Abstract
Mutational activation of BRAF is the most prevalent genetic alteration in human melanoma, with ≥ 50% of tumours expressing the BRAF(V600E) oncoprotein1,2. Moreover, the marked tumour regression and improved survival of late-stage BRAF-mutated melanoma patients in response to treatment with vemurafenib demonstrates the essential role of oncogenic BRAF in melanoma maintenance3,4. However, as most patients relapse with lethal drug-resistant disease, understanding and preventing mechanism(s) of resistance is critical to providing improved therapy5. Here we investigate the cause and consequences of vemurafenib resistance using two independently derived primary human melanoma xeno-graft models in which drugresistanceisselected by continuous vemurafenib administration. In one of these models, resistant tumours show continued dependency on BRAF(V600E) → MEK → ERK signalling owing to elevated BRAF(V600E) expression. Most importantly, we demonstrate that vemurafenib-resistant melanomas become drug dependent for their continued proliferation, such that cessation of drug administration leads to regression of established drug-resistant tumours. We further demonstrate that a discontinuous dosing strategy, which exploits the fitness disadvantage displayed by drug-resistant cells in the absence of the drug, forestalls the onset of lethal drug-resistant disease. These data highlight the concept that drug-resistant cells may also display drug dependency, such that altered dosing may prevent the emergence of lethal drug resistance. Such observations may contribute to sustaining the durability of the vemurafenib response with the ultimate goal of curative therapy for the subset of melanoma patients with BRAF mutations.
To model the emergence of drug resistance, we developed an early passage, vemurafenib-naive, primary human-patient-derived xeno-graft (PDX) BRAFT1799A-mutated melanoma model, HMEX1906 (Supplementary Table1), which was continuously treated with vemurafenib in immunocompromised mice. This system models the emergence of drug-resistant melanoma in response to drug exposures similar to those in patients. Furthermore, this model permits the sampling of serial biopsies from a single tumour, allowing us to investigate the presence of more than one clonally derived mechanism of resistance within the original tumour.
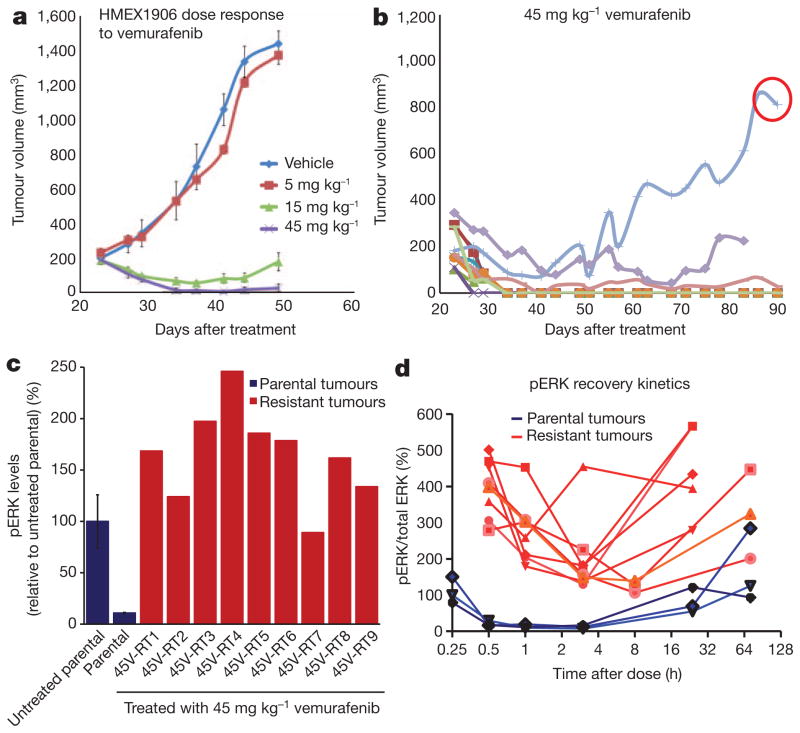
HMEX1906 melanomas are highly sensitive to vemurafenib, with tumour regression observed at clinically relevant drug exposures (Fig. 1a and Supplementary Fig. 1a–c). To generate drug-resistant melanomas, tumour-bearing mice were dosed for 8 weeks with 45 mg kg−1 vemurafenib. This dose resulted in over 80% inhibition of phosphorylated (p) ERK1 and ERK2 (also known as MAPK3 and MAPK1, respectively) (Supplementary Fig. 1d) for up to 24 h, a degree of inhibition previously associated with tumour regression in clinical trials4,5. Approximately 56 days after dosing was initiated, drug-resistant tumours emerged in 2 out of 10 mice (Fig. 1b). One such tumour (45V-RT) was harvested, fragmented and re-implanted into a new cohort of mice, which were then treated with 45 mg kg−1 vemurafenib to generate drug-resistant tumours for exploration of mechanisms of resistance (Supplementary Fig. 1e, f).
Figure 1. Resistance to vemurafenib in a primary human melanoma xenograft model.
a, Mice bearing subcutaneous HMEX1906 tumours were dosed with vehicle (n = 10), 5 mg kg−1 (n = 8), 15 mg kg−1 (n = 8) or 45 mg kg−1 (n = 10) vemurafenib twice daily (mean tumour volume ± s.e.m.). b, Continuous dosing of tumour-bearing mice over an extended time leads to the emergence of resistant tumours. The tumour circled in red was excised, subdivided and re-implanted to be used for further analysis. c, Parental tumours (n = 3 untreated and treated, mean pERK levels ± s.e.m. for the three different tumours) and resistant tumours were treated with 45 mg kg−1 vemurafenib, and lysates were collected 3 h after the drug dose to measure pathway inhibition using pERK levels. d, The pharmacodynamics of pERK1/2 were evaluated over multiple time points for eight resistant tumours (red) and three parental tumours (blue).
Next, we assessed differences in the response to vemurafenib between sensitive parental HMEX1906 and resistant 45V-RT tumours by measuring pERK1 and pERK2 levels 3 h after drug dosing (Fig. 1c). Whereas pERK1/2 and the expression of ERK1/2 target genes such as DUSP6 and SPRY4 were strongly suppressed in sensitive parental HMEX1906 tumours, they were largely unaffected in drug-resistant 45V-RT tumours (Fig. 1c and Supplementary Fig. 2a, b). Analysis of fine needle aspirates (FNAs) of eight resistant tumours over a 72-h time course revealed higher pERK1/2 levels compared to parental tumours 30 min after drug administration, with the nadir of pERK1/2 consistently higher than that observed in parental drug-sensitive tumours (Fig. 1d). Hence, resistant tumours do respond to drug treatment, but the degree of pERK1/2 inhibition was less profound compared to sensitive melanomas. These data suggest that BRAF(V600E) remains essential for sustaining MEK→ERK pathway activation. One explanation for such observations is that BRAF was mutated to a vemurafenib-resistant state. Alternatively, upstream (for example, NRAS) or downstream (for example, MEK1; also known as MAP2K1) nodes in the RAS→RAF→MEK→ERK pathway may be mutationally activated, as described recently6,7. However, exome sequence analysis failed to reveal secondary mutations in the coding sequences of BRAF, NRAS, KRAS, HRAS or MEK1 in resistant tumours (data not shown).
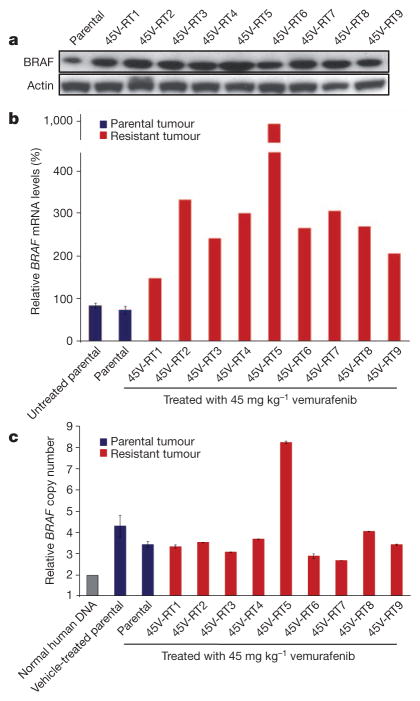
To determine whether BRAF overexpression or alternative splicing might account for vemurafenib resistance8,9, BRAF(V600E) expression was measured in sensitive and resistant tumours. Immunoblot analysis indicated that both sensitive and resistant tumours expressed an 85 kilodalton (kDa) isoform of BRAF(V600E) (Fig. 2a). However, compared to sensitive tumours, all nine resistant tumours expressed elevated levels of BRAF messenger RNA and protein, with the 45V-RT5 tumour showing the highest levels (Fig. 2a, b). Taqman analysis of BRAF copy number indicated that the parental HMEX1906 tumour contained approximately six copies of BRAFT1799A. Although eight out of nine of the resistant tumours showed no additional BRAF copy number gain, the 45V-RT5 tumour was found to have ~14 copies of BRAFT1799A (Fig. 2c), consistent with BRAF amplification as a mechanism of vemurafenib resistance9. These data suggest that the parental tumour contains heterogeneous vemurafenib-resistant cells, all of which show elevated BRAF mRNA/protein expression but only a subpopulation further amplify BRAF10. Finally, we did not detect evidence of alternatively spliced isoforms of BRAF(T1799A) or BRAF(V600E) by mRNA or protein analysis8.
Figure 2. Resistant tumours show increased BRAF(V600E) expression.
a, BRAF protein level was determined by western blot (with actin as a loading control) in parental and resistant tumours (all lysates were collected 3 h after the drug dose). b, BRAF mRNA was measured by quantitative polymerase chain reaction with reverse transcription (RT–qPCR), (n = 3 untreated and treated independent parental tumours, BRAF mRNA levels ± s.e.m.). c, BRAF copy number was determined by qPCR of genomic DNA (n = 3 untreated and treated independent parental tumours, BRAF copy number levels ± s.e.m.).
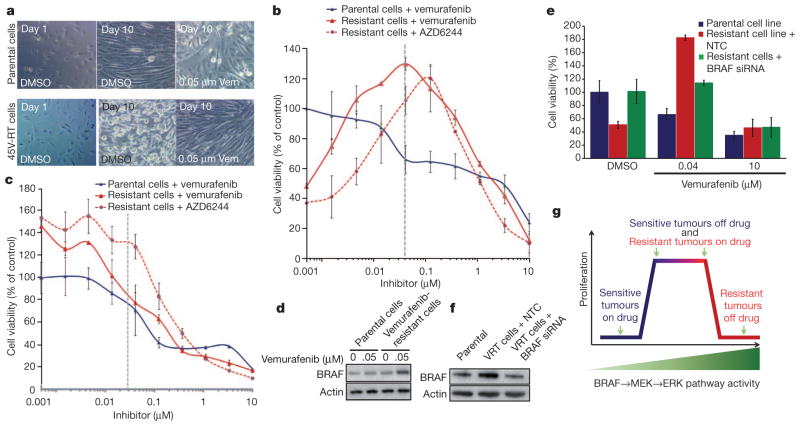
To confirm elevated levels of BRAF protein as a resistance mechanism, melanoma cell lines were derived from the parental HMEX1906 and the vemurafenib-resistant 45V-RT tumour (Fig. 1b). We noted difficulty in establishing cultures of drug-resistant cells unless the media contained ~50 nM vemurafenib. This observation is consistent with reports that vemurafenib-resistant variants of BRAF(V600E)-expressing M288, SK-MEL28 or M14 melanoma cell lines require vemurafenib for continuous proliferation11. In addition, HMEX1906 melanoma cells grown in the absence of drug and 45V-RT melanoma cells grown in the presence of drug showed similar morphology (Fig. 3a, top middle and bottom right). However, culturing 45V-RT melanoma cells in the absence of vemurafenib for 10 days resulted in marked alterations in cell morphology. Cells appeared rounded, refractile and spindle shaped, features characteristic of cells with elevated RAF→MEK→ERK signalling12–14 (Fig. 3a, middle). Furthermore, a cell-proliferation assay conducted with a range of drug concentrations indicated a bell-shaped response to vemurafenib, with peak proliferation in the resistant cells occurring at 50 nM vemurafenib, and with diminished cell proliferation noted at lower and higher drug concentrations (Fig. 3b). A similar curve was observed with the MEK inhibitor AZD6244, with a shift in peak proliferation consistent with the compound’s decreased potency. Analysis of BRAF(V600E)→MEK→ERK signalling indicated that reducing the concentration of vemurafenib led to elevated pERK1/2 levels in the resistant 45V-RT cells. Moreover, the level of pERK1/2 in resistant 45V-RT cells cultured in 50 nM vemurafenib was similar to that detected in parental HMEX1906 cells cultured in the absence of vemurafenib (Fig. 3c, dotted line). After 10 days of culture in the presence of 50 nM vemurafenib, 45V-RT cells showed elevated BRAF(V600E) protein expression similar to resistant tumours in mice (Figs 2a and 3d). To confirm that resistant cells remained oncogene dependent, we inhibited BRAF(V600E) expression by RNA interference (using siBRAF short interfering RNA). Complete knockdown of BRAF(V600E) expression in resistant 45V-RT cells resulted in suppression of proliferation; hence, resistant cells remain dependent on oncogenic BRAF(V600E) signalling for proliferation (Supplementary Fig. 3a, b). However, partial suppression of BRAF(V600E) to levels detected in parental cells (Fig. 3f) re-sensitized resistant cells to both vemurafenib and AZD6244 (Fig. 3e and Supplementary Fig. 3c). These data confirm that resistant tumour cells remain oncogene dependent and that drug resistance is due to elevated expression of BRAF(V600E). Moreover, the fitness benefit given to resistant cells by elevated BRAF(V600E) in the presence of vemurafenib becomes a fitness deficit when the drug is removed. To test this hypothesis, we expressed a conditional BRAF(V600E)–oestrogen receptor (ER; also known as ESR1) fusion protein in parental HMEX1906 cells, such that addition of 4-hydroxytamoxifen (4-HT) leads to increased BRAF(V600E) signalling12–14 (Supplementary Fig. 4a). As predicted, elevated BRAF(V600E) activity in the parental cells led to increased pERK levels but decreased proliferation (Supplementary Fig. 4a, b). These data indicate that HMEX1906 cells are responding to both the quality and quantity of BRAF(V600E)→MEK→ERK signalling such that either reduced (in response to vemurafenib) or enhanced (in response to BRAF(V600E)–ER activation) pathway activation has a deleterious effect on their proliferation12–14.
Figure 3. Vemurafenib-resistant tumour cells require continuous exposure to vemurafenib.
a, Parental (top) and vemurafenib-resistant (bottom) tumour-derived cells were imaged after 1 day (left), after 10 days (middle) in culture (0.05% dimethylsulphoxide (DMSO)), and after 10 days of culture in 0.05 μM vemurafenib (Vem; right). Original magnification, ×40. b, Parental and vemurafenib-resistant cells were treated with the indicated concentrations of vemurafenib and AZD6244 for 72 h, and viability was determined using the Cell Titer-Glo ATP-based luminescence assay, with DMSO-treated parental cells set as the control. c, A parallel plate similar to b was set up and corresponding pERK1/2 levels were measured from samples. d, BRAF protein level was determined in parental and resistant tumour cells by western blot. e, Resistant and parental tumour cells were subjected to BRAF siRNA, treated with vemurafenib or control (DMSO), and cell viability was determined by Cell Titer-Glo assay after 3 days of culture. NTC, non-targeting control. f, BRAF knockdown efficiency was determined by western blot with actin as loading control. g, Model correlating BRAF→MEK→ERK pathway activity and tumour-cell proliferation. b, c and e show mean percentage ± s.e.m., n = 6.
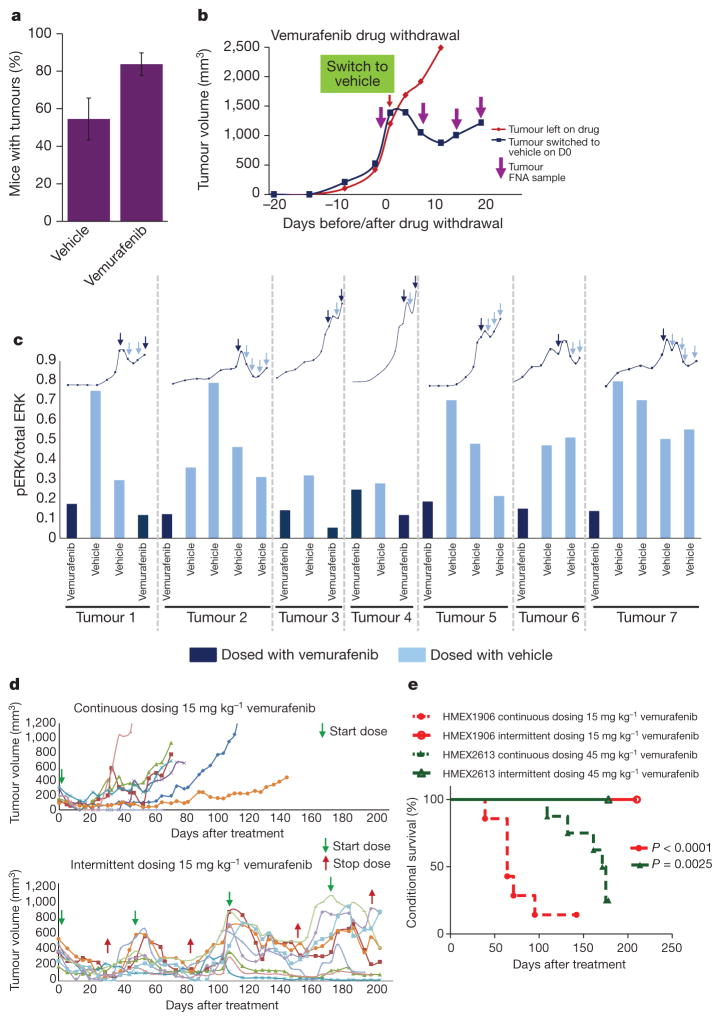
To test whether observations made with cultured melanoma cells are relevant to tumorigenesis in vivo, we evaluated the effects of cessation of drug administration on vemurafenib-resistant tumours in mice. Initially we noted that significantly fewer drug-resistant tumours grew in vehicle-treated mice as compared to vemurafenib-treated mice (Fig. 4a). Furthermore, cessation of drug treatment of mice carrying vemufarenib-resistant melanomas led to clear signs of regression within 10 days after drug withdrawal (Fig. 4b). Consistent with in vitro observations, immunoblot analysis of melanoma specimens collected by serial FNAs from each tumour indicated that drug withdrawal led to elevated pMEK1/2→pERK1/2 signalling, concomitant with tumour regression (Fig. 4c and Supplementary Fig. 5a). However, following an initial period of tumour regression after drug withdrawal, tumours showed re-growth—at which time pERK1/2 levels in the vehicle-treated tumours (Fig. 4c and Supplementary Fig. 5a, light blue bars) returned to their original levels (Fig. 4c and Supplementary Fig. 5a, dark blue bars). These data support the hypothesis that vemurafenib-resistant tumours suffer a fitness deficit in the absence of vemurafenib. On the basis of molecular analysis of the HMEX1906 model, we propose that resistance to BRAF inhibitors is due to increased BRAF(V600E) expression. To expand on observations made using the HMEX1906 model, we tested whether this phenomenon might hold true in additional models of vemurafenib resistance. First, we assessed the effect of vemurafenib withdrawal from SK-MEL239-C3 cells, in which resistance is due to expression of a 61-kDa splice variant of BRAF(V600E) (Supplementary Fig. 6b)8. In a clonogenicity assay, we observed significantly fewer SK-MEL239-C3 cell colonies when cultured in the absence of vemurafenib (Supplementary Fig. 6c). In addition, we tested the effects of vemurafenib withdrawal from a second BRAF-mutated PDX (M120214) (Supplementary Table 1) isolated from a patient whose melanoma already showed vemurafenib resistance. This PDX was established in mice dosed with 45 mg kg−1 vemurafenib (twice daily) immediately after tumour implantation. After 59 days of drug treatment, drug administration was ceased in four out of five tumour-bearing mice. All four tumours demonstrated clear signs of drug-withdrawal-induced tumour regression (Supplementary Fig. 6c), consistent with observations in the HMEX1906 model (Fig. 4c). These models support the observation that vemurafenib-resistant 45V-RT melanomas show a fitness deficit in the absence of vemurafenib.
Figure 4. Intermittent dosing of vemurafenib can be exploited to forestall the development of drug resistance in vivo.
a, Vemurafenib-resistant tumours were implanted and then mice were dosed with either vehicle or 45 mg kg−1 vemurafenib twice daily (mean percentage ± s.e.m., n = 30) and monitored for tumour establishment over a period of 100 days. b, Vemurafenib-resistant tumours were implanted into nude mice and dosed with 45 mg kg−1 vemurafenib twice daily immediately after implant. Once tumours reached a volume of ~1,500 mm3, mice were switched from vemurafenib to vehicle control (blue line), while one mouse remained on vemurafenib (red line). FNAs (purple arrows) were taken from the tumours before and after drug withdrawal to evaluate pERK. c, Lysates collected from the FNA were used to measure pERK, bars represent the pERK1/2 levels from seven different tumours (separated by dotted grey lines), while mice were dosed with vemurafenib (dark blue bars) or vehicle (light blue bars). The growth kinetics for each tumour is represented by the line graph above the pERK1/2 bars and FNA sampling is depicted by arrows (dark blue, on drug; light blue, off drug). d, Tumour growth kinetics of naive parental HMEX1906 tumours with seven tumours dosed continuously (top) and nine tumours dosed intermittently (bottom). Intermittent dosing of vemurafenib was carried out on a 4-week on drug (green arrow) and 2-week off drug (red arrow) schedule with 15 mg kg−1 vemurafenib twice daily. e, Kaplan–Meier curve of data in d (n = 7, continuous dosing and n = 9, intermittent dosing) and Supplementary Fig. 6a (n = 7, continuous dosing and n = 8, intermittent dosing), shows that there is a significant survival advantage with an intermittent dosing (solid lines) compared to a continuous dosing schedule (dashed lines). The end point for euthanasia was predetermined as a tumour size of 1,200 mm3.
One prediction of this model is that, whereas continuous vemurafenib treatment will inevitably select for drug-resistant tumour cells, discontinuous dosing would create a disadvantageous environment for drug-resistant cells—thereby forestalling the onset of lethal drug resistance. To that end, mice were implanted with parental HMX1906 tumours and treated either continuously or intermittently (4 weeks on, 2 weeks off) with 15 mg kg−1 vemurafenib (twice daily), such that mice on the intermittent dosing schedule received the same or a greater cumulative drug dose as mice on the continuous schedule over the entire treatment period. As predicted, mice continuously dosed with vemurafenib developed lethal drug-resistant disease within 100 days after initiation of drug administration. By contrast, none of the mice on the intermittent dosing schedule developed drug-resistant disease over the course of 200 days (Figs 1b and 4d, and Supplementary Fig. 5b). In addition, a similar intermittent versus continuous dosing experiment was conducted in another early passage PDX expressing BRAF(V600E), HMEX2613 (Supplementary Fig. 6a and Supplementary Table1). In this case, the intermittent dosing schedule was individualized for each tumour-bearing mouse. As in the HMEX1906 model, HMEX2613 tumours treated continuously with 45 mg kg−1 vemurafenib (twice daily) developed lethal drug-resistant disease, whereas mice dosed intermittently with vemurafenib did not (Fig. 4e). Irrespective of the underlying mechanism of resistance in the two models, these results indicate that intermittent dosing significantly delays the onset of drug resistance by exploiting the fitness deficit shown by drug-resistant tumour cells in the absence of drug. Furthermore, although we observed that counter-selection against resistant cells by cessation of vemurafenib administration allowed drug-sensitive tumours to restart their growth, these cells remained responsive to the antitumour effects of vemurafenib re-administration.
Although vemurafenib can inhibit BRAF(V600E)→MEK→ERK signalling sufficiently to elicit marked tumour regression, the durability of vemurafenib responses is limited by acquired drug resistance6–9,15,16. Our results suggest that the proliferation of vemurafenib-resistant cells can be dependent on the continuous presence of the drug, such that tumour growth is inhibited after cessation of drug administration. These data are consistent with previous results indicating that both normal and tumour cells can be sensitive to both the quality (that is, which pathways are activated) and the quantity (that is, magnitude of pathway activation) of signal pathway activation12–14,17–19. Furthermore, we show that discontinuous dosing forestalls the onset of drug resistance in two primary human xenograft models. Our observations, and those of others, suggest that the majority of BRAF(V600E) melanomas remain reliant on the reactivation of ERK despite ongoing inhibition of BRAF(V600E). In these cases, drug resistance is achieved via elevated signalling through receptor tyrosine kinases, mutational activation of NRAS or MEK, amplification of BRAF, or alternative splicing of the BRAFT1799A precursor mRNA to yield aberrant forms of BRAF(V600E)6–9,15,16. Our data indicate that some mechanisms of vemurafenib resistance confer a fitness deficit upon the tumour cells in the absence of the drug. This is probably due to elevated ERK1/2 activation that leads to arrest of the cell division cycle or the onset of apoptosis. Indeed, established literature indicates that deliberate elevation of RAF→MEK→ERK signalling in bona fide human cancer cells can have antiproliferative effects11–13. Furthermore, these data suggest that the durability of responses to agents like vemurafenib may be improved through alterations in the dosing schedule, a phenomenon consistent with a recent case report of two melanoma patients with BRAF mutations who demonstrated a secondary antitumour response to BRAF inhibition after cessation of BRAF-inhibitor treatment owing to acquired drug resistance20. Moreover, these results may have implications for other targeted cancer therapies, especially those that target RAF→MEK→ERK signalling. Although published clinical observations are still lacking, we suggest that dose regimens that exceed the daily maximum tolerated dose could be used to induce rapid tumour regression, followed by a drug holiday to prevent the onset of toxicities observed with chronic daily dosing and the emergence of drug-resistant tumour cells. Whereas continuous dosing promotes the clonal expansion of drug-resistant cells12–14,21,22, intermittent dosing could serve to eliminate the fitness advantage of the resistant cells and delay the onset of drug-resistant disease. Hence, our results could have an impact on the use of pathway-targeted therapies to treat at least the subset of melanomas in which BRAF is mutated.
METHODS
Copy number assay
DNA from tumour tissue was extracted using the DNeasy Blood and Tissue Kit (69504, Quiagen) according to the manufacturer’s protocol. Quantitative PCRwas carried outusing the Taqman genotypingmaster mix, Taqman assay (Hs04949885_cn, Hs05005955_cn or Hs04949201_cn; Applied Biosystems), and RnaseP was used as a normalization control for DNA content. Quantitative analysis was carried out using the 7500 Real-time PCR system (Applied Biosystems).
RT–qPCR
RNA was extracted using the RNeasy Kit (74104, Quiagen) according to the manufacturer’s protocol. One-step RT–qPCR reactions were carried out in triplicate using the Quantitect Multiple RT–PCR master mix, Taqman Gene expression assay primer and 18S probe, Taqman Gene expression assay primer and BRAF probe (4331182, Applied Biosystems), and Quantitect RT mix. Quantitative measurements were collected using the 7500 Realtime PCR system (Applied Biosystems). Endogenous control 18S was used as a normalization control for RNA content.
pERK1/2 and pMEK measurements
Meso Scale Discovery plates were used for pERK1/2 (K111DWD-2), total ERK (K111DXD-2), pMEK (K111DUD-2) or total MEK (K111CWD-2) analysis according to the manufacturer’s protocol. Plates were analysed on the SECTOR Imager. Both pERK1/2 and pMEK readings were normalized to the total ERK and total MEK levels, respectively. Data for Supplementary Figs 1d, e, 2 and 4a, c were collected using this method.
Cell viability assay
HMEX1906 cells were split 1:2 the day before seeding. Cells were plated in 100 μl of media the next day at 2,000 cells per well onto black-walled, clear-bottom 96-well plates (3904, Corning Costar). Cells were incubated for 3 days with or without compound at 37 °C before carrying out the viability assay. Using the Cell Titer-Glo Luminescent Cell Viability assay kit (Promega) and instructions, luminescent measurements were taken on Trilux MicroBeta2. The graphically represented values are means ± s.d. for three independent samples.
Cell culture
HMEX1906 cell lines were generated using FNAs from either the parental or the resistant tumours. The FNA was then directly flushed out into EGM media (CC-3124, Lonza) and transferred onto collagen-coated plates (BD Biosciences). Media was changed every day until all tumour debris was gone. Once cell lines were established the plates were maintained at about 50–80% confluence and with media change twice a week. Resistant tumour cell lines were maintained in 50 nM vemurafenib.
BRAF siRNA and western blots
HMEX1906 cells were plated 1 day before transfection at 70% confluency. The next day parallel plates were left untreated, or were transfected with a non-targeting pool of siRNA or BRAF14 on target plus siRNA (target sequence, AGACGGGACUCGAGUGAUG, J-003460-14; Dharmacon). For the transfections, 1,000 μl of Opti-MEM was mixed with 17.5 μl of siRNA to give a final concentration of 50 nM; this was then combined with 1,000 μl Opti-MEM and 21 μl of Dharmafect 1. After a 20-min incubation at room temperature (25 °C) on a shaker, the transfection mix was applied drop-wise to the cells and incubated overnight. Protein lysates, pERK1/2 assays and cell viability assays were then carried out 72 h after transfection. BRAF immunoblot was carried out with RAF-B (F-7) antibody (sc-5284; Santa Cruz).
Accumen pERK1/2 assay
HMEX1906 cells were plated at 2,000 cells per well onto black-walled, clear-bottom Corning Costar 96-well plates (#3904) and incubated for 72 h in varying drug concentrations. On day 3, the media was discarded and the cells were fixed using 100 ml of 4% paraformaldehyde in PBS for 15–20 min. The wells were washed with PBS and then permeabilized with PBS plus 0.1%Triton for 10–15 min at room temperature. The plates were then blocked with 5% normal goat serum for 1 h, after which pERK1/2 (4370; Santa Cruz) primary antibody was applied to the cells at a 1:200 dilution in PBS with 0.1% Triton and 1% BSA. Plates were left overnight on a shaker at 4 °C. Plates were washed and secondary antibody (Invitrogen Alexa 488) and Hoechst stain (34580; Invitrogen) were applied at 1:1,000 and 1:2,500, respectively, for 1–1.5 h. The plates were washed and sealed to scan on the Acumen EX3. PERK1/2 levels were normalized to cell numbers for data analysis.
Clonogenic assay
SK-Mel-239-C3 vemurafenib-resistant cells were plated at the indicated cell density in 2 μM vemurafenib. The next day, half the plates were washed and re-fed with media lacking vemurafenib (day 0). Plates were stained with crystal violet on the days indicated. Plates were re-fed with the appropriate media plus or minus vemurafenib every 3 days.
Supplementary Material
Acknowledgments
We thank the members of the Novartis Institutes for BioMedical Reseach (NIBR) Pharmacology department for technical support, comments and discussions during the course of this work. We thank C. Voliva, N. Aziz and E. Collisson for discussions. We thank B. Weisburd and the rest of the NIBR Bioinformatics department for assistance with exome sequencing data analysis. We thank S. Kaufman for sharing her knowledge of cell-based assays. We thank V. Marsh, N. Rosen, P. Poulikakos and D. Solit for providing additional advice and reagents. M.D.T. was supported by an NIBR Presidential Postdoctoral Fellowship. M.M. acknowledges support from the Melanoma Research Alliance and the National Cancer Institute (R01-CA176839). A.S.L. was supported by a National Research Service Award T32 training grant HL007185.
Footnotes
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Author Contributions M.D.T., M.M. and D.D.S. designed all experiments. M.D.T. performed in vivo and in vitro experiments and collected data. M.D.T., M.M. and D.D.S. analysed data, wrote the paper and guided the manuscript through review. F.S. assisted in performing in vivo experiments. A.S.L. carried out the clonogenic assay with the SK-Mel-239-C3 cells. M.P.L. and R.D. provided the human patient biopsy samples, and R.D. assisted with data analysis and interpretation. W.R.S. and N.K.P. provided input on the experimental approach and on the manuscript. M.M. and D.D.S. are co-senior authors of this manuscript.
Supplementary Information is available in the online version of the paper.
References
- 1.Fecher LA, Amaravadi RK, Flaherty KT. The MAPK pathway in melanoma. Curr Opin Oncol. 2008;20:183–189. doi: 10.1097/CCO.0b013e3282f5271c. [DOI] [PubMed] [Google Scholar]
- 2.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 3.Sosman JA, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bollag G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagle N, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poulikakos PI, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi H, et al. Melanoma whole-exome sequencing identifies V600EB-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nature Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett DC. How to make a melanoma: what do we know of the primary clonal events? Pigment Cell Melanoma Res. 2008;21:27–38. doi: 10.1111/j.1755-148X.2007.00433.x. [DOI] [PubMed] [Google Scholar]
- 11.Petti C, et al. Coexpression of NRASQ61R and BRAFV600E in human melanoma cells activates senescence and increases susceptibility to cell-mediated cytotoxicity. Cancer Res. 2006;66:6503–6511. doi: 10.1158/0008-5472.CAN-05-4671. [DOI] [PubMed] [Google Scholar]
- 12.Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woods D, et al. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5598–5611. doi: 10.1128/mcb.17.9.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 15.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villanueva J, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/ PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tap WD, et al. Pharmacodynamic characterization of the efficacy signals due to selective BRAF inhibition with PLX4032 in malignant melanoma. Neoplasia. 2010;12:637–649. doi: 10.1593/neo.10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoeflich KP, et al. Oncogenic BRAF is required for tumor growth and maintenance in melanoma models. Cancer Res. 2006;66:999–1006. doi: 10.1158/0008-5472.CAN-05-2720. [DOI] [PubMed] [Google Scholar]
- 19.Hingorani SR, Jacobetz MA, Robertson GP, Herlyn M, Tuveson DA. Suppression of BRAF(V599E) in human melanoma abrogates transformation. Cancer Res. 2003;63:5198–5202. [PubMed] [Google Scholar]
- 20.Neyns B, Seghers AC, Wilgenhof S, Lebbe C. Successful rechallenge in two patients with BRAF-V600-mutant melanoma who experienced previous progression during treatment with a selective BRAF inhibitor. Melanoma Res. 2012;22:466–472. doi: 10.1097/CMR.0b013e3283541541. [DOI] [PubMed] [Google Scholar]
- 21.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chmielecki J, et al. EGFR-mutant lung adenocarcinomas treated first-line with the novel EGFR inhibitor, XL647, can subsequently retain moderate sensitivity to erlotinib. J Thorac Oncol. 2012;7:434–442. doi: 10.1097/JTO.0b013e31823c5aee. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.