackerly DD. Trait evolution, community assembly, and the phylogenetic structure of ecological communities. Am Nat. 2007;170:271–283. doi: 10.1086/519400. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Shikano S, Toda M. Trade-off between interspecific competitive ability and growth rate in bacteria. Ecology. 1990;71:645–650. [Google Scholar]
- Lennon JT, Khatana SA, Marston MF, Martiny JB. Is there a cost of virus resistance in marine cyanobacteria. ISME J. 2007;1:300–312. doi: 10.1038/ismej.2007.37. [DOI] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Losos JB, Leal M, Glor RE, Queiroz KD, Hertz PE, Schettino LR, et al. Niche lability in the evolution of a Caribbean lizard community. Nature. 2003;424:542–545. doi: 10.1038/nature01814. [DOI] [PubMed] [Google Scholar]
- Losos JB. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol Lett. 2008;11:995–1003. doi: 10.1111/j.1461-0248.2008.01229.x. [DOI] [PubMed] [Google Scholar]
- MacArthur R, Levins R. The limiting similarity, convergence, and divergence of coexisting species. Am Nat. 1967;101:377–385. [Google Scholar]
- Mayfield MM, Levine JM. Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol Lett. 2010;13:1085–1093. doi: 10.1111/j.1461-0248.2010.01509.x. [DOI] [PubMed] [Google Scholar]
- Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- Newton RJ, Jones SE, Helmus MR, McMahon KD. Phylogenetic ecology of the freshwater actinobacteria acI lineage. Appl Environ Microbiol. 2007;73:7169–7176. doi: 10.1128/AEM.00794-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontarp M, Canback B, Tunlid A, Lundberg P. Phylogenetic analysis suggests that habitat filtering is structuring marine bacterial communities across the globe. Microb Ecol. 2012;64:8–17. doi: 10.1007/s00248-011-0005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis A, Gittleman JL, Cardillo M. Global patterns in the phylogenetic structure of island mammal assemblages. Proc R Soc B. 2008;275:1549–1556. doi: 10.1098/rspb.2008.0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- Rice NH, Martinez-Meyer E, Peterson AT. Ecological niche differentiation in the Aphelocoma jays: a phylogenetic perspective. Biol J Linn Soc. 2003;80:369–383. [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Toward a Census of Bacteria in Soil. PLoS Comput Biol. 2006;2:e92. doi: 10.1371/journal.pcbi.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvertown J, Dodd M, Gowing D, Lawson C, McConway K. Phylogeny and the hierarchical organization of plant diversity. Ecology. 2006;87:39–49. doi: 10.1890/0012-9658(2006)87[39:pathoo]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- Staley JT. The bacterial species dilemma and the genomic-phylogenetic species concept. Proc R Soc B. 2006;361:1899–1909. doi: 10.1098/rstb.2006.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegen JC, Lin X, Konopka AE, Fredrickson JK. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012;6:1653–1664. doi: 10.1038/ismej.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MB, Waterbury JB, Chisholm SW. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature. 2003;424:1047–1051. doi: 10.1038/nature01929. [DOI] [PubMed] [Google Scholar]
- Sun S, Chen J, Li W, Altinatas I, Lin A, Peltier S, et al. Community cyberinfrastructure for Advanced Microbial Ecology Research and Analysis: the CAMERA resource. Nucleic Acids Res. 2010;39:D546–D551. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson NG. Phylogenetic resolution and quantifying the phylogenetic diversity and dispersion of communities. PLoS One. 2009;4:e4390. doi: 10.1371/journal.pone.0004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt DE, Benoit J, et al. Genotypic diversity within a natural coastal bacterioplankton population. Science. 2005;307:1311–1313. doi: 10.1126/science.1106028. [DOI] [PubMed] [Google Scholar]
- Tilman D. Niche tradeoffs, neutrality, and community structure: a stochastic theory of resource competition, invasion, and community assembly. Proc Natl Acad Sci USA. 2004;101:10854–10861. doi: 10.1073/pnas.0403458101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamosi SM, Heard SB, Vamosi JC, Webb CO. Emerging patterns in the comparative analysis of phylogenetic community structure. Mol Ecol. 2009;18:572–592. doi: 10.1111/j.1365-294X.2008.04001.x. [DOI] [PubMed] [Google Scholar]
- Verdú M, Rey PJ, Alcántara JM, Siles G, Valiente-Banuet A. Phylogenetic signatures of facilitation and competition in successional communities. J Ecol. 2009;97:1171–1180. [Google Scholar]
- Wang J, Soininen J, He J, Shen J. Phylogenetic clustering increases with elevation for microbes. Environ Microbiol Rep. 2012;4:217–226. doi: 10.1111/j.1758-2229.2011.00324.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DM, Cohan FM, Bhaya D, Heidelberg JF, Kuhl M, Grossman A. Genomics, environmental genomics and the issue of microbial species. Heredity. 2008;100:207–219. doi: 10.1038/sj.hdy.6801011. [DOI] [PubMed] [Google Scholar]
- Ward DM, Bateson MM, Ferris MJ, Kuhl M, Wieland A, Koeppel A, et al. Cyanobacterial ecotypes in the microbial mat community of Mushroom Spring (Yellowstone National Park, Wyoming) as species-like units linking microbial community composition, structure and function. Proc R Soc B. 2006;361:1997–2008. doi: 10.1098/rstb.2006.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CO, Ackerly DD, Kembel SW. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics. 2008;24:2098–2100. doi: 10.1093/bioinformatics/btn358. [DOI] [PubMed] [Google Scholar]
- Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Phylogenies and community ecology. Annu Rev Ecol Syst. 2002;33:475–505. [Google Scholar]
- Weiher E, Keddy PA. The assembly of experimental wetland plant communities. Oikos. 1995;73:323. [Google Scholar]
- Wiedenbeck J, Cohan FM. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol Rev. 2011;35:957–976. doi: 10.1111/j.1574-6976.2011.00292.x. [DOI] [PubMed] [Google Scholar]
- Wiens JJ, Ackerly DD, Allen AP, Anacker BL, Buckley LB, Cornell HV, et al. Niche conservatism as an emerging principle in ecology and conservation biology. Ecol Lett. 2010;13:1310–1324. doi: 10.1111/j.1461-0248.2010.01515.x. [DOI] [PubMed] [Google Scholar]
- Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.