Abstract
Background
The cause of corticosteroid resistant asthma is unknown.
Objective
To perform gene microarray analyses using BAL cells from well-characterized corticosteroid resistant (CR) and sensitive (CS) asthmatics to elucidate the differential expression of genes that contribute to the development of corticosteroid resistance.
Methods
The patients were characterized as CR or CS based on FEV1% predicted improvement after one week course of oral prednisone. Expression of selected gene targets was verified by real time PCR and by ELISA.
Results
Microarray analyses demonstrated significantly higher levels (over three-fold increase, p<0.05) of transcripts for TNFα, IL-1α, IL-1β, IL-6, CXCL1, CXCL2, CXCL3, CXCL8 (IL-8), CCL3, CCL4, CCL20 in BAL cells of CR asthmatics. These findings, confirmed by RT-PCR in additional BAL samples, were consistent with classical macrophage activation by bacterial products. In contrast, markers of alternatively-activated macrophages, Arginase I and CCL24, were decreased. Genes associated with activation of the LPS signaling pathway (EGR1, DUSP2, MAIL, TNFAIP3) were significantly elevated in CR BAL samples (p<0.05). These patients had significantly higher amounts (1444.0±457.3 pg per mg of total protein) of LPS in BAL fluid than CS asthmatics (270.5±216.0 pg; p<0.05) as detected by LAL assay and confirmed by gas chromatography mass spectrometry analysis. Pronged exposure to LPS induced functional steroid resistance to dexamethasone (DEX) in normal monocytes, demonstrated by persistently elevated IL-6 levels in the presence of DEX.
Conclusions
Classical macrophage activation and induction of LPS signaling pathways along with high endotoxin levels detected in BAL fluid from CR asthmatics suggest that LPS exposure may contribute to CR asthma.
Keywords: corticosteroids, asthma, resistance, genes, endotoxin
INTRODUCTION
Corticosteroids are highly effective in the treatment of inflammation and are currently recommended for management of persistent asthma as the preferred therapy for control of airway inflammation.1 However, a subset of asthmatics does not respond to steroid therapy2–6 and is therefore at risk for escalation of disease severity. Corticosteroid resistance presents a major management problem and accounts for a disproportionate amount of healthcare costs in this group of patients.7,8 The studies suggest that there is a substantial need for further improvement in asthma therapy because a large group of asthmatic patients remain symptomatic despite optimal therapy.
The data collected to date suggest that, in most cases, corticosteroid resistant (CR) asthma is an acquired condition.3 However, the environmental factors that contribute to CR asthma are unknown. Use of new technologies such as gene microarray analyses which can examine the complete gene expression profile of inflammatory airway cells from CR vs. corticosteroid sensitive (CS) asthmatics have the potential to identify new hypotheses concerning the cause of this increasingly recognized challenge in the treatment of asthma. The gene microarray approach is a comprehensive technique that is not limited by a preconceived hypothesis. The present study was undertaken to identify genes that are differentially expressed in BAL cells from CR and CS asthmatics to determine what may contribute to CR asthma and the potential role that endotoxin (LPS) may have in this condition.
MATERIALS AND METHODS
Subjects
Patients with a diagnosis of asthma according to American Thoracic Society criteria9 were selected for evaluation. Asthmatics had a baseline FEV1% predicted of 55% to 85% of predicted, a β2-adrenergic response of ≥12% of baseline FEV1% predicted and/or a methacholine PC20 value of ≤8 mg/ml. Asthma patients were further subdivided in CR and CS groups, based on their response to steroids. The definition was based on change in FEV1% predicted over a one-week course of 40 mg/day of oral prednisone. Asthmatic patients were defined as CS if they had an increase in FEV1% predicted of ≥15% after a one-week course of prednisone, and as CR if ≤12% change in FEV1% predicted was observed. None of the asthmatic subjects recruited for this study had evidence for other types of lung diseases. None of the subjects had received systemic corticosteroid therapy for at least one month prior to bronchoscopy. Normal healthy adult donors with no history of chronic respiratory disorders and normal spirometry were recruited for the study as well. All subjects provided written informed consent to participate in the study. The study was approved by the Institutional Review Board at National Jewish Medical and Research Center, Denver, CO.
Ten CR and eight CS asthma patients were recruited for the study and underwent bronchoscopy with collection of BAL. Gene array studies of BAL macrophages were performed in 3 CR and 3 CS asthmatics. To confirm gene array results real-time PCR was performed using BAL samples from an additional 7 CR and 5 CS asthmatics. Two CS asthma patients underwent bronchoscopy twice, first time the RNA from cells was used for gene array study; second time the RNA from BAL cells was used for real-time PCR. No difference in BAL differential counts in between two bronchoscopies was found for these 2 CS asthmatics (data not shown). Blood monocytes for evaluation of steroid response after pre-incubation with LPS were obtained from 6 normal control subjects.
Preparation of BAL Cells and Fluids
Fiberoptic bronchoscopies with BAL were performed according to the guidelines of the American Thoracic Society.10 BAL cells were filtered through a 70-μm Nylon cell strainer (Becton Dickinson Labware, Franklin Lakes, NJ), spun at 200g for 10 min, washed two times, and resuspended in HBSS. After filtration of BAL cells, BAL fluid was aliquoted and stored at −80°C. BAL differentials were obtained on cytospin preparations by using a Diff-Quick (Scientific Products, McGraw Park, IL) stain, counting a minimum of 500 cells. RNA from freshly isolated 3×106 BAL cells was preserved in the RLT buffer and stored at −80°C for further RNA extraction.
Gene Expression Profile Analysis
The gene microarray studies were performed by the Gene Microarray Core Lab at the University of Colorado at Denver and Health Sciences Center (UCDHSC), Denver, CO, as described by us earlier11.12 (for details see online supplemental data file).
Real-time PCR was used to confirm expression of selected genes in BAL cells. RNA was transcribed into cDNA, and analyzed by real-time PCR using the dual-labeled fluorigenic probe method on an ABI Prism 7000 sequence detector (Applied Biosystems, Foster City, CA) as described.11,13 All primers and probes for the target genes and housekeeping genes (GAPDH, 18s RNA) were purchased from Applied Biosystems. Standard curves for all targets were generated using the fluorescent data from ten-fold serial dilutions of total cDNA of the highest expression sample. Quantities of target genes in test samples were normalized to the corresponding housekeeping gene levels in each sample. A blinded investigator who was unaware of the clinical status of the patients performed the assays.
Multiplex ELISA Assay
A Human 17-plex panel ELISA (171-A11127) (Bio-Rad, Hercules, CA) was performed to analyze human cytokine/chemokine expression in BAL fluid samples from 6 CR and 4 CS asthmatics. To account for the variability in protein content in BAL fluid due to the differences in the recovered BAL fluid volume, the data was normalized to the total protein amounts in the samples detected by the Bradford assay (Bio-Rad, Hercules, CA).
Preparation of Peripheral Blood Monocytes (PBMC) and Culture
Human PBMC were isolated by Ficoll-Hypaque® density gradient centrifugation of heparinized venous blood. Monocytes were isolated from PBMC using magnetic bead negative selection (Monocyte isolation kit II, Miltenyi Biotec, Auburn, CA), and resuspended in RPMI 1640 (BioWhittaker) containing 10% charcoal filtered heat inactivated steroid-free FCS (Gemini Bio Products, Calabasas, CA), 40 μmol/l L-glutamine, 100 U/ml penicillin, 100 U/ml streptomycin, and 20 mmol/l HEPES (GIBCO BRL Life Technologies) at a concentration of 5×105/ml. Monocytes were pretreated with 10 ng/ml LPS for 1 or 24h, and then cultured for an additional 24h in the absence or presence of 10−9 to 10−6M dexamethasone (DEX). Culture supernatants were collected and stored at −20°C until analysis for IL-6 production by ELISA, using a human IL-6 ELISA kit (R&D Systems, Inc., Minneapolis, MD), according to the manufacturer’s instructions.
Measurement of LPS in BAL Fluid
The content of LPS in BAL fluid was analyzed by the chromogenic limulus amebocyte lysate test (Cambrex Bio Science Walkersville, Inc., Walkersville, MD) according to the manufacturer’s instructions and normalized to the total protein level in each sample.
Gas Chromatography/Electron Ionization-Mass Spectrometry (GS/EI-MS) analysis of BAL fluid
This analysis was performed by Analytical Toxicology Laboratory, Department of Environmental and Radiological Health Sciences, Colorado State University, Fort Collins, CO based on quantification of 3-hydroxy fatty acids (3-OHFA) as a chemical marker of LPS (for detailes see online supplemental data file).
Statistical Methods
Patient clinical variables were analyzed using descriptive statistics, including proportions or means with ranges when appropriate. For details on gene microarray analysis look at the online supplemental data file.
Analysis for statistical significant differences for the data gathered by ELISAs and real-time PCR was performed with unpaired two-tailed t–test and nonparametric Mann-Whitney test, respectively. The Pearson’s correlation analysis was performed with log transformed data that assumed Gaussian distribution. The Graph Pad Prism, version 4.01 (San Diego, CA) was used for all statistical calculations. P values ≤0.05 were considered significant.
RESULTS
Patient characteristics
Eighteen asthmatics (10 CR and 8 CS) were recruited for this study and underwent bronchoscopy with collection of BAL. Patients were divided into CR and CS groups based on FEV1% predicted responses after a one week course of oral prednisone. As shown in Supplementary Table I, patients in the CR group did not show any improvement in pre-bronchodilator FEV1% predicted after exposure to prednisone (mean±SEM 67.0±2.7% and 65.8±2.5% before vs. after steroid burst, respectively; p=0.54). In contrast, patients in the CS group showed significant improvement in their lung function after steroid burst (68.6±2.7% and 83.1±2.8%, before vs. after steroid burst, respectively; p<0.0001, as compared to FEV1% predicted before steroid burst). During the study, the patients continued to use inhaled steroids, but were asked to withhold them 24h prior to bronchoscopy. Patients that had respiratory infection and/or used antibiotics within four weeks of scheduled bronchoscopy were excluded from the study. There was no evidence of active infection during the time of bronchoscopy in either of the asthma groups studied.
The number of total white cells in BAL samples (see Supplementary Table II) collected was significantly lower in samples from CR as compared CS asthma group (p=0.002) and also varied between patients (mean±SEM total white cell count for CR (n=10) and CS (n=8) groups was 9.3±1.7×106 and 17.3±1.2×106, respectively). This difference is due to the differences in the volume of BAL fluid (% BAL fluid return) recovered after bronchoscopy (BAL fluid % return was 41.3±5.7% vs. 62.8±2.6% in CR and CS asthma groups, respectively (p=0.008)). However, the percentage of macrophages, lymphocytes, neutrophils and eosinophils did not differ between the two groups. Equal amounts of BAL cells were processed from each patient for RNA isolation. Macrophages were the major cell population in BAL samples: macrophages composed a mean percentage of 91.3±3.9% and 93.2±1.3% BAL samples from CR (n=10) and CS (n=8) asthma study groups, respectively.
Gene microarray analysis of BAL samples reveals classical macrophage activation in samples from CR asthmatics
BAL samples collected from three CR and three CS Caucasian asthma patients were initially analyzed to screen for potential gene candidates with gene microarray. None of the patients selected were current or former smokers. In terms of asthma severity, both groups were equivalent: baseline pre-bronchodilator FEV1% predicted was 66.6±3.9% in the CR group and 69.4±5.9% in the CS group, p=0.71. Post bronchodilator FEV1% predicted was 80.0±8.5% and 85.0±8.1%, respectively in the CR vs. CS groups, p=0.69.
By using unpaired t-test with Weltch’s correction, we found significant (p<0.05) up-regulation (over three-fold) of 38 probe sets representing 30 genes in CR asthma samples compared with CS asthma samples. Of these 30 genes, 8 were transcribed sequences and hypothetical proteins of unknown function. Three genes were significantly down-regulated (over three-fold) in CR asthma samples compared with CS asthma samples.
It was found that BAL cells from CR asthmatics are remarkably more activated than CS asthmatics. As shown in Table I, a number of pro-inflammatory genes demonstrated significantly greater up-regulation in CR, as compared to CS, asthma; this pattern of activation was confirmed with MAS 5.0 and RMA algorithms of data analysis in GeneSpring™ software.
Table I.
Gene profiling of BAL cells from CR vs. CS asthma
| Classification | Affymetrix U133 + 2.0 | Gene Accession# | Mean CR asthma±SE* | Mean CS asthma±SE* | Fold induction in CR/CS asthma (MAS 5.0) | Fold induction in CR/CS asthma (RMA) |
|---|---|---|---|---|---|---|
| Molecules associated with classical pattern of macrophage activation | ||||||
|
| ||||||
| IL-6 | 205207_at | NM_000600 | 8199±2183 | 231±52 | 35.54† | 34.22† |
| TNF alpha | 207113_s_at | NM_000594 | 4640±593 | 431±114 | 10.76† | 8.9† |
| IL-1 beta | 205067_at | NM_000576 | 20205±6204 | 1947±351 | 10.38† | 7.38† |
| IL-1 alpha | 210118_s_at | M15329 | 8921±4015 | 1053±327 | 8.47† | 6.5† |
| CCL20 | 205476_at | NM_004591 | 15591±3047 | 580±56 | 26.90† | 21.7† |
| CXCL8 (IL-8) | 202859_x_at | NM_000584 | 30651±1958 | 2014±837 | 15.22† | 24.05† |
| CCL3 | 205114_s_at | NM_002983 | 6374±2466 | 752±184 | 8.48† | 11.09† |
| CXCL3 | 207850_at | NM_002090 | 20093±5258 | 2590±569 | 7.72† | 4.92† |
| CCL4 | 204103_at | NM_002984 | 9648±747 | 1372±277 | 7.03† | 5.89† |
| CXCL2 | 209774_x_at | M57731 | 22556±2913 | 3738±1174 | 6.03† | 7.35† |
| CXCL1 | 204470_at | NM_001511 | 663±161 | 117±19 | 5.66† | 7.34† |
|
| ||||||
| Molecules associated with alternative pattern of macrophage activation | ||||||
|
| ||||||
| CCL24 | 221463_at | NM_00299 | 330±58 | 2388±290 | 0.14† | 0.16† |
| arginase 1 | 231662_at | AV652232 | 9±10 | 37±18 | absent | absent |
|
| ||||||
| LPS/TNF related signaling molecules | ||||||
|
| ||||||
| early growth response 1 | 227404_s_at | AI459194 | 39963±13583 | 2696±299 | 14.82† | 12.45† |
| molecule possessing ankyrin repeats induced by LPS (MAIL) | 223217_s_at | BE646573 | 23852±4436 | 3040±1170 | 7.85† | 7.15† |
| dual specificity phosphatase 2 | 204794_at | NM_004418 | 1643±83 | 212±18 | 7.73† | 10.52† |
| TNF alpha-induced protein 3 | 202643_s_at | AI738896 | 11911±4214 | 1849±389 | 6.44† | 4.92† |
Mean signal intensity±SEM of three BAL samples from CR asthmatics and three BAL samples from CS asthmatics
p<0.05 comparing data sets from CR asthma vs. CS asthma groups analyzed with two-tailed unpaired t test
Since BAL samples were mostly composed of macrophages (Supplementary Table II), a majority of genes identified by gene microarray were related to macrophage function and activation. Macrophages are known to express different functional programs in response to microenvironmental signals.14,15 It is known that cytokines and microbial products profoundly affect the function of these cells. Recently, the concept of classical (activation by microbial products and pro-inflammatory cytokines) and alternative activation pattern of macrophages (Th2) has been proposed.14,15 These stimuli induce distinct cell activation programs that differ with regard to receptor expression, effector function, produce a distinct pattern of cytokines and chemokines.14
Interestingly, our microarray analysis of BAL cells obtained from CR and CS patients suggest that macrophages from CR patients have undergone classical activation, expressing significantly higher levels of TNFα, IL-6, IL-1α and IL-1β mRNA compared to CS patients (Table I). As well, a distinct set of chemokines (CXCL1, CXCL2, CXCL3, CXCL8 (IL-8), CCL3, CCL4, CCL20) was found to be significantly activated in BAL cells from CR patients. LPS is an important stimulus for triggering classical macrophage activation.14 Along with cytokines and chemokines, several other genes linked to the LPS signaling pathway were found to be significantly up-regulated in BAL samples from CR asthmatics - EGR1,16–18 MAIL,19,20 DUSP2, TNFα-induced protein 321 (Table I). This indicates the potential exposure of macrophages to LPS in the airways of CR asthmatics. In contrast, markers of alternatively-activated macrophages (Arg I and CCL24), that are reported to be up-regulated by Th2 cytokines in allergic asthma15,22,23 were significantly down-regulated in macrophages from CR asthmatics (Table I). Analysis of gene array data revealed no difference in the expression of Th2 cytokines and their receptors in BAL samples from CR and CS asthmatics (data not shown). These findings suggest that CR macrophages exhibit a pro-inflammatory pattern of gene expression and have undergone classical macrophage activation in contrast to the CS macrophages that typically exhibit a pattern of gene expression characteristic of Th2 exposure and alternative activation.
Confirmation of gene microarray results by real-time PCR and ELISA
To rule out the possibility of false positives when running single microarrays on a sample, we confirmed the gene array data by real-time PCR and ELISA using 14 additional BAL samples (7 CR and 7 CS asthmatics; 2 of the CS asthmatics were repeat bronchoscopies involving CS asthmatics studied in gene microarrays). As shown in Figure 1, real-time PCR amplification of TNFα, IL-8, IL-1β and CCL20 (MIP3α) mRNA confirmed our gene array findings that airway BAL cells from CR asthmatics are associated with significantly greater pro-inflammatory cytokine and chemokine gene expression than found in CS asthmatics (p≤0.01). As for markers of alternative macrophage activation, significantly higher (p<0.05) levels of CCL24 mRNA expression were found in BAL samples from CS asthmatics, but no difference in CCL22 and SOCS1 expression was observed (Figure 2). No mannose receptor C type 1, another gene associated with alternative activation of macrophages14 - was detected in either groups of asthmatics. The summarized data concerning additional targets associated with classical macrophage activation analyzed by real-time PCR is presented in Table II.
Figure 1.
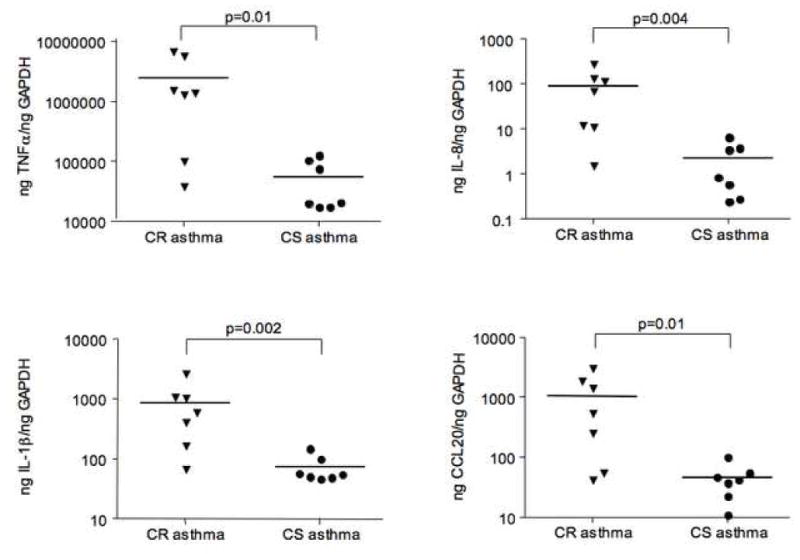
Significant increase in expression of markers associated with classical macrophage activation in BAL cells from CR as compared to CS asthmatics as shown by real-time PCR.
Figure 2.
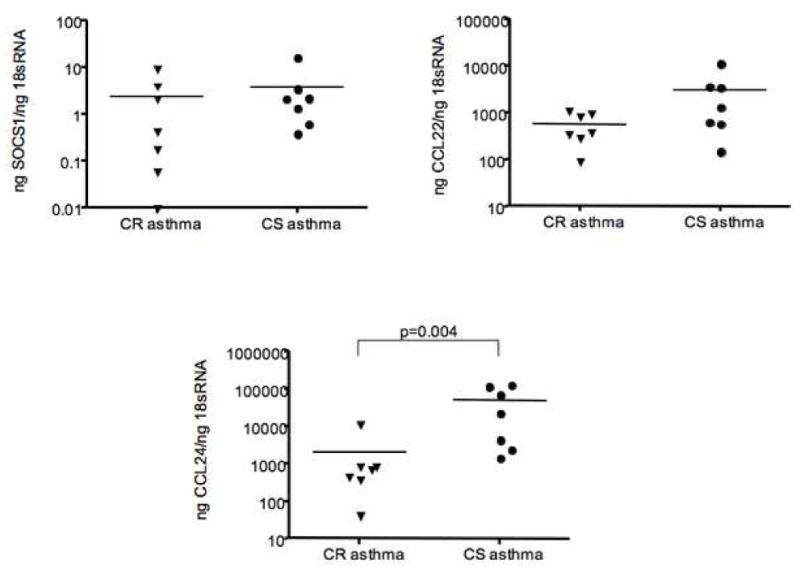
Expression of markers of alternative macrophage activation in BAL cells from CR as compared to CS asthmatics as shown by real-time PCR.
Table II.
Gene profiling of BAL cells from CR vs. CS asthma by real-time PCR
| Classification | Mean CR asthma±SE* | Mean CS asthma±SE* | Fold induction in CR/CS asthma | P value |
|---|---|---|---|---|
| IL-6 | 503±172 | 7.2±1.9 | 69.9 | 0.0007 |
| CXCL8 (IL-8) | 2740±847 | 10.4±1.6 | 263.5 | 0.0006 |
| CCL3 | 25.9±9.1 | 4.2±1.7 | 6.2 | 0.0379 |
| CXCL3 | 14.2±3.3 | 3.8±1.3 | 3.74 | 0.0175 |
| CCL4 | 13.2±5.8 | 3.3±1.3 | 4.0 | 0.0379 |
| CXCL2 | 13.2±3.5 | 4.1±1.2 | 3.22 | 0.0530 |
| CXCL1 | 8.4±4.0 | 2.5±1.3 | 3.36 | 0.1282 |
| early growth response 1 | 14.8±3.9 | 2.2±0.6 | 6.73 | 0.0006 |
| molecule possessing ankyrin repeats induced by LPS (MAIL) | 10.5±3.5 | 2.8±0.8 | 3.75 | 0.0262 |
| dual specificity phosphatase 2 | 34.3±17.0 | 0.8±0.4 | 42.88 | 0.0041 |
| TNF alpha-induced protein 3 | 11911±4214 | 1849±389 | 2.37 | 0.0379 |
Mean signal intensity±SEM ng of the target mRNA/ng 18s RNA of 7 additional BAL samples from CR asthmatics and 7 additional BAL samples from CS asthmatics
To determine whether these differences in cytokine production between the groups can also be seen in BAL fluid, a subset of samples (6 CR and 4 CS asthmatic BAL fluids) was screened by a 17-plex ELISA assay. Clinical features of the subset of patients selected for the BAL fluid cytokine analysis were similar to those described in patients characteristics table (Supplementary Table 1). No difference in the total protein concentration was observed between BAL fluids from 2 groups of patients studied. Significantly higher levels of TNFα, IL-1β and IL-8 were found in BAL fluid from CR, as compared to CS, asthmatics (Figure 3). No difference in Th2 cytokine levels in BAL fluid was found (Figure 3).
Figure 3.
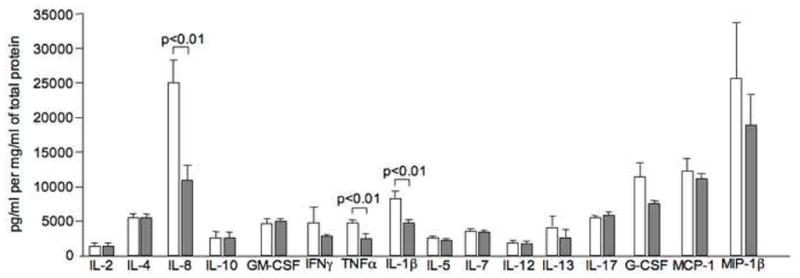
Increased expression of pro-inflammatory cytokines in the BAL fluid samples from CR as compared to CS asthmatics as detected by the 17-plex panel ELISA (n=6 CR asthma group (open bars), n=4 CS asthma group (filled bars)). Expression of 17 different targets was analyzed in 100 μl of BAL fluid and normalized to the total protein amount in each sample.
Presence of LPS in the BAL fluid of CR asthmatics
Since LPS signaling genes were activated on gene microarray, we evaluated the levels of LPS in BAL fluid. The chromogenic limulus amebocyte lysate test was performed and the data obtained were normalized to total protein in BAL fluid. It was found that BAL fluid from 9 out of 10 CR asthmatics tested contained substantial amounts of LPS. In contrast, only trace amounts of LPS could be detected in BAL fluid of CS asthma patients (Figure 4A). 1444.0±457.3 pg LPS per mg of total protein was detected in BAL fluid from CR asthma patients as compared to 270.5±216.0 pg LPS per mg of total protein found in BAL fluid of CS asthmatics, p≤0.05 (Figure 4A).
Figure 4.
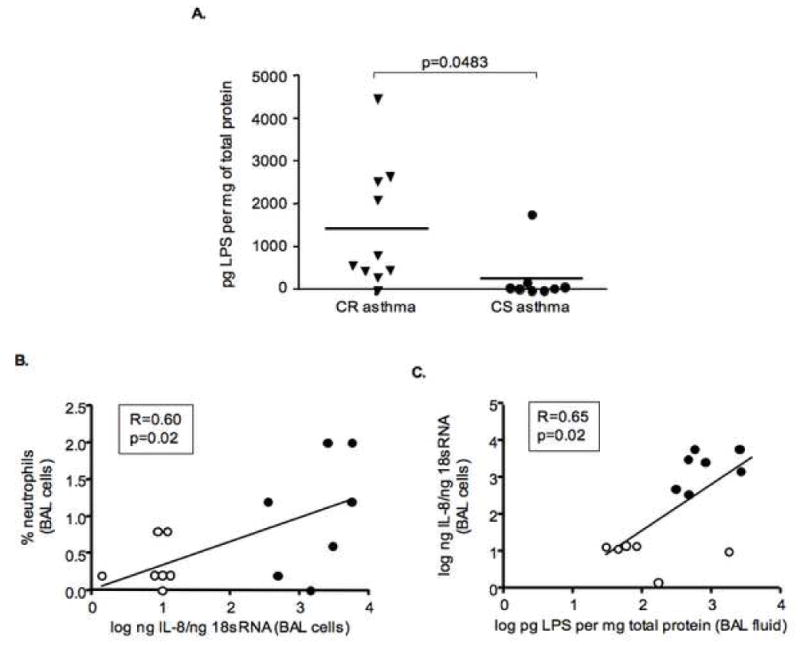
Presence of high amounts of LPS in CR asthma BAL fluid. (A) The content of LPS in BAL fluid was analyzed by the chromogenic limulus amebocyte lysate test and normalized to the total protein level in each sample. IL-8 mRNA expression by BAL cells from CR and CS asthmatics positively correlates with the % of neutrophils in BAL samples (B) and amounts of LPS in BAL fluid (C) (filled circles – CR asthmatics, opened circles – CS asthmatics).
To rule out false positive readings of the LAL assay due to the potential impact of other biologically active components in BAL fluid samples additional experiments were performed with four selected BAL fluid samples where serial dilutions of the BAL fluid samples were tested by LAL assay or the samples were boiled, spiked with known amounts of LPS or measured by LAL assay in presence of the proteinase inhibitor (to rule out confounding results of colorimetric endpoint of the LAL assay due to potential presence of active proteases in BAL fluid samples). No significant impact on the LPS detection was noted (data not shown).
We confirmed presence of LPS in a set of several BAL fluid samples by LPS gas chromatography mass spectrometry analysis based on quantification of 3-hydroxy fatty acids (3-OHFA) as a chemical marker of LPS24.25 (Table III). A significant correlation between the amounts of LPS detected by LAL assay and LPS mass spectrometry assay was noted (R=0.9949, p=0.0051). As well BALF3 and BALF4 were split in half (samples A and B) to assess variability of endotoxin detection by mass spectrometry and based on the data (Table III) the reproducibility of the results was satisfactory.
Table III.
Endotoxin analysis for C8-C17 3-OH fatty acids in BAL fluid samples
| Sample ID | 3-OH fatty acids | LAL assay results |
|---|---|---|
|
| ||
| ng/ml | ng/ml | |
|
| ||
| BALF1 | 1.33 | 2.67 |
| BALF2 | 1.85 | 4.48 |
| BALF3 (A) | 34.76 | 92.25 |
| BALF3 (B) | 25.02 | 92.25 |
| BALF4 (A) | 18.05 | 39.02 |
| BALF4 (B) | 14.17 | 39.02 |
The biologic significance of these observations were supported by evidence for a significant correlation between the % of neutrophils in BAL samples and the amount of IL-8 mRNA expression in BAL cells (R=0.60, p=0.02), as well as between the amount of LPS in BAL fluid and IL-8 mRNA expression by BAL cells (R=0.65, p=0.02) (Figure 4B,C).
Prolonged exposure to LPS alters normal steroid responses of human monocytes in vitro
To assess the effects of LPS on cellular steroid responses, normal human monocytes (n=6) isolated from PBMC using magnetic bead negative selection, were stimulated with LPS alone for 1h or 24h and then various concentrations of DEX were added and the cells were cultured for an additional 24h in the continued presence of LPS. Cell supernatants were collected and analyzed for IL-6 by ELISA. It was found that the duration of LPS exposure determined steroid responsiveness of the cells. IL-6 production by human monocytes was significantly suppressed by steroids when the cells were pretreated with LPS for 1h and then cultured with DEX in the continued presence of LPS (Figure 5A). Pretreatment of cells with LPS for 24h resulted in loss of the ability of DEX to suppress IL-6 production by human monocytes (Figure 5B). Similar effects had been observed when TNFα was measured in cell culture supernatants from monocytes treated with LPS (Figure 5C,D). Thus, LPS re-exposure induced functional steroid resistance.
Figure 5.
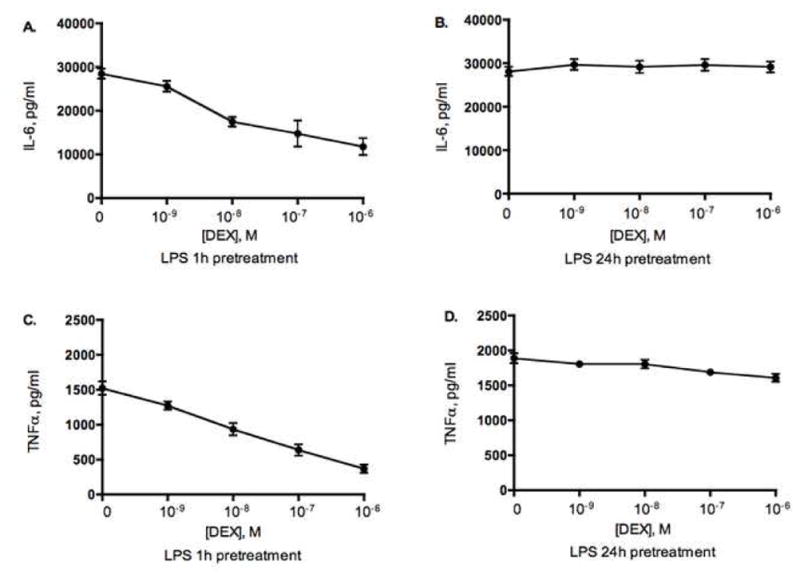
LPS induces functional steroid resistance in normal human monocytes in vitro. Monocytes were cultured with 10ng/ml of LPS for 1h or 24h prior addition of DEX for additional 24h. IL-6 (A,B) and TNFα (C,D) was measured in cell culture supernatants by ELISA. (n=6).
DISCUSSION
The current study determined gene expression profiles of BAL cells collected from CR and CS asthmatics. As demonstrated by activation of multiple pro-inflammatory cytokines and chemokines accompanied by activation of LPS signaling pathways, BAL macrophages were remarkably more skewed towards a pro-inflammatory, anti micrbiol, classically activated phenotype in CR, as compared to CS asthmatics. These data gathered by gene microarray were further confirmed in a separate group of CR and CS asthmatics by real-time PCR and ELISA. The significance of the gene expression profiles presented in Table I is supported not only by the large observed between-group differences, but also by their interrelationships (i.e., a related set of genes were activated). A set of related pro-inflammatory cytokine and chemokine genes (TNFα, IL-6, IL-1β, IL-1α, CXCL1, CXCL2, CXCL3, CXCL8 (IL-8), CCL3, CCL4, CCL20) with strong observed differences reflects a more convincing trend than a strong difference from just one gene and supports a programmed pattern of cell activation.
Macrophages are known to express different functional programs in response to unique microenvironmental signals.14 It has been recognized that macrophages respond to microbial products, such as LPS26 by undergoing classical macrophage activation. This response is associated with increased expression of multiple genes including pro-inflammatory chemokines (CXCL8), cytokines (TNFα, IL-1β, IL-6) and regulators of host defense.27 Early studies have suggested that classical macrophage activation is inhibited by Th2 molecules including IL-4, IL-13, and IL-10. More recently, it has been recognized that these molecules do more that simply down-regulate classically-activated macrophages, but they promote a distinct response, termed alternative macrophage activation. Gene profiling studies have shown that alternatively-activated macrophages are characterized by a different pattern of gene expression than classically-activated macrophages15,12,13 and include genes involved in phagocytosis (mannose receptor, dectin-1, scavenger receptor A), tissue remodeling (arginase I and IGF-I) and with the termination of inflammation (CCL24 and CCL16).
There had been only two studies that attempted to identify novel pathways involved in the pathogenesis of steroid responses in asthma.28,29 One study has performed gene expression profiling in inflammatory cells of CR and CS asthma patients and this was done using peripheral blood mononuclear cells in search of genes that accurately predict responders and nonresponders to inhaled steroids.28 Their analysis focused on genes that were up- or down-regulated by in vitro IL-1β/TNFα stimulation and reversed with steroid treatment in the CS asthma group. Via analysis of the most extreme phenotypes of patients, 15 genes were selected that predicted steroid responses of CR and CS patients with 84% accuracy.28 Our current study was performed with inflammatory cells from airways, thereby allowing us, for the first time, to investigate corticosteroid resistance at the target organ level.
Another recent study29 compared gene expression in epithelial brushings from subjects with asthma before and after treatment with either an inhaled corticosteroid (fluticasone) or placebo in a double blind randomized controlled trial. Also gene expression in epithelial brushings from two control groups (healthy nonsmokers and habitual smokers) was analyzed to identify epithelial abnormalities that are asthma specific. The patients in this study demonstrated mild airway obstruction with FEV1% predicted 87±12%, no patients with asthma were using corticosteroids or long-acting beta agonists prior to enrollment. All patients enrolled into our study had FEV1% predicted lower then 80% and many patients were on inhaled corticosteroids. All three studies (including ours) yielded nonoverlapping datasets of genes that were differentially expressed likely due to the differences in tissue and cell components analyzed, disease severity, differences in sample acquisition. Nevertheless, we believe that these studies provided useful information and insights into contribution of various cell types in pathogenesis of steroid resistance in asthma.
In the current study, we demonstrated a significant increase in TNFα mRNA and protein production by BAL cells of CR patients. This observation is consistent with two recent studies that also demonstrated evidence of up-regulation of the TNFα axis in refractory asthma.30,31 It has been shown that these patients had increased expression of membrane-bound TNFα, TNFα receptor 1 and TNFα-converting enzyme by peripheral blood monocytes.30 As well, significant increases of TNFα were reported in BAL fluid, with a significant change in the number of TNFα producing cells in endobronchial biopsies of severe asthmatics that were refractory to corticosteroids.31 Two clinical trials confirmed that treatment of this group of patients with etanercept (soluble TNFα receptor IgG1 Fc fusion protein) resulted in significant improvement in asthma symptoms, lung function and bronchial hyperresponsiveness,30,31 suggesting that TNFα is a potential factor that requires intervention in CR asthma. There is mounting evidence that corticosteroid insensitivity is associated but not limited to severe asthma, and given the heterogeneity of asthma, treatment resistance may not represent a distinct phenotype in children and adults32–34. Thus, it is difficult to directly extrapolate the results of the studies that demonstrate association of TNFα with refractory asthma, since these studies defined their study population by severity of disease, but not by degree of reversibility following steroid use.
Nothing is known about the phenotypic characteristics of airway macrophages of CS vs. CR asthmatics. Studies in the allergen driven models of airway hyperresponsiveness suggest that the Th2 environment of the sensitized and antigen-challenged lung promotes the expression of genes associated with alternative macrophage activation, including arginase.23 In addition, increased expression of arginase has been detected in BAL cells from asthmatics suggesting that macrophages may undergo alternative macrophage activation in asthma. However, our current studies involving microarray analyses of macrophages obtained from CS and CR patients suggest that CR macrophages exhibit a pro-inflammatory pattern of gene expression and may have deviated towards a gene expression profile similar to that seen during classical microbial macrophage activation. This contrasts to CS macrophages found in most asthmatics that typically exhibit a pattern of gene expression characteristic of alternative macrophage activation. The genes activated, including cytokines and chemokines and molecules in the LPS/NFκB signaling pathway [EGR1,11,13 MAIL (14, 15), DUSP2, TNFα-induced protein 316] indicate the potential exposure of inflammatory cells to LPS in the airways of CR asthmatics. Of interest, no arginase and mannose receptor C type 1 - genes associated with alternative activation of macrophages - were detected in either groups of asthmatics. A recent paper detected arginase activity/protein only in lung lysates and positive cells (macrophages) located deep in the tissue, suggesting that alternatively activated macrophages are not well lavagable.28
Our current study detected the presence of high endotoxin levels in BAL fluid samples of CR asthma. Presence of LPS in BAL fluid was confirmed by two independent methods, i.e. LAL assay and gas chromatography mass spectrometry. Importantly, a significant correlation between the % of neutrophils in BAL samples and the amount of IL-8 mRNA expression by BAL cells, as well as between the amount of LPS in BAL fluid and IL-8 mRNA expression by BAL cells was found, suggesting a functional relationship between LPS exposure and production of IL-8 by BAL macrophages and neutrophil infiltration.
Interestingly, the total number of white cells in BAL samples from CR asthma patients was significantly lower then is CS patients. There may be fewer macrophages in CR group because the cells are more activated and adhesive and therefore resistant to lavage due to LPS presence, since endotoxin is known to stimulate monocyte adhesion.35
Endotoxin, an LPS that comprises most of the outer layer of the bacterial membrane of gram-negative bacteria, is a potent immunomodulator capable of inducing activation of numerous immune cells and cytokine secretion. Exposure to environmental endotoxin is a leading cause of occupational asthma.36 Higher exposure to house-dust endotoxin has also been associated with increased asthma severity in several studies.37–39 The National Survey of Endotoxin in U.S. Housing demonstrated significant relationships between household endotoxin and diagnosed asthma, recent asthma symptoms, current use of asthma medications and wheezing.40
Cellular responses to endotoxin are dependent on its membrane receptor, toll-like receptor (TLR) 4, and are enhanced by CD14.41 Intracellular signaling mediated by endotoxin shares a common MyD88-dependent pathway with other TLRs, and also has a MyD88-independent pathway that is particular to TLR4 and TLR4-mediated activation. Our in vitro experiments indicate that exposure of monocytes to LPS induces cellular steroid resistance. Further studies are needed to delineate the exact activation pathways involved in LPS-induced steroid resistance.
The cause of increased LPS in the airways of CR asthmatics is currently unknown. Bacterial colonization in the airways,42,43 environmental and occupational exposures44,45 should be further explored to assess their contribution to high endotoxin presence in the airways of CR asthma. A recent study by Bisgaard at el.42 reported that neonates colonized in the hypopharyngeal region with S. pneumoniae, H. influenzae or M. catarrhalis, or with combination of these organisms are at risk for recurrent wheeze and asthma early in life, suggesting that microbial colonization of the airways early in life may predispose to more severe asthma. As well, several studies demonstrated anti-inflammatory and steroid-sparing effects of macrolides as add-on therapy for patients with refractory asthma43,46–48. They had been shown to improve pulmonary function, asthma quality of life score and reduce noneosinophilic airway inflammation, particulary neutrophil inflammation, in asthma47,48. Recently, Platts-Mills et al. showed that living with a dog, as compared to living with a cat, significantly increases exposure to airborne endotoxin44, thus creating an environment for worsening asthma. Since dog ownership was reported to be associated with increased endotoxin in house dust44,45, we have administered a questionnaire to CR and CS asthmatics on dog ownership and allergic sensitivity to dogs. Using an established cohort of CR vs. CS asthmatics49,50, we gathered preliminary data demonstrating a significantly increased prevalence of dog ownership among CR asthmatics51. In this regard, we found that 12 out of 18 (66.6%) CR asthmatics lived with at least one dog at home, as compared to 6 out of 16 (37.5%) of CS asthmatics (p≤0.05). As well, it was noted that dog allergy is more prevalent among CR asthmatics. To date, 13 out of 18 CR asthmatics studied were sensitized to dog allergen vs. 5 out of 16 CS asthmatics (p≤0.01). No differences in cat exposures between the two groups were noted.
Thus, understanding what conditions will lead to LPS presence in the airways will help to develop alternative approaches in treatment of CR asthmatics.
Supplementary Material
Clinical Implications statements.
Classical macrophage activation and induction of LPS signaling pathways in BAL cells along with high endotoxin levels in BAL fluid were found in CR asthmatics, suggesting contribution of endotoxin exposure to corticosteroid resistance.
Acknowledgments
This work was supported by NIH grants AI070140, HL37260, General Clinical Research Center grant MO1 RR00051 from the Division of Research Resources, the Edelstein Family Chair in Pediatric Allergy and Immunology, and the University of Colorado Cancer Center.
The authors thank Maureen Sandoval for her help in preparing this manuscript. We would like to thank Brian Cranmer and Stephen Reynolds (Analytical Toxicology Laboratory, Department of Environmental and Radiological Health Sciences, Colorado State University, Fort Collins, CO) for their assistance in the LPS detection in BAL fluid by Gas Chromatography/Electron Ionization-Mass Spectrometry (GS/EI-MS) analysis.
ABBREVIATIONS
- BAL cells
bronchoalveolar lavage cells
- CR asthma
corticosteroid resistant asthma
- CS asthma
corticosteroid sensitive asthma
- DEX
dexamethasone
- FEV1
forced expiratory volume in one second
- LPS
lipopolysaccharide or endotoxin
- PBMC
peripheral blood mononuclear cells
Footnotes
Data deposition footnote: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession # GSE7368).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.National Asthma Education and Prevention Program. Expert Panel Report: Guidelines for the Diagnosis and Management of Asthma Update on Selected Topics--2002. J Allergy Clin Immunol. 2002;110:S141–219. [PubMed] [Google Scholar]
- 2.Szefler SJ, Martin RJ, King TS, Boushey HA, Cherniack RM, Chinchilli VM, et al. Significant variability in response to inhaled corticosteroids for persistent asthma. J Allergy Clin Immunol. 2002;109:410–8. doi: 10.1067/mai.2002.122635. [DOI] [PubMed] [Google Scholar]
- 3.Sher ER, Leung DY, Surs W, Kam JC, Zieg G, Kamada AK, et al. Steroid-resistant asthma. Cellular mechanisms contributing to inadequate response to glucocorticoid therapy. J Clin Invest. 1994;93:33–9. doi: 10.1172/JCI116963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leung DY, Bloom JW. Update on glucocorticoid action and resistance. J Allergy Clin Immunol. 2003;111:3–22. doi: 10.1067/mai.2003.97. [DOI] [PubMed] [Google Scholar]
- 5.Ito K, Chung KF, Adcock IM. Update on glucocorticoid action and resistance. J Allergy Clin Immunol. 2006;117:522–43. doi: 10.1016/j.jaci.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 6.Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br Med Bull. 2000;56:1054–70. doi: 10.1258/0007142001903535. [DOI] [PubMed] [Google Scholar]
- 7.Barnes PJ, Jonsson B, Klim JB. The costs of asthma. Eur Respir J. 1996;9:636–42. doi: 10.1183/09031936.96.09040636. [DOI] [PubMed] [Google Scholar]
- 8.Vollmer WM, Markson LE, O’Connor E, Frazier EA, Berger M, Buist AS. Association of asthma control with health care utilization: a prospective evaluation. Am J Respir Crit Care Med. 2002;165:195–9. doi: 10.1164/ajrccm.165.2.2102127. [DOI] [PubMed] [Google Scholar]
- 9.Standards for the diagnosis and care of patients with chronic obstructive pulmonary disease (COPD) and asthma. This official statement of the American Thoracic Society was adopted by the ATS Board of Directors, November 1986. Am Rev Respir Dis. 1987;136:225–44. doi: 10.1164/ajrccm/136.1.225. [DOI] [PubMed] [Google Scholar]
- 10.Guidelines for fiberoptic bronchoscopy in adults. American Thoracic Society, Medical Section of the American Lung Association. Am Rev Respir Dis. 1987;136:1066. doi: 10.1164/ajrccm/136.4.1066. [DOI] [PubMed] [Google Scholar]
- 11.Nomura I, Goleva E, Howell MD, Hamid QA, Ong PY, Hall CF, et al. Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J Immunol. 2003;171:3262–9. doi: 10.4049/jimmunol.171.6.3262. [DOI] [PubMed] [Google Scholar]
- 12.Nomura I, Gao B, Boguniewicz M, Darst MA, Travers JB, Leung DY. Distinct patterns of gene expression in the skin lesions of atopic dermatitis and psoriasis: a gene microarray analysis. J Allergy Clin Immunol. 2003;112:1195–202. doi: 10.1016/j.jaci.2003.08.049. [DOI] [PubMed] [Google Scholar]
- 13.Goleva E, Li LB, Eves PT, Strand MJ, Martin RJ, Leung DY. Increased glucocorticoid receptor beta alters steroid response in glucocorticoid-insensitive asthma. Am J Respir Crit Care Med. 2006;173:607–16. doi: 10.1164/rccm.200507-1046OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–55. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 15.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 16.Sweet MJ, Hume DA. Endotoxin signal transduction in macrophages. J Leukoc Biol. 1996;60:8–26. doi: 10.1002/jlb.60.1.8. [DOI] [PubMed] [Google Scholar]
- 17.Yao J, Mackman N, Edgington TS, Fan ST. Lipopolysaccharide induction of the tumor necrosis factor-alpha promoter in human monocytic cells. Regulation by Egr-1, c-Jun, and NF-kappaB transcription factors. J Biol Chem. 1997;272:17795–801. doi: 10.1074/jbc.272.28.17795. [DOI] [PubMed] [Google Scholar]
- 18.Guha M, O’Connell MA, Pawlinski R, Hollis A, McGovern P, Yan SF, et al. Lipopolysaccharide activation of the MEK-ERK1/2 pathway in human monocytic cells mediates tissue factor and tumor necrosis factor alpha expression by inducing Elk-1 phosphorylation and Egr-1 expression. Blood. 2001;98:1429–39. doi: 10.1182/blood.v98.5.1429. [DOI] [PubMed] [Google Scholar]
- 19.Kitamura H, Kanehira K, Okita K, Morimatsu M, Saito M. MAIL, a novel nuclear I kappa B protein that potentiates LPS-induced IL-6 production. FEBS Lett. 2000;485:53–6. doi: 10.1016/s0014-5793(00)02185-2. [DOI] [PubMed] [Google Scholar]
- 20.Ito T, Morimatsu M, Oonuma T, Shiina T, Kitamura H, Syuto B. Transcriptional regulation of the MAIL gene in LPS-stimulated RAW264 mouse macrophages. Gene. 2004;342:137–43. doi: 10.1016/j.gene.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 21.O’Reilly SM, Moynagh PN. Regulation of Toll-like receptor 4 signalling by A20 zinc finger protein. Biochem Biophys Res Commun. 2003;303:586–93. doi: 10.1016/s0006-291x(03)00389-9. [DOI] [PubMed] [Google Scholar]
- 22.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–92. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmermann N, King NE, Laporte J, Yang M, Mishra A, Pope SM, et al. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest. 2003;111:1863–74. doi: 10.1172/JCI17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reynolds SJ, Milton DK, Heederik D, Thorne PS, Donham KJ, Croteau EA, et al. Interlaboratory evaluation of endotoxin analyses in agricultural dusts--comparison of LAL assay and mass spectrometry. J Environ Monit. 2005;7:1371–7. doi: 10.1039/b509256f. [DOI] [PubMed] [Google Scholar]
- 25.Alwis KU, Milton DK. Recombinant factor C assay for measuring endotoxin in house dust: comparison with LAL, and (1 – > 3)-beta-D-glucans. Am J Ind Med. 2006;49:296–300. doi: 10.1002/ajim.20264. [DOI] [PubMed] [Google Scholar]
- 26.Chen ZT, Li SL, Cai EQ, Wu WL, Jin JS, Zhu B. LPS induces pulmonary intravascular macrophages producing inflammatory mediators via activating NF-kappaB. J Cell Biochem. 2003;89:1206–14. doi: 10.1002/jcb.10590. [DOI] [PubMed] [Google Scholar]
- 27.Locati M, Deuschle U, Massardi ML, Martinez FO, Sironi M, Sozzani S, et al. Analysis of the gene expression profile activated by the CC chemokine ligand 5/RANTES and by lipopolysaccharide in human monocytes. J Immunol. 2002;168:3557–62. doi: 10.4049/jimmunol.168.7.3557. [DOI] [PubMed] [Google Scholar]
- 28.Hakonarson H, Bjornsdottir US, Halapi E, Bradfield J, Zink F, Mouy M, et al. Profiling of genes expressed in peripheral blood mononuclear cells predicts glucocorticoid sensitivity in asthma patients. Proc Natl Acad Sci U S A. 2005;102:14789–94. doi: 10.1073/pnas.0409904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A. 2007;104:15858–63. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berry MA, Hargadon B, Shelley M, Parker D, Shaw DE, Green RH, et al. Evidence of a role of tumor necrosis factor alpha in refractory asthma. N Engl J Med. 2006;354:697–708. doi: 10.1056/NEJMoa050580. [DOI] [PubMed] [Google Scholar]
- 31.Howarth PH, Babu KS, Arshad HS, Lau L, Buckley M, McConnell W, et al. Tumour necrosis factor (TNFalpha) as a novel therapeutic target in symptomatic corticosteroid dependent asthma. Thorax. 2005;60:1012–8. doi: 10.1136/thx.2005.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiley J, Smith R, Noel P. Asthma phenotypes. Curr Opin Pulm Med. 2007;13:19–23. doi: 10.1097/MCP.0b013e328011b84b. [DOI] [PubMed] [Google Scholar]
- 33.Moore WC, Bleecker ER, Curran-Everett D, Erzurum SC, Ameredes BT, Bacharier L, et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119:405–13. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore WC, Peters SP. Update in asthma 2006. Am J Respir Crit Care Med. 2007;175:649–54. doi: 10.1164/rccm.200701-099UP. [DOI] [PubMed] [Google Scholar]
- 35.Doherty DE, Zagarella L, Henson PM, Worthen GS. Lipopolysaccharide stimulates monocyte adherence by effects on both the monocyte and the endothelial cell. J Immunol. 1989;143:3673–9. [PubMed] [Google Scholar]
- 36.Liu AH. Endotoxin exposure in allergy and asthma: reconciling a paradox. J Allergy Clin Immunol. 2002;109:379–92. doi: 10.1067/mai.2002.122157. [DOI] [PubMed] [Google Scholar]
- 37.Michel O, Ginanni R, Duchateau J, Vertongen F, Le Bon B, Sergysels R. Domestic endotoxin exposure and clinical severity of asthma. Clin Exp Allergy. 1991;21:441–8. doi: 10.1111/j.1365-2222.1991.tb01684.x. [DOI] [PubMed] [Google Scholar]
- 38.Michel O, Kips J, Duchateau J, Vertongen F, Robert L, Collet H, et al. Severity of asthma is related to endotoxin in house dust. Am J Respir Crit Care Med. 1996;154:1641–6. doi: 10.1164/ajrccm.154.6.8970348. [DOI] [PubMed] [Google Scholar]
- 39.Rizzo MC, Naspitz CK, Fernandez-Caldas E, Lockey RF, Mimica I, Sole D. Endotoxin exposure and symptoms in asthmatic children. Pediatr Allergy Immunol. 1997;8:121–6. doi: 10.1111/j.1399-3038.1997.tb00164.x. [DOI] [PubMed] [Google Scholar]
- 40.Thorne PS, Kulhankova K, Yin M, Cohn R, Arbes SJ, Jr, Zeldin DC. Endotoxin exposure is a risk factor for asthma: the national survey of endotoxin in United States housing. Am J Respir Crit Care Med. 2005;172:1371–7. doi: 10.1164/rccm.200505-758OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 42.Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bonnelykke K, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. 2007;357:1487–95. doi: 10.1056/NEJMoa052632. [DOI] [PubMed] [Google Scholar]
- 43.Sutherland ER, Martin RJ. Asthma and atypical bacterial infection. Chest. 2007;132:1962–6. doi: 10.1378/chest.06-2415. [DOI] [PubMed] [Google Scholar]
- 44.Platts-Mills JA, Custis NJ, Woodfolk JA, Platts-Mills TA. Airborne endotoxin in homes with domestic animals: implications for cat-specific tolerance. J Allergy Clin Immunol. 2005;116:384–9. doi: 10.1016/j.jaci.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 45.Gereda JE, Klinnert MD, Price MR, Leung DY, Liu AH. Metropolitan home living conditions associated with indoor endotoxin levels. J Allergy Clin Immunol. 2001;107:790–6. doi: 10.1067/mai.2001.115245. [DOI] [PubMed] [Google Scholar]
- 46.Spahn JD, Fost DA, Covar R, Martin RJ, Brown EE, Szefler SJ, et al. Clarithromycin potentiates glucocorticoid responsiveness in patients with asthma: results of a pilot study. Ann Allergy Asthma Immunol. 2001;87:501–5. doi: 10.1016/S1081-1206(10)62264-8. [DOI] [PubMed] [Google Scholar]
- 47.Gotfried MH. Macrolides for the treatment of chronic sinusitis, asthma, and COPD. Chest. 2004;125:52S–60S. doi: 10.1378/chest.125.2_suppl.52s. [DOI] [PubMed] [Google Scholar]
- 48.Simpson JL, Powell H, Boyle MJ, Scott RJ, Gibson PG. Clarithromycin targets neutrophilic airway inflammation in refractory asthma. Am J Respir Crit Care Med. 2008;177:148–55. doi: 10.1164/rccm.200707-1134OC. [DOI] [PubMed] [Google Scholar]
- 49.Hamid QA, Wenzel SE, Hauk PJ, Tsicopoulos A, Wallaert B, Lafitte JJ, Chrousos GP, Szefler SJ, Leung DY. Increased glucocorticoid receptor beta in airway cells of glucocorticoid-insensitive asthma. Am J Respir Crit Care Med. 1999;159(5 Pt 1):1600–1604. doi: 10.1164/ajrccm.159.5.9804131. [DOI] [PubMed] [Google Scholar]
- 50.Leung DY, Hamid Q, Vottero A, Szefler SJ, Surs W, Minshall E, Chrousos GP, Klemm DJ. Association of glucocorticoid insensitivity with increased expression of glucocorticoid receptor beta. J Exp Med. 1997;186(9):1567–1574. doi: 10.1084/jem.186.9.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hauk P, Goleva E, Liu AH, Hall CF, Martin RJ, Leung DYM. Dog ownership: A risk factor of corticosteroid-resistant (CR) asthma? J Allergy Clin Immunol. 2007;119(1 Suppl):S164. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


