Abstract
MicroRNAs (miRNAs) are critical regulators of gene expression, yet much remains unknown regarding their changes resulting from environmental exposures as they influence cellular signaling across various tissues. We set out to investigate miRNA responses to formaldehyde, a critical air pollutant and known carcinogen that disrupts miRNA expression profiles. Rats were exposed by inhalation to either 0 or 2 ppm formaldehyde for 7, 28, or 28 days followed by a 7-day recovery. Genome-wide miRNA expression profiles were assessed within the nasal respiratory epithelium, circulating white blood cells (WBC), and bone marrow (BM). miRNAs showed altered expression in the nose and WBC but not in the BM. Notably in the nose, miR-10b and members of the let-7 family, known nasopharyngeal carcinoma players, showed decreased expression. To integrate miRNA responses with transcriptional changes, genome-wide messenger RNA profiles were assessed in the nose and WBC. Although formaldehyde-induced changes in miRNA and transcript expression were largely tissue specific, pathway analyses revealed an enrichment of immune system/inflammation signaling in the nose and WBC. Specific to the nose was enrichment for apoptosis/proliferation signaling, involving let-7a, let-7c, and let-7f. Across all tissues and time points assessed, miRNAs were predicted to regulate between 7% and 35% of the transcriptional responses and were suggested to play a role in signaling processes including immune/inflammation-related pathways. These data inform our current hypothesis that formaldehyde-induced inflammatory signals originating in the nose may drive WBC effects.
Key Words: epigenetics, gene expression, formaldehyde, inhalation, microRNA.
MicroRNAs (miRNAs) are key players in cellular responses to toxic agents in the environment, including carcinogens (Bailey and Fry, 2013). miRNAs are short (~21 nucleotides in length), noncoding RNAs that regulate gene expression by base pairing to target transcripts and causing transcript degradation and/or translational repression (Filipowicz et al., 2008). Estimated to regulate the expression of approximately 30% of all mammalian protein-coding genes (Filipowicz et al., 2008), miRNAs are critical mediators of gene expression. Because they play such pivotal roles in transcriptional regulation, miRNA expression profiles have been evaluated for associations with diseases such as nasopharyngeal carcinoma (Chen et al., 2009) and leukemia (Garzon et al., 2008).
Toxicological studies evaluating the effects of air toxicants on miRNA expression have been performed. For example, diesel exhaust particles have been shown to disrupt miRNA expression profiles in human airway cells (Jardim et al., 2009). Only a few studies have assessed temporal changes in miRNA expression after exposure to environmental toxicants. For example, time series have been examined in response to lipopolysaccharide (Moschos et al., 2007). miRNA expression profiles have been shown to change over time after an exposure ends (Moschos et al., 2007); however, differences in their responses resulting from various exposure durations and from other toxicants are understudied. Moreover, comparisons of exposure-induced miRNA expression changes across several tissues/organs of the body have not been done.
In previous research, we showed that gaseous formaldehyde exposure modifies miRNA expression profiles in cultured human airway cells (Rager et al., 2011). Following up on the in vitro study, we examined in vivo the nasal epithelial responses of nonhuman primates (Rager et al., 2013). Formaldehyde is of high environmental interest, as it is a common indoor and outdoor air pollutant (IARC, 2012; NTP, 2011). It is also a high-volume industrial chemical used in a large number of consumer and industrial applications (IARC, 2012; NTP, 2011). Additionally, formaldehyde is formed endogenously through normal cellular metabolism (IARC, 2012; NTP, 2011). Because of the ubiquitous nature of both endogenous and exogenous formaldehyde exposures, formaldehyde has been studied extensively since the 1980s. Pioneering studies demonstrated that formaldehyde inhalation exposure caused nasal cancer in rats (Kerns et al., 1983; Swenberg et al., 1980) and, to a lesser extent, in mice (Kerns et al., 1983). Since then, hundreds of reports have been published focusing on the health effects of formaldehyde inhalation exposure (IARC, 2012; NTP, 2011).
Formaldehyde inhalation exposure has been associated with several detrimental health effects including increased risk of childhood asthma (McGwin et al., 2010), upper respiratory tract infections (Lyapina et al., 2004), nasopharyngeal cancer (Hauptmann et al., 2004), and leukemia (Beane Freeman et al., 2009). Formaldehyde is currently classified as a known human carcinogen by the International Agency for Research on Cancer (IARC, 2012) and the National Toxicology Program (NTP, 2011). Both agencies view that there is sufficient evidence that formaldehyde causes cancer of the nasopharynx and leukemia, particularly myeloid leukemia (IARC, 2012; NTP, 2011). The conclusion that formaldehyde is a leukemogen has been controversial, as the mechanisms by which formaldehyde could cause toxicity at sites distal to the respiratory tract are unknown (NTP, 2011).
Toxicological investigations have shown clear associations between formaldehyde exposure and tissue damage (Chang et al., 1983; Monticello et al., 1991), increases in cell proliferation (Chang et al., 1983; Monticello et al., 1991), DNA damage (Lu et al., 2010, 2011; Moeller et al., 2011), inflammation (Andersen et al., 2008, 2010), changes in miRNA expression (Rager et al., 2013), and changes in gene expression signatures (Andersen et al., 2010) in direct target regions of exposure. However, there is currently a lack of knowledge regarding inhaled formaldehyde’s ability to influence gene expression and miRNA expression profiles in tissues distant from the site of contact.
In this study, the effects of formaldehyde inhalation exposure were assessed by examining miRNA endpoints across time and throughout multiple tissues using the rodent model. Specifically, genome-wide miRNA expression profiles were compared throughout 3 tissues: (1) the nasal epithelium, (2) circulating white blood cells (WBC), and (3) the bone marrow (BM). The miRNA responses were then integrated with formaldehyde-induced transcriptomic changes, where miRNA-messenger RNA (mRNA) interactions were predicted and responses assessed at the systems level. This study contributes critical knowledge toward the understanding of an epigenetic mechanism that may underlie formaldehyde-induced responses, as well as a broader understanding on how miRNAs respond over time and across varied tissues.
MATERIALS AND METHODS
Ethics statement and animals.
Animals were exposed, sedated, and euthanized using protocols approved by the Lovelace Respiratory Research Institute’s animal care and use committee. Male Fischer rats (Charles River, Wilmington, Massachusetts) were selected from the Lovelace Respiratory Research Institute colony. Animals were approximately 6 to 8 weeks of age and weighed between 150 and 250g. Animals were housed, fed, and cared for as previously described (Lu et al., 2011). Prior to the exposures, animals were conditioned to nose-only exposure tubes for 6h/day over 4 conditioning sessions.
Formaldehyde exposures.
Rats received nose-only inhalation exposures of 2 ppm formaldehyde. To generate the exposure conditions, deuterated/13C labeled paraformaldehyde (Cambridge Isotope Laboratories, Inc.) was vaporized and directed through a delivery line and into a Tedlar bag. During each of the exposures, 1 Tedlar bag was diluted with prefiltered air and delivered to the inhalation chambers at a target formaldehyde concentration of 2 ppm. Nose-only chamber concentrations were monitored by collecting breathing port samples using Waters XpoSure Aldehyde Sampler cartridges. Cartridges were analyzed by ultraviolet high-performance liquid chromatography.
Three exposure durations were investigated. First, the 7-day group received formaldehyde exposure for 7 days (6h/day), where formaldehyde exposures averaged to 2.00 (± 0.08 SD). Second, the 28-day group received formaldehyde exposure for 28 days (6h/day), where formaldehyde exposures averaged to 2.03 (± 0.06 SD). Third, a 28-day plus recovery group received formaldehyde exposure for 28 days (6h/day) with a 7-day recovery period following the last exposure, where formaldehyde exposures averaged to 2.03 (± 0.06 SD). Control (unexposed) rats were placed in a nose-only exposure chamber containing room air for matched durations.
Sample collection.
After the last exposure or recovery period, animals were euthanized using an IP injection of pentobarbital-based solution (Euthasol, Virbac Corp., Fort Worth, Texas). Sample collection started immediately after the last exposure in order to parallel time of death and sample collection times used in our previous studies (Lu et al., 2011; Moeller et al., 2011). Nasal epithelial tissue from the nasal respiratory epithelium was collected as previously described (Lu et al., 2011) and stored in RNAlater (Qiagen, Valencia, California). Whole blood was collected by cardiac stick using a heparin-laced syringe, and WBC were isolated using Vacutainer CPT cell preparation tubes (Becton Dickinson, Franklin Lakes, New Jersey) and stored in TRIzol LS (Life Technologies, Grand Island, New York). To collect BM cells, femurs were flushed with saline and BM cells stored in RNAlater (Qiagen). All samples were stored at −80°C and shipped by overnight courier on dry ice to the University of North Carolina at Chapel Hill for further processing.
RNA isolation.
Nasal epithelial tissue samples were disrupted and homogenized using a TissueRuptor (Qiagen) and RNA isolated using the miRNeasy kit (Qiagen). For the WBC, samples were homogenized in TRIzol (Life Technologies) and RNA isolated according to the standard TRIzol protocol. The BM samples were filtered through 70-μm nylon mesh filters (Fischer Scientific, Waltham, Massachusetts) to remove bone fragments. Filtered BM samples were then homogenized in TRIzol (Life Technologies) and RNA isolated according to the standard TRIzol protocol. Extracted RNA was quantified with a Nanodrop 1000 spectrophotometer (Thermo Scientific, Waltham, Massachusetts) and its integrity verified with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, California) using both the Nano and Small RNA kits. The quality control incorporated RNA integrity number assessment and electrophoretogram peak assessment of small RNA molecules.
miRNA microarray analysis.
RNA samples were labeled and hybridized to the Agilent Rat miRNA Microarray based on miRBase v16.0. For the nose and BM samples, exposed and unexposed samples were assessed in biological triplicate. For the WBC samples, exposed samples were assessed in biological quadruplicate and unexposed samples were assessed in biological triplicate. Microarray data have been submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus repository (Edgar et al., 2002) and are available under accession number GSE42393 (www.ncbi.nlm.nih.gov/geo). For miRNA analysis, data were normalized and background noise was eliminated, and data were statistically assessed as previously detailed (Rager et al., 2013). In the case where multiple microarray chips were used, batch effect was identified and removed (Partek Genomics Suite, St Louis, Missouri). Differential expression was defined as a significant difference in miRNA levels between exposed versus unexposed samples, where 3 statistical requirements were set: (1) fold change of ≥ 1.5 or ≤ −1.5 (average exposed versus average unexposed); (2) p < .05 (ANOVA); and (3) a false discovery rate corrected q value < .1.
Transcript (mRNA) microarray analysis.
Nose and WBC RNA samples from the 7-day and 28-day groups were labeled and hybridized to the Affymetrix GeneChip Rat Gene 1.0 ST Array (Santa Clara, California). Exposed and unexposed samples were assessed in biological triplicate. Microarray data have been submitted to the NCBI Gene Expression Omnibus repository (Edgar et al., 2002) and are available under accession number GSE42394. To analyze the mRNA microarray results, data were first normalized by robust multichip average and background noise was eliminated by removing mRNA probes with signal intensities less than the median signal across all replicates. Differential expression was defined as a significant difference in mRNA levels between exposed versus unexposed samples, where 2 statistical requirements were set: (1) fold change of ≥ 1.5 or ≤ −1.5 (average exposed vs average unexposed) and (2) p < .01 (ANOVA).
To assess whether changes in transcript levels reflected potential shifts in cell population (eg, inflammatory cell infiltrate), the formaldehyde-responsive transcripts were compared against a database of immune cell–specific transcripts. The formaldehyde-associated transcripts identified in this study was compared to 1135 transcripts previously identified as specifically expressed in blood cell subsets (eg, B-cell lymphocytes, T-cell lymphocytes, CD8+ T-cell lymphocytes, and granulocytes) (Palmer et al., 2006).
Predicting miRNA transcriptional targets.
Transcriptional targets of formaldehyde-responsive miRNAs were predicted in silico. The Ingenuity Knowledge Database was queried for experimentally validated miRNA-mRNA interactions, as well as predicted interactions based on TargetScan algorithms that identify potential matches between 3′-untranslated mRNA regions and miRNA seed sequences (http://www.targetscan.org). The resulting predicted miRNA-mRNA interactions were filtered for high confidence, defined as those with total context plus scores < −0.4. The total context plus score controls for factors influencing miRNA targeting, including miRNA binding site type and location, local adenine and uracil content, supplementary pairing, target site abundance, and seed-pairing stability (Garcia et al., 2011). Interactions were also directionally filtered to include matches between miRNAs with formaldehyde-induced increased expression and transcripts with formaldehyde-induced decreased expression, and vice versa.
Pathway enrichment analysis of transcriptional responses.
The Comparative Toxicogenomics Database (CTD) was used to identify biological pathways associated with formaldehyde-induced changes in transcriptional profiles (http://ctdbase.org). Using the Set Analyzer tool in CTD, the hypergeometric distribution was used to calculate the probability that the fraction of genes associated with a biological pathway was significantly higher than by chance alone (Boyle et al., 2004). The lists of formaldehyde-associated transcripts were assessed using 2 biological pathway databases within CTD, KEGG, and REACTOME. Pathways were filtered for those with p ≤ .01.
Confirming miRNA microarray results using reverse transcriptase PCR.
To confirm the miRNA microarray results, formaldehyde-induced changes in miRNA expression were validated using real-time reverse transcriptase PCR (RT-PCR). The same sample replicates used for the microarray analysis were used for RT-PCR, plated in technical triplicate. TaqMan MicroRNA Primer Assays for hsa-miR-31 (ID 000185) and the U6 housekeeping miRNA (ID 001973) were used with the TaqMan Small RNA Assays PCR kit (Applied Biosystems, Carlsbad, California). The MyCyler Thermal Cycler (Bio-Rad, Hercules, California) was used for the reverse transcription step, and the Lightcycler 480 (Roche, Indianapolis, Indiana) was used for the real-time step. The resulting RT-PCR cycle times were normalized against the U6 housekeeping miRNA, and fold changes in expression were calculated based off delta delta cycle time values, as previously described (Rager et al., 2013). Statistical significance of the difference in miRNA expression levels between the formaldehyde-exposed and -unexposed samples was calculated using ANOVA.
Confirming transcript microarray results using RT-PCR.
To verify the results from the transcript microarray analysis, RT-PCR was performed at the gene expression level using nose samples from the 28-day rat group. The same sample replicates used for the microarray analysis were used for RT-PCR, plated in technical triplicate. QuantiTect Primer Assays were used with QuantiTect SYBR Green PCR kits (Qiagen) and the Stratagene Mx3005P QPCR System (Agilent Technologies). Specifically, chloride channel calcium activated 3 (Clca3) (Cat. No. QT01570870), C-type lectin domain family 11, member a (Clec11a) (Cat. No. QT00407043), dosage suppressor of mck1 homolog, meiosis-specific homologous recombination (Dmc1) (Cat. No. QT01627241), and secreted frizzled-related protein 4 (Sfrp4) (Cat. No. QT00179830) were evaluated for changes in transcript levels induced by formaldehyde exposure. Resulting RT-PCR cycle times were normalized against the β-actin housekeeping gene, and fold changes in expression were calculated used delta delta cycle time values, as previously described (Rager, et al., 2013). Statistical significance comparing the expression levels between exposed and unexposed samples was calculated using ANOVA.
RESULTS
Study Design
In this study, we investigated formaldehyde-induced changes in miRNA expression profiles at 3 different exposure times and 3 different tissues. Specifically, male Fischer rats received nose-only inhalation exposures of 2 ppm formaldehyde (6h/day) for (1) 7 days, (2) 28 days, or (3) 28 days followed by a 7-day recovery (Fig. 1). Control (unexposed) rats were placed in nose-only exposure tubes containing room air for the same duration. In order to examine the effects of formaldehyde across multiple tissues, samples were collected from the nasal epithelium, circulating mononuclear WBC, and BM cells immediately after the last exposure. An exposure of 2 ppm formaldehyde was used, as this exposure level has been shown to alter gene expression profiles (Andersen et al., 2010) and DNA adduct levels (Lu et al., 2011) in sites of direct contact in rats. Isotope-labeled [13CD2] formaldehyde was used to allow for the detection of exogenously produced DNA adducts in a parallel experiment, as in previous investigations of our research team (Lu et al., 2011; Moeller et al., 2011).
FIG. 1.
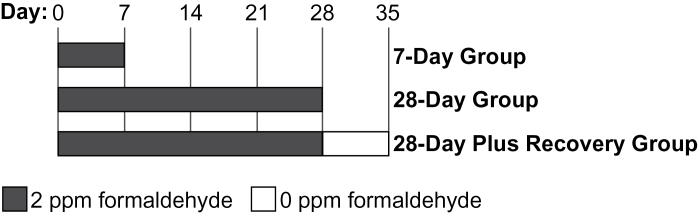
Design of rat formaldehyde exposure conditions over time. Control (unexposed) rats were treated using matched conditions.
Formaldehyde Alters miRNA Expression in the Nose and WBC
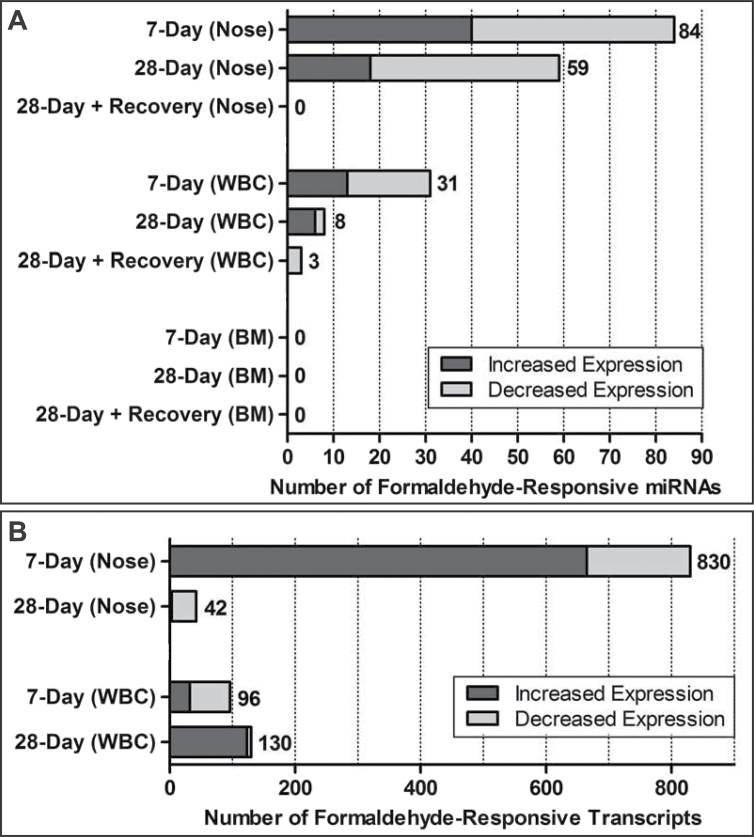
To determine whether formaldehyde inhalation exposure modifies the expression levels of miRNAs within the rat nose, WBC, and BM, small RNAs from these samples were assessed using the Agilent Rat miRNA Microarray, release 16.0. This array measures the expression levels of 695 rat miRNAs. Formaldehyde exposure was found to alter the expression levels of 108 miRNAs. Specifically, 84, 59, and 0 miRNAs were altered in the nose in the 7-day, 28-day, and 28-day plus recovery groups, respectively. Some of the formaldehyde-responsive miRNAs overlapped between exposure durations, as detailed in the next section. In the WBC, formaldehyde exposure altered the expression of 40 miRNAs. Specifically, 31, 8, and 3 miRNAs were altered in the 7-day, 28-day, and 28-day plus recovery groups, respectively. In the BM, no miRNAs were identified as significantly altered (Fig. 2A; Supplementary Table 1).
FIG. 2.
Distribution of (A) formaldehyde-responsive microRNAs and (B) mRNA transcripts in the rat.
Formaldehyde-Responsive miRNAs are Tissue and Time Specific
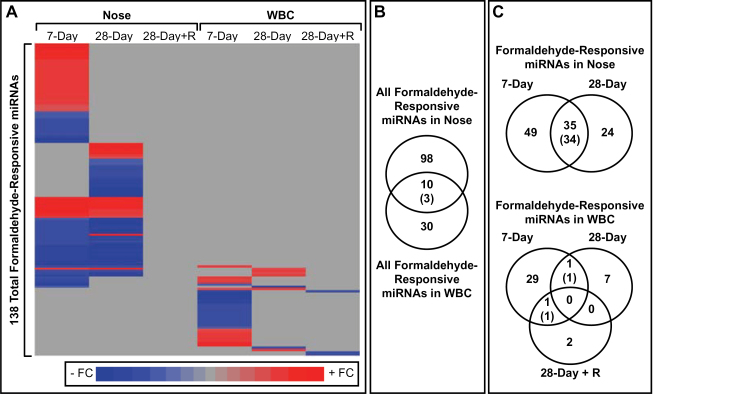
Comparing the formaldehyde-responsive miRNAs across the different tissues revealed largely distinct miRNA responses. Out of a total 108 formaldehyde-responsive miRNAs within the nose, only 10 were also responsive within the WBC. Most (7/10) of these overlapping miRNAs displayed different directions of altered expression (Figs. 3A and B).
FIG. 3.
Formaldehyde-responsive miRNAs in the nose and white blood cells. A, A heat map displays formaldehyde-responsive miRNAs, shaded according to fold change value (FC = exposed/unexposed). B and C, Venn diagrams show how formaldehyde-responsive miRNAs overlap between tissues and exposure conditions. Overlapping miRNAs are enumerated in the centers of the venn diagrams, and the number of overlapping miRNAs with the same direction of altered expression is shown within parentheses. Abbreviation: miRNA, microRNA.
Comparing the formaldehyde-responsive miRNAs across the different exposure durations revealed that many miRNAs show similar expression changes over time in the nose but not in the WBC (Fig. 3C). A total of 35 miRNAs had altered expression levels in the nose in both the 7-day and 28-day exposure groups. Furthermore, 34 of these 35 miRNAs showed altered expression with similar directionality. Seven of the 34 miRNAs with sustained decreased expression over time, let-7a, let-7c, let-7f, miR-10b, miR-126, miR-21, and miR-23a, have previously been shown to have decreased expression in cultured lung cells exposed to 1 ppm formaldehyde (Rager et al., 2011). One of the 34 miRNAs with sustained decreased expression over time, miR-203, has also previously been identified as decreased in expression within the nasal epithelium of nonhuman primates exposed to 2 and 6 ppm formaldehyde over 2 days (Rager et al., 2013). Notably, these miRNA expression changes did not persist in the rat nose after 7 days of recovery.
Formaldehyde-responsive miRNAs were less sustained over time within the WBC. Only 2 miRNAs had significantly altered expression when comparing across the 7-day, 28-day, and 28-day plus recovery groups (Fig. 3C). MiR-326 showed increased expression in both the 7-day and 28-day groups. MiR-212 showed decreased expression in both the 7-day and 28-day plus recovery groups. MiR-212 also showed decreased expression (FC = −2.34, p = .008, q value = .155) in formaldehyde-exposed WBC samples from the 28-day group, but it did not pass the stringent multiple test correction/q value requirement.
Formaldehyde Alters Transcript Levels in the Nose and WBC
To assess the influence of formaldehyde inhalation exposure at the mRNA level, a transcriptomics-based analysis was performed using the nose and WBC samples from the 7-day and 28-day exposure groups. Only the nose and WBC samples were assessed at the transcript level, as no changes in miRNA expression were detected in the BM.
Transcript levels were measured using the Affymetrix GeneChip Rat Gene 1.0 ST Array, which assesses the expression levels of 27342 genes. Formaldehyde exposure was found to cause the differential expression of 830 and 42 genes in the nose in the 7-day and 28-day groups, respectively (Fig. 2B; Supplementary Table 2). Together, these represent 864 formaldehyde-responsive transcripts in the nose. Notably, 209 of the 830 formaldehyde-responsive transcripts (25%) in the nose of the 7-day group represent olfactory receptors. In the WBC, formaldehyde exposure altered the expression levels of 96 and 130 genes in the 7-day and 28-day groups, respectively (Fig. 2B; Supplementary Table 2). These represent 222 total formaldehyde-responsive transcripts in the WBC.
Formaldehyde-Responsive Transcripts are Tissue and Time Specific
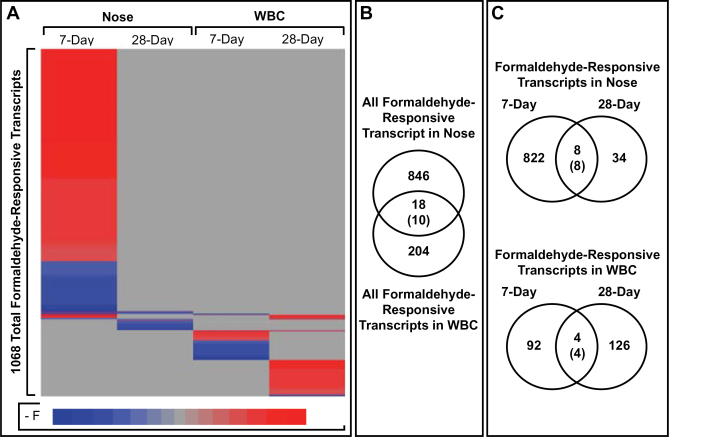
Comparing the formaldehyde-responsive transcripts across the different tissues revealed largely distinct transcriptional responses. Out of a total 864 formaldehyde-responsive transcripts within the nose, only 18 (2%) were also responsive to formaldehyde exposure within the WBC (Figs. 4A and B). Only 10 of these overlapping mRNAs displayed the same direction of altered expression.
FIG. 4.
Formaldehyde-responsive transcripts in the nose and white blood cells. A, A heat map displays formaldehyde-responsive transcripts, shaded according to fold change value (FC = exposed/unexposed). B and C, Venn diagrams show how formaldehyde-responsive transcripts overlap between tissues and exposure conditions. Overlapping transcripts are enumerated in the centers of the venn diagrams, and the number of overlapping transcripts with the same direction of altered expression is shown within parentheses.
Very few transcripts show similar expression patterns over time. Only 8 transcripts were altered in the nose in both the 7-day and 28-day exposure groups (Fig. 4C). All 8 of these transcripts notably showed altered expression with similar directionality in both exposure groups. These transcripts include one that showed decreased expression, namely dosage suppressor of mck1 homolog, meiosis-specific homologous recombination (Dmc1), and 7 transcripts that showed increased expression, namely CD34 molecule (Cd34), C-type lectin domain family 7, member A (Clec7a), Fc fragment of IgG, low affinity IIb, receptor (CD32) (Fcgr2b), lipoprotein lipase (Lpl), lymphocyte-specific protein 1 (Lsp1), musculoskeletal, embryonic nuclear protein 1 (Mustn1), and oncostatin M receptor (Osmr).
In the WBC, only 4 transcripts were altered in both the 7-day and 28-day exposure groups (Fig. 4C). One of these transcripts was decreased in expression by formaldehyde in both exposure groups, namely ubiquitin protein ligase E3 component n-recognin 4 (Ubr4). The remaining transcripts were increased in expression in both exposure groups and included CD160 molecule (Cd160), granzyme K (Gzmk), and killer cell lectin-like receptor subfamily G, member 1 (Klrg1).
A Small Proportion of Formaldehyde-Responsive Transcripts in the Nose Are Immune Cell Specific
Formaldehyde exposure has been shown to cause inflammatory cell infiltrates in the nasal epithelium of rats, specifically at exposure concentrations ≥ 6 ppm formaldehyde (Andersen et al., 2008, 2010; Pavkov et al., 1982). Although inflammatory cell infiltrates resulting from 2 ppm formaldehyde are either unobservable or minimal (Andersen et al., 2008, 2010; Pavkov et al., 1982), inflammation-related genomic signaling has been identified following 2 ppm formaldehyde in the rat nose (Andersen et al., 2010). Therefore, it is important to relate formaldehyde-induced changes in mRNA transcriptional profiles to potential shifts in the inflammatory cell population.
To gain more information on how our results relate to potential inflammatory cell infiltrates, the 864 formaldehyde-associated transcripts identified within the rat nose were compared with 1135 transcripts previously identified as expressed specifically in immune cell subsets (eg, B-cell lymphocytes, T-cell lymphocytes, CD8+ T-cell lymphocytes, and granulocytes) (Palmer et al., 2006). This comparison revealed that only 35 of the 864 (4%) formaldehyde-responsive transcripts in the nose are immune cell specific (Supplementary Table 3). Mainly 2 immune cell subsets are represented by these 35 formaldehyde-responsive transcripts: granulocytes and B-cell lymphocytes. Therefore, although the assessed nasal tissues likely contained a pool of epithelial cells and immune-related cells, the estimated genomic signaling from B cells and granulocytes represents a very small fraction of the total transcriptional response.
Formaldehyde-Responsive miRNAs May Mediate a Fraction of the Transcriptional Effects
Interactions between formaldehyde-responsive miRNAs and differentially expressed transcripts were predicted based on previous experimental findings as well as computational predictions based on sequence matches between 3′-untranslated mRNA regions and miRNA seed sequences. These predictions revealed that, in the nose, 294 of the 830 (35%) differentially expressed transcripts are likely regulated by 50 miRNAs in the 7-day group and 9 of the 42 (21%) differentially expressed transcripts are likely regulated by 5 miRNAs in the 28-day group. In the WBC, 29 of the 96 (30%) differentially expressed transcripts were predicted to be regulated by 12 miRNAs in the 7-day group and 9 of the 130 (7%) differentially expressed transcripts were predicted to be regulated by 4 miRNAs in the 28-day group (Supplementary Table 4).
Pathways Are Associated With Transcriptional Responses to Formaldehyde
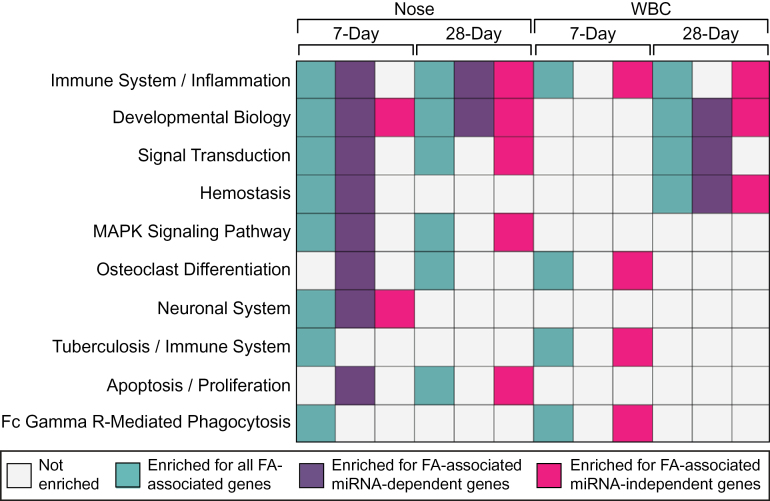
Transcripts were next assessed for relationships to biological pathways. Analyses were carried out for the 7-day and 28-day nose groups and 7-day and 28-day WBC groups using the following lists: (1) all formaldehyde-associated transcripts, (2) formaldehyde-associated transcripts predicted as regulated by formaldehyde-responsive miRNAs (ie, miRNA-dependent transcripts), and (3) formaldehyde-responsive transcripts not predicted to be regulated by miRNAs (ie, miRNA-independent transcripts). For inclusion, the formaldehyde-associated pathways had to be enriched within at least 1 list containing all formaldehyde-associated transcripts.
A total of 45 pathways were identified as associated with formaldehyde-induced transcriptomic changes (Supplementary Table 5). The top 10 most significant pathways associated with formaldehyde-induced transcriptional responses were identified to include immune system/inflammation, signal transduction, mitogen-activated protein kinase (MAPK) pathway, neuronal system, and apoptosis/proliferation, among others (Fig. 5).
FIG. 5.
Ten pathways most associated with formaldehyde-induced transcriptional responses. Abbreviations: FA, formaldehyde; miRNA, microRNA; WBC, white blood cells.
Microarray Results Were Confirmed Using RT-PCR
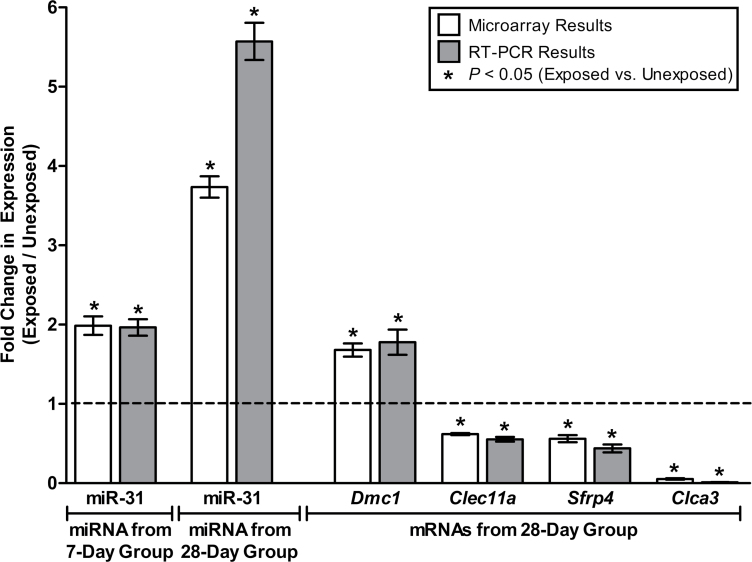
Formaldehyde-induced changes in miRNA and transcript expression levels identified through microarray analysis were validated using RT-PCR. RT-PCR was performed on miR-31, the miRNA with the greatest increase in expression in the nose of the 28-day group and also an increase in expression in the nose of the 7-day group. Supporting the microarray findings, miR-31 showed significantly increased expression in the formaldehyde-exposed nose samples of the 7-day and 28-day groups through RT-PCR (Fig. 6). A subset of genes differentially expressed by formaldehyde in the nose of the 28-day group was also verified using RT-PCR. Specifically, Dmc1 and Clca3 were tested, as they showed the greatest increase and decrease in expression resulting from exposure, respectively. Two other genes, Clec11a and Sfrp4, were evaluated using RT-PCR in order to test genes that were not as drastically altered at the expression level but still identified as differentially expressed (Fig. 6).
FIG. 6.
RT-PCR results alongside microarray results in the rat nose. Abbreviations: miRNA, microRNA; mRNA, messenger RNA; RT-PCR, reverse transcriptase PCR.
DISCUSSION
In this study, a multitiered approach was employed to enable an understanding of the genome-wide miRNA responses to formaldehyde and to establish how these responses relate to alterations in transcriptional profiles over time and in various tissues. Formaldehyde is a common air toxicant that we previously have shown to significantly disrupt miRNA expression profiles in direct target tissues in vitro (Rager et al., 2011) and in vivo (Rager et al., 2013). For the current study, the effects of formaldehyde were further evaluated in vivo across several tissues, where rats received nose-only inhalation exposures of 2 ppm formaldehyde for 7 days, 28 days, or 28 days followed by a 7-day recovery without formaldehyde exposure and compared with controls.
This investigation is the first to simultaneously examine exposure-induced perturbations in miRNA expression profiles throughout the nasal epithelium, circulating WBC, and BM. The nose was the site of greatest change in formaldehyde-induced miRNA expression profiles (n = 108 miRNAs) followed by the WBC (n = 40 miRNAs) after 7 and 28 days of exposure. There were no miRNAs that showed significantly altered expression in the BM. These findings suggest that cells in direct contact with formaldehyde exposure are more responsive at the miRNA level than cells distant from exposure contact sites.
Within the rat nose, formaldehyde-induced miRNA changes showed high stability over the exposure time points. Of the 59 formaldehyde-responsive miRNAs in the nose of the 28-day group, 34 (58%) were also altered with similar directionality in the 7-day group. Many of these miRNAs with sustained expression alterations have been shown to be altered in cultured human lung cells exposed to formaldehyde (Rager et al., 2011). Specifically, let-7a, let-7c, let-7f, miR-10b, miR-126, miR-21, and miR-23a were all significantly decreased in expression in formaldehyde-exposed lung cells (Rager et al., 2011) and in the nose of rats exposed to formaldehyde for 7 and 28 days. Of these miRNAs, let-7a, let-7c, let-7f, and miR-10b were also decreased in expression in nasopharyngeal carcinoma tissue in comparison to healthy tissue (Li et al., 2011). The downregulation of the let-7 family of miRNAs (eg, let-7a, let-7c, let-7f) is of particular importance, as these miRNAs are implicated as tumor suppressors that act on apoptosis/cell proliferation pathways (Shenouda and Alahari, 2009). These common patterns of miRNA expression associated with both exposure and disease could represent etiologic relationships and warrant further investigation.
One of the goals of this study was to compare formaldehyde-altered miRNA expression to mRNA transcript levels. Genome-wide transcriptional profiles were assessed in the nose and WBC of the 7-day and 28-day group but not in the BM, as this was not a site of formaldehyde-altered miRNA expression. Within the nose, formaldehyde exposure resulted in the differential expression of 830 transcripts in rats exposed for 7 days and 42 transcripts in rats exposed for 28 days. Seven transcripts showed formaldehyde-induced expression in both exposure groups, namely Cd34, Clec7a, Fcgr2b, Lpl, Lsp1, Mustn1, and Osmr. One transcript was increased in expression by formaldehyde in both exposure groups, namely Dmc1. Some of these transcripts have previously been identified as differentially expressed by formaldehyde exposure. For instance, Lsp1 had altered expression in a previous study evaluating formaldehyde-induced changes in the rat nasal epithelium (Andersen et al., 2010).
In order to assess which of the formaldehyde-induced transcriptional changes may be attributable to miRNAs, potential miRNA-mRNA interactions that occur upon exposure to formaldehyde were predicted. For this analysis, interactions between formaldehyde-responsive miRNAs and differentially expressed transcripts were predicted in the nose and WBC of the 7-day and 28-day groups. It was predicted that between 7% and 35% of the formaldehyde-associated transcripts were regulated by formaldehyde-responsive miRNAs. These data suggest that the miRNAs may be operating through mechanisms other than mRNA degradation (Bailey and Fry, 2013; Filipowicz et al., 2008) and that other transcriptional regulators, such as DNA methylation and histone modifications (Cedar and Bergman, 2009), may play a role in formaldehyde-induced genomic responses. Our group has shown in vitro that formaldehyde binds to lysine molecules in histone 4 (Lu et al., 2008). These findings support the potential role of histone modification in formaldehyde-induced responses.
Because the analyzed nasal tissue contained mixed cell populations (ie, epithelial and immune/inflammation-related cells), the formaldehyde-responsive transcripts were compared against a list of immune cell–specific transcripts to test for potential cell population shifts. Only a small proportion (4%) of the formaldehyde-responsive transcripts in the nose is specifically expressed in immune cells, mostly comprising granulocytes and B-cell lymphocytes. Therefore, potential signaling from B cells and granulocytes represents a small fraction of the total transcriptional response observed in the nose.
A large number of formaldehyde-associated transcripts in the nose from the 7-day group (n = 209) encode olfactory receptors. The olfactory receptor family initiates signal transduction pathways leading to the perception of smell (Young et al., 2003). Because formaldehyde can have a pungent irritating odor at high levels (NTP, 2011), this transcriptional response is not unexpected. Notably, the olfactory receptor transcripts were not identified as differentially expressed by formaldehyde after 28 days of exposure, suggesting a transient time-dependent response.
In addition to the olfactory-associated signaling, the transcriptional response in the nose included immune system/inflammation and MAPK signaling-associated genes. For instance, LSP1 plays a role in MAPK signaling, particularly in neutrophils, and regulates leukocyte recruitment to sites of inflammation (Jongstra-Bilen and Jongstra, 2006). The inflammatory-related signaling response observed in this study was acute, in which more inflammation-related miRNAs and transcripts were altered after 7 days in comparison to 28 days of exposure. As in the nose, genes associated with immune system/inflammation and signal transduction were altered in the WBC. For example, CD160 is expressed in T cells and serves as a negative regulator of CD4+ T-cell activation (Cai et al., 2008). Additionally, GZMK and KLRG1 are expressed in natural killer cells and T cells and also play roles in T-cell activation (Cai et al., 2008; Wang et al., 2008). Not surprising given the mRNA shifts, inflammation and immune-related miRNAs were also altered in expression by formaldehyde exposure within WBC. Altered here, miR-143, miR-150, and miR-342 have previously been shown to have altered expression in circulating leukocytes that are involved in acute inflammation triggered by E. coli lipopolysaccharide infusion in humans (Goncharova et al., 2010).
Parallel to the genomic and epigenomics endpoints assessed in this study, endogenous and exogenous hydroxymethyl-dG adducts were measured in the same tissues. Although exogenous DNA adducts from the rat nose increased with increasing days of exposure, no exogenous DNA adducts were detected in the WBC following up to 28 days of exposure, measured with a detection capability of 1.3 adducts per 10 billion dG (Yu and Swenberg, in preparation). Because inhaled formaldehyde does not reach the circulating blood (Casanova et al., 1988; Heck et al., 1985; Lu et al., 2010), our research team hypothesizes that the inflammation-associated signaling within the nose may in fact drive changes in WBC signaling observed in this study. Our future research will investigate inflammatory response pathways (eg, cytokine signaling) as potential mediators of formaldehyde-induced genomic and epigenomic changes in the nose and WBC.
The relationship between formaldehyde inhalation exposure and effects on the immune system has been studied to a limited extent. For example, formaldehyde exposure has been linked to both suppression and stimulation of the immune system, as evidenced by its association with increased risk of childhood asthma (McGwin et al., 2010) and increased risk of upper respiratory tract infections in humans (Lyapina et al., 2004). In formaldehyde-exposed rodents, immunomodulation has been observed through the assessment of antibody production in plasma (Fujimaki et al., 2004). Our findings associate formaldehyde exposure to transcriptional changes related to immune system/inflammation signaling in both the nose and WBC samples that is not simply a result of the presence of immune-related cells. The data from this study suggest that miRNAs may, in part, regulate these immunomodulation and/or inflammatory responses.
To date, there are inconsistent findings regarding formaldehyde’s influence on gene expression within circulating blood. The expression levels of 6 transcripts in human peripheral whole blood samples have been shown to correlate with formaldehyde exposure, as evaluated using urinary concentrations of a formaldehyde adduct, thiazolidine-4-carboxylate (Li et al., 2007). In contrast, formaldehyde inhalation exposure of approximately 0.7 ppm for 5 days (4h/day) did not cause significant changes in gene expression patterns within peripheral WBC of humans (Zeller et al., 2011). None of these aforementioned transcripts were represented by the 222 total transcripts identified in this study as differentially expressed in the WBC. The data provided here are critical to elucidating systemic effects of formaldehyde inhalation exposure.
In addition to the inflammatory-associated signaling in the nose, formaldehyde exposure was found to alter the expression of genes involved in apoptosis/proliferation-related signaling. These genes included Dmc1 and Sfrp4, which were also verified using gene-specific alternative approaches. DMC1 is essential for meiotic recombination, where targeted Dmc1 disruption causes apoptosis (Yoshida et al., 1998). SFRP4 is a secreted-type WNT signaling inhibitor which plays an important role in the regulation of cell proliferation and cell death (Katoh and Katoh, 2007). The association to cell death and proliferation-related signaling in the nose is supportive of previous findings that formaldehyde inhalation exposure causes cell death and cytotoxicity-induced cell proliferation (Monticello et al., 1991) and altered genomic signaling related to apoptosis/proliferation (Andersen et al., 2010) in the rat nasal epithelium. Cell death and proliferation responses are an integral part of the mode of action linking formaldehyde inhalation exposure to cancer of the upper respiratory tract (NTP, 2011; Swenberg et al., 2013). Our findings provide further evidence of transcriptional responses related to cell death and proliferation in tissues of direct contact to formaldehyde inhalation exposure.
CONCLUSIONS
Taken together this study advances the growing body of knowledge on miRNAs and their relationship to formaldehyde. This study presents 3 major findings: First, formaldehyde inhalation exposure induces tissue and time-dependent responses at the genomic and epigenomic level. Here, formaldehyde exposure significantly disrupts miRNA expression profiles within the rat nose and WBC but not within the BM. Although miRNAs are but 1 regulator of gene expression, this finding could inform the understanding of diseases associated with formaldehyde. Second, miRNAs are predicted to regulate up to 35% of formaldehyde-induced transcriptional responses. Third, exposure-responsive miRNAs are likely regulators of critical pathways, including those related to immune/inflammation response and apoptosis/proliferation. These results increase the understanding of mechanisms and biological pathways potentially underlying formaldehyde-induced health effects and also broaden the current knowledge on miRNA responses across time and in different tissues.
SUPPLEMENTARY MATERIAL
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institutes of Health (P30-ES010126, P42-ES005948, ES019315, T32-ES007018, T32-ES007126); Texas Commission for Environmental Quality (582-12-21861). The Research Foundation for Health and Environmental Effects, a 501(c)(3) organization, provided funding for the animal exposures conducted at the Lovelace Respiratory Research Institute. Support was also provided by the Society of Toxicology’s graduate student fellowships sponsored by Novartis and Syngenta.
Supplementary Material
ACKNOWLEDGMENTS
The authors disclose no competing conflicts of interest.
REFERENCES
- Andersen M. E., Clewell H. J., 3rd, Bermudez E., Dodd D. E., Willson G. A., Campbell J. L., Thomas R. S. (2010). Formaldehyde: Integrating dosimetry, cytotoxicity, and genomics to understand dose-dependent transitions for an endogenous compound. Toxicol. Sci. 118, 716–731 [DOI] [PubMed] [Google Scholar]
- Andersen M. E., Clewell H. J., 3rd, Bermudez E., Willson G. A., Thomas R. S. (2008). Genomic signatures and dose-dependent transitions in nasal epithelial responses to inhaled formaldehyde in the rat. Toxicol. Sci. 105, 368–383 [DOI] [PubMed] [Google Scholar]
- Bailey K. A., Fry R. C. (2013). Environmental toxicants and perturbation of miRNA signaling. In: microRNAs in Toxicology and Medicine (ed Sahu S. C.), John Wiley & Sons, Ltd, Chichester, UK. 10.1002/9781118695999.ch2 [Google Scholar]
- Beane Freeman L. E., Blair A., Lubin J. H., Stewart P. A., Hayes R. B., Hoover R. N., Hauptmann M. (2009). Mortality from lymphohematopoietic malignancies among workers in formaldehyde industries: The National Cancer Institute Cohort. J. Natl. Cancer Inst. 101, 751–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle E. I., Weng S., Gollub J., Jin H., Botstein D., Cherry J. M., Sherlock G. (2004). GO::TermFinder–Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20, 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G., Anumanthan A., Brown J. A., Greenfield E. A., Zhu B., Freeman G. J. (2008). CD160 inhibits activation of human CD4+ T cells through interaction with herpesvirus entry mediator. Nat. Immunol. 9, 176–185 [DOI] [PubMed] [Google Scholar]
- Casanova M., Heck H. D., Everitt J. I., Harrington W. W., Jr, Popp J. A. (1988). Formaldehyde concentrations in the blood of rhesus monkeys after inhalation exposure. Food Chem. Toxicol. 26, 715–716 [DOI] [PubMed] [Google Scholar]
- Cedar H., Bergman Y. (2009). Linking DNA methylation and histone modification: Patterns and paradigms. Nat. Rev. Genet. 10, 295–304 [DOI] [PubMed] [Google Scholar]
- Chang J. C., Gross E. A., Swenberg J. A., Barrow C. S. (1983). Nasal cavity deposition, histopathology, and cell proliferation after single or repeated formaldehyde exposures in B6C3F1 mice and F-344 rats. Toxicol. Appl. Pharmacol. 68, 161–176 [DOI] [PubMed] [Google Scholar]
- Chen H. C., Chen G. H., Chen Y. H., Liao W. L., Liu C. Y., Chang K. P., Chang Y. S., Chen S. J. (2009). MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. Br J Cancer 100, 1002–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- Fujimaki H., Kurokawa Y., Kunugita N., Kikuchi M., Sato F., Arashidani K. (2004). Differential immunogenic and neurogenic inflammatory responses in an allergic mouse model exposed to low levels of formaldehyde. Toxicology 197, 1–13 [DOI] [PubMed] [Google Scholar]
- Garcia D. M., Baek D., Shin C., Bell G. W., Grimson A., Bartel D. P. (2011). Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 18, 1139–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R., Volinia S., Liu C. G., Fernandez-Cymering C., Palumbo T., Pichiorri F., Fabbri M., Coombes K., Alder H., Nakamura T., et al. (2008). MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 111, 3183–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncharova E. A., Lim P. N., Chisolm A., Fogle H. W., Taylor J. H., Goncharov D. A., Eszterhas A., Panettieri R. A., Krymskaya V. P. (2010). Interferons modulate mitogen-induced protein synthesis in airway smooth muscle. Am J Physiol Lung Cell Mol Physiol 299, L25–L35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptmann M., Lubin J. H., Stewart P. A., Hayes R. B., Blair A. (2004). Mortality from solid cancers among workers in formaldehyde industries. Am. J. Epidemiol. 159, 1117–1130 [DOI] [PubMed] [Google Scholar]
- Heck H. D., Casanova-Schmitz M., Dodd P. B., Schachter E. N., Witek T. J., Tosun T. (1985). Formaldehyde (CH2O) concentrations in the blood of humans and Fischer-344 rats exposed to CH2O under controlled conditions. Am. Ind. Hyg. Assoc. J. 46, 1–3 [DOI] [PubMed] [Google Scholar]
- IARC (2012). IARC Monographs on the Evaluation of Carcinogenic Risks to Humans: Volume 100F, A Review of Human Carcinogens: Chemical Agents and Related Occupations. International Agency for Research on Cancer.
- Jardim M. J., Fry R. C., Jaspers I., Dailey L., Diaz-Sanchez D. (2009). Disruption of microRNA expression in human airway cells by diesel exhaust particles is linked to tumorigenesis-associated pathways. Environ. Health Perspect. 117, 1745–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongstra-Bilen J., Jongstra J. (2006). Leukocyte-specific protein 1 (LSP1): A regulator of leukocyte emigration in inflammation. Immunol. Res. 35, 65–74 [DOI] [PubMed] [Google Scholar]
- Katoh M., Katoh M. (2007). WNT signaling pathway and stem cell signaling network. Clin. Cancer Res. 13, 4042–4045 [DOI] [PubMed] [Google Scholar]
- Kerns W. D., Pavkov K. L., Donofrio D. J., Gralla E. J., Swenberg J. A. (1983). Carcinogenicity of formaldehyde in rats and mice after long-term inhalation exposure. Cancer Res. 43, 4382–4392 [PubMed] [Google Scholar]
- Li G. Y., Lee H. Y., Shin H. S., Kim H. Y., Lim C. H., Lee B. H. (2007). Identification of gene markers for formaldehyde exposure in humans. Environ. Health Perspect. 115, 1460–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Chen J. X., Fu X. P., Yang S., Zhang Z., Chen K. h. H., Li Y. (2011). microRNA expression profiling of nasopharyngeal carcinoma. Oncol. Rep. 25, 1353–1363 [DOI] [PubMed] [Google Scholar]
- Lu K., Boysen G., Gao L., Collins L. B., Swenberg J. A. (2008). Formaldehyde-induced histone modifications in vitro. Chem. Res. Toxicol. 21, 1586–1593 [DOI] [PubMed] [Google Scholar]
- Lu K., Collins L. B., Ru H., Bermudez E., Swenberg J. A. (2010). Distribution of DNA adducts caused by inhaled formaldehyde is consistent with induction of nasal carcinoma but not leukemia. Toxicol. Sci. 116, 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., Moeller B., Doyle-Eisele M., McDonald J., Swenberg J. A. (2011). Molecular dosimetry of N2-hydroxymethyl-dG DNA adducts in rats exposed to formaldehyde. Chem. Res. Toxicol. 24, 159–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina M., Zhelezova G., Petrova E., Boev M. (2004). Flow cytometric determination of neutrophil respiratory burst activity in workers exposed to formaldehyde. Int. Arch. Occup. Environ. Health 77, 335–340 [DOI] [PubMed] [Google Scholar]
- McGwin G., Lienert J., Kennedy J. I. (2010). Formaldehyde exposure and asthma in children: A systematic review. Environ. Health Perspect. 118, 313–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller B. C., Lu K., Doyle-Eisele M., McDonald J., Gigliotti A., Swenberg J. A. (2011). Determination of N2-hydroxymethyl-dG adducts in the nasal epithelium and bone marrow of nonhuman primates following 13CD2-formaldehyde inhalation exposure. Chem. Res. Toxicol. 24, 162–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticello T. M., Miller F. J., Morgan K. T. (1991). Regional increases in rat nasal epithelial cell proliferation following acute and subchronic inhalation of formaldehyde. Toxicol. Appl. Pharmacol. 111, 409–421 [DOI] [PubMed] [Google Scholar]
- Moschos S. A., Williams A. E., Perry M. M., Birrell M. A., Belvisi M. G., Lindsay M. A. (2007). Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics 8, 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NTP (2011). Report on Carcinogens, 12th ed U.S. Department of Health and Human Services, Public Health Service, National Toxicology Program, Research Triangle Park, NC [Google Scholar]
- Palmer C., Diehn M., Alizadeh A. A., Brown P. O. (2006). Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics 7, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavkov K. L., Kems W. D., Mitchell R. I., Connell M. M., Donofrio D. J., Harroff H. H. (1982). Final Report on A Chronic Inhalation Toxicology Study in Rats and Mice Exposed to Formaldehyde. Chemical Industry Institute of Toxicology, Columbus, OH [Google Scholar]
- Rager J. E., Moeller B. C., Doyle-Eisele M., Kracko D., Swenberg J. A., Fry R. C. (2013). Formaldehyde and epigenetic alterations: MicroRNA changes in the nasal epithelium of nonhuman primates. Environ. Health Perspect. 121, 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager J. E., Smeester L., Jaspers I., Sexton K. G., Fry R. C. (2011). Epigenetic changes induced by air toxics: Formaldehyde exposure alters miRNA expression profiles in human lung cells. Environ. Health Perspect. 119, 494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenouda S. K., Alahari S. K. (2009). MicroRNA function in cancer: Oncogene or a tumor suppressor? Cancer Metastasis Rev. 28, 369–378 [DOI] [PubMed] [Google Scholar]
- Swenberg J. A., Kerns W. D., Mitchell R. I., Gralla E. J., Pavkov K. L. (1980). Induction of squamous cell carcinomas of the rat nasal cavity by inhalation exposure to formaldehyde vapor. Cancer Res. 40, 3398–3402 [PubMed] [Google Scholar]
- Swenberg J. A., Moeller B. C., Lu K., Rager J. E., Fry R. C., Starr T. B. (2013). Formaldehyde carcinogenicity research: 30 years and counting for mode of action, epidemiology, and cancer risk assessment. Toxicol. Pathol. 41, 181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Windgassen D., Papoutsakis E. T. (2008). Comparative analysis of transcriptional profiling of CD3+, CD4+ and CD8+ T cells identifies novel immune response players in T-cell activation. BMC Genomics 9, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Kondoh G., Matsuda Y., Habu T., Nishimune Y., Morita T. (1998). The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol. Cell 1, 707–718 [DOI] [PubMed] [Google Scholar]
- Young J. M., Shykind B. M., Lane R. P., Tonnes-Priddy L., Ross J. A., Walker M., Williams E. M., Trask B. J. (2003). Odorant receptor expressed sequence tags demonstrate olfactory expression of over 400 genes, extensive alternate splicing and unequal expression levels. Genome Biol 4, R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller J., Neuss S., Mueller J., Kühner S., Holzmann K., Högel J., Klingmann C., Bruckner T., Triebig G., Speit G. (2011). Assessment of genotoxic effects and changes in gene expression in humans exposed to formaldehyde by inhalation under controlled conditions. Mutagenesis 26, 555–561 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.