Abstract
Natural regulatory T (nTreg) cells generated in the thymus are essential throughout life for the maintenance of T-cell homeostasis and the prevention of autoimmunity. T-cell receptor (TCR)/CD28-mediated activation of nuclear factor-κB and (J)un (N)-terminal kinase pathways is known to play a key role in nTreg cell development but many of the predicted molecular interactions are based on extrapolations from non-Treg cell TCR stimulation with non-physiological ligands. For the first time, we provide strong genetic evidence of a scaffold function for the Caspase Recruitment Domain (CARD) of the TCR signalling protein CARD-MAGUK1 (CARMA1) in nTreg cell development in vivo. We report two, new, N-ethyl-N-nitrosourea-derived mutant mice, Vulpo and Zerda, with a profound block in the development of nTreg cells in the thymus as well as impaired inducible Treg cell differentiation in the periphery. Despite independent heritage, both mutants harbour different point mutations in the CARD of the CARMA1 protein. Mutations in vulpo and zerda do not affect expression levels of CARMA1 but still impair signalling through the TCR due to defective downstream Bcl-10 recruitment by the mutated CARD of CARMA1. Phenotypic differences observed between Vulpo and Zerda mutants suggest a role for the CARD of CARMA1 independent of Bcl-10 activation of downstream pathways. We conclude that our forward genetic approach demonstrates a critical role for the CARD function of CARMA1 in Treg cell development in vivo.
Keywords: regulatory T cells, signalling/signal transduction, thymus
Introduction
Regulatory T (Treg) cells are a subpopulation of T cells that modulate the immune system and maintain tolerance to self-antigens.1 They maintain allogeneic transplant tolerance in experimental models,2 but also facilitate tumour escape from immune monitoring.3 These cells are defined by their expression of the transcription factor, FOXP3 in addition to surface phenotypic markers not restricted to Treg cells, such as CD4, CD25, glucocorticoid-induced tumour necrosis factor receptor (GITR) and cytotoxic T-lymphocyte antigen 4 (CTLA-4).4 Their importance is underscored by the lymphoproliferative and multi-organ autoimmunity phenotypes of Scurfy mice and the human condition immunodysregulation polyendocrinopathy and enteropathy, X-linked (known as IPEX) syndrome, which result from mutations in the Foxp3 gene and consequent lack of Treg cells.5 The majority of Treg cells develop naturally in the thymus (nTreg cells) as a subpopulation of CD4+ T cells. They are positively selected and migrate to the periphery. In addition to these naturally occurring nTreg cells, transforming growth factor-β (TGF-β) and interleukin-2 (IL-2) are able to promote, in response to T-cell receptor (TCR)/CD28 co-stimulation, the differentiation of peripheral naive CD4+ T cells into CD4+ CD25+ FOXP3+ T cells, otherwise known as inducible Treg (iTreg) cells, that possess T-cell suppressive properties akin to those of nTreg cells.6–8
The signalling events necessary for nTreg development in the thymus are not fully understood. Three main signals have been identified. First, it is thought that self-peptide–MHC recognition by the TCR is required with an avidity intermediate between that necessary for positive selection of conventional T effector (Tconv) cells and that needed for the deletion of autoreactive T cells.9 Second, co-stimulatory signals play an important role: we know that mice deficient in CD28, CD40 or lymphocyte function-associated antigen 1 all display a marked reduction in nTreg cells;1,10–12 and third, IL-2 is also known to be crucial for nTreg cell development.4
Based on these and other data, a two-step model of nTreg cell development has been projected in which an initial TCR/CD28-mediated signal is required for activation of the nuclear factor-κB (NF-κB) and Jun N-terminal kinase (JNK) pathways, and the consequent induction of common γ-chain cytokine-responsive FOXP3− Treg precursor cells from immature thymocytes.1 Two precursor populations of nTreg cells have been described: CD4+ CD8+ thymocytes that express FOXP3,13,14 and a more distal CD4+ CD8− CD25+ CD122+ GITR+ Foxp3− thymocyte population.15 Whether the proximal precursor gives rise to the distal precursor before mature nTreg cell generation is not known,4 but in the second step of nTreg cell development these distal, now γ-chain cytokine-responsive, FOXP3− Treg cells are exposed to IL-2 in the absence of new TCR/CD28 ligation, resulting in their differentiation into mature FOXP3+ nTreg cells.15,16
How the signals arising from multiple receptors are integrated to drive the development of nTregs in vivo is far from resolved and much of what we do know is based on data extrapolated from co-transfection studies in T-cell lines.17,18 To address this issue we used an unbiased forward genetic approach to identify dominant genes required for the development of nTreg cells in the thymus. We report two new, independently derived, N-ethyl-N-nitrosourea (ENU)-mutants, Vulpo and Zerda, which have a near complete (> 90%) block in nTreg cell development, as well as impaired iTreg cell differentiation in the periphery. Both culpable mutations are amino acid (aa) substitutions in the Caspase Recruitment Domain (CARD) of the CARD-MAGUK1 (CARMA1) scaffold protein. Despite Vulpo and Zerda mutant mice expressing normal levels of CARMA1, signalling through the TCR is impaired because of defective downstream B-cell CLL/lymphoma 10 (Bcl-10) protein recruitment by the mutated CARD of CARMA1. However, the zerda mutation resulted in a greater block in nTreg cell development and impaired TCR signalling. Our study provides new in vivo evidence of a critical role for the scaffold function of the CARD of CARMA1 in Treg cell development and suggests that it has an in vivo function independent of Bcl-10 recruitment.
Materials and methods
Mice and mutagenesis
N-Ethyl-N-nitrosourea mutagenesis was used to generate random mutations in the sperm of C57BL/6J (Charles River sub-strain) male mice, generation 0 (G0). The G0 mice were bred to C57BL/6J wild-type (Wt) females to obtain G1 mice derived from an ENU-mutated sperm with a unique spectrum of point mutations (˜ 2500 heterozygous mutations per G1).19 Mice were bred in Central Biomedical Services (Imperial College London, UK) and kept under specific pathogen-free conditions. All animal work performed as part of this project was authorized by the Home Office under the Animals (Scientific Procedures) Act 1986, License PPL 70/6490. Tail vein blood samples (50 μl) were taken from 6-week-old heterozygous, G1 ENU mutant mice and analysed by flow cytometry to identify mutants with abnormal nTreg cell development.
The MRL/Mp.lpr/lpr mouse serum samples were provided by M. Botto (Imperial College, London, UK).
Homozygous mutants were used for all experiments unless otherwise stated.
Cell preparation and flow cytometry
Cells were treated with red blood cell lysis buffer (90% volume/volume 0.16 m NH4Cl, 10% volume/volume 0.17 m Tris–HCl pH 7.65, final pH 7.2) and then 25 μl of anti-mouse Fcγ-R (CD16/32) antibody (eBioscience, San Diego, CA) added at a final concentration of 1/20 for 10 min at room temperature, to block non-specific antibody binding. Cells were subsequently stained with conjugated monoclonal antibodies (mAbs) for 30 min at 4°. All antibodies were applied to a 1/200 final dilution and from eBioscience, unless stated: peridinin chlorophyll protein-Cy5.5-conjugated anti-CD4, Pacific Blue-anti-CD8 (BD Biosciences, San Jose, CA), phycoerythrin-Cy7-anti-CD25, FITC-anti-B220 (BD Biosciences), allophycocyanin-anti-CD44, allophycocyanin-anti-GITR, FITC-anti-CD62L (BD Biosciences), phycoerythrin-anti-CD69 (PharMingen, San Jose, CA). Cells were washed by centrifugation in FACS buffer for 2 min at 3000 g and 100 μl Cytofix/Cytoperm™ (BD Biosciences) was applied for 20 min at 4° to permeabilize cells before intracellular staining with FITC-anti-Foxp3, as per manufacturer's protocol. All samples were acquired on a DAKO Cyan 9 flow cytometer and data were analysed with Summit™ software (Dako, Stockport, UK).
In vitro iTreg cell differentiation and CD4+ T-cell activation
To measure iTreg cell differentiation, MACS® purified splenic CD4+ T cells were sorted as naive CD4+ CD25− CD44lo cells, then stimulated with plate-bound anti-CD3 (0–10 μg/ml) and anti-CD28 (1 μg/ml) for 3 days in the presence of recombinant IL-2 (100 U/ml) and recombinant TGF-β1 (2.5 ng/ml) (Cell Signaling, Beverly, MA) and stained for FOXP3 expression. MACS® (Miltenyi Biotec, Bergisch Gladbach, Germany) purified splenic CD4+ T cells were used to test T-cell activation as previously described.20 To measure up-regulation of activation markers by flow cytometry, cells were activated for 48 hr with plate-bound anti-CD3 (1 μg/ml) and anti-CD28 (2 μg/ml) in the presence of recombinant IL-2 (30 U/ml) (eBioscience), then stained for CD25, CD62L, CD69 and CD44.
Genomic linkage analysis
Heterozygote G1 phenodeviants with abnormal nTreg cell development were examined to determine whether the phenotype was heritable: G1 phenodeviants were crossed with Wt mice and the G2 population was screened for 50% expression of deviant phenotype. Once transmissibility was confirmed, each nTreg cell mutation was bred to homozygosity and mapped. Heterozygotes (G1) were mated to Wt 129Sv1 mJ mice; the affected hybrid (H) mice (H1) were then crossed with the Wt 129Sv1 mJ animal again. The H2 offspring were phenotyped and tail DNA was prepared from each mouse.21 Samples were sent to LGC Genomics (Hoddesdon, UK) for genotyping against 363 single nucleotide polymorphisms, which differ between C57BL/6 and 129/SvJ mice. The meiotic recombination frequency between the single nucleotide polymorphisms and the mutation was calculated and a logarithm of odds (LOD) score was found, where a 22 LOD score > 3 strongly indicates linkage.
Carma1 sequencing
All coding exons from carma 1 (2–25) were amplified from genomic DNA by PCR using the JumpStart REDtaq ReadyMix (Sigma, St Louis, MO). The PCR samples were purified by QIAquick PCR Purification Kit (Qiagen, Venlo, Limburg, the Netherlands) then sequenced (MRC CSC Genomics Core Laboratory, Imperial College London) and the data were analysed with Sequencher software (Gene Codes Corporation, Ann Arbor, MI). Primers are listed in Table 1.
Table 1.
Carma 1 primers used to sequence Carma 1 in mutant and wild-type genomic DNA
| Exon | Forward sequence | Reverse sequence |
|---|---|---|
| Non-coding | TCATTTCGCATCACAACAAGCACTG | TGGCAACCAGAGGTTTATTTCCAGG |
| Non-coding | AGTGAACACAGAGCCCCAGTATGA | TTCCATCTGGTGATGGCTCCTGA |
| Non-coding | TGCTCAGTTAGAAGAAACAAGGAACCC | CAGGTTCTGCTCAGAATAAAGCAAAAGG |
| Non-coding | TCCAATGGATAAGCACTGCCTCCC | GCGAGGTTGCTTTAGATGCTATCCC |
| 2 | CTTATGCCAGGCAAACTGCACTTTC | AGACAGAGACTCACTGCTCAGGAC |
| Non-coding | CCAGAGCACAGTTGGTGACATGAAA | AGAAAACTGACATGATAGTGTGTCCCTTG |
| 3 | TGTATGGAACCTCTCACCCACACTC | CCCCAATGACAGGACTTTCCATCTC |
| 4 | GGCAGCCCAAGAGTTCAGCA | ACACCATTTAAACCCCTCTGAATGAGAC |
| 5 | AGGTCCCCTAACTTTCCATGAGCC | GGGAACTAAGAGTTCAGTAAACGCTGC |
| 6 | TGTGCCAACAGCATGGTTTGTAGTC | GGCATTGCATGTGTCAGGAAATGG |
| 6 | CTTTTCTCCCAGAGCTAGGGCATTG | AGCTCAAGAATGACATCGAGAACCG |
| 7 | ACGGTGGAGATACATCCTTGGTGG | GGCTCAGTGTTCTTGAAGCAGAGAC |
| 8 | GAATGCATCTCTGAGCTGGGGAC | CATGAGCTAACTGTAGCCACTAAGCAA |
| 9 | AAACAACACAGACACACATACAGACATACA | AGACAGAAACAGGAGGATCATAAGCTCA |
| 10 | TCCCTGGGTAGCAACCAAAGTGAC | ACCCTGGAGATGGTATGAGCCAAG |
| 11 | AGCATATACATGCGGGCATCACAC | CCTGCCTCTCTGTGCTAATGGAAG |
| Non-coding | GGAAGCCTTGCGTTATACTTCCACC | TTCAGCCACAAACTCCTGGCTC |
| 12 | TGTAACAGAAGGCTGGGACCCTAC | ACACAAAAGTTGAGAGCCCCTGAAG |
| 13 | CATGTTCAGATGACTCTGAGGCCG | CCAAATCCAAGGTCCTGTTCCATCC |
| 14 | GAGAGTGTGTCAGTGTCCCCAATG | GCACCCTTCTGAACAGCTTAGACC |
| 15 | AACTGATTTGTGACTGAGACTCTGGC | TGTAGTAAGAGCTGTAGGGTTCTCACC |
| 16 | ACCTTGTAATGCAGAGTGATGAGGC | AGAGGACAGGGCGGTAGTTATTCC |
| 17 | CTTGTAGCAGACTGGGAAGCATGAC | GCACACAACTCTGAATGGCGATGG |
| 18 | GGGCAGGATTGCAAGTTCAATTTTAGAC | ACATTGTGGCTCAGCACAGATGTTAG |
| 19 and 20 | ACCTGTCACTGCGGGGATAAAATG | GCTCTCAGCCAATCAGATGGTCAG |
| 21 | AGGGCTTCAGTCAATGTGGGCATC | TCTGTGGGAACCTGGATTCTGTACC |
| 22 | AACATTACGAGGTGGCCGAGTG | GTTCAGCAGGTTTCTGAGACACCC |
| 23 | TTGGGGAGTCACAGCTTGGAAAC | TGGTCTGGCAGAGCAATCATGAAC |
| 24 | GGTCAAGGACCACACATGACGG | GTGAGTGTCCATTGCAAGCCCC |
| 25 | ACTGTGCTTGATGGAAACACCTGC | TCGAGCCCAAGAATGTAAGAAGCC |
Immunological assays
Anti-ssDNA IgG antibodies were measured by ELISA as previously described.23 Results were expressed as arbitrary units relative to a standard positive sample derived from an MRL/Mp.lpr/lpr mice pool. For Western blots, detergent extracts were resolved by 10% SDS–PAGE and electrophorectically transferred.24 The primary antibodies used were: rabbit mAb to CARD11 (1D12) (Cell Signaling) and mouse mAb to β-actin (Abcam, Cambridge, UK). The secondary antibodies used were: polyclonal goat anti-rabbit immunoglobulin/horseradish peroxidase (Dako) and polyclonal goat anti-mouse immunoglobulin/horseradish peroxidase (Dako). Wild-type, Vulpo and Zerda thymus lysates were run in duplicate. For immunoprecipitations, MACS®-purified splenic CD4+ T cells from three mouse spleens were activated in 1.5 ml RPMI-1640, PMA (200 ng/ml) and ionomycin (300 ng/ml) for 6 hr at 37°. Cells were harvested with PBS and lysed in 1 ml RIPA buffer plus protease inhibitor cocktail (Sigma-Aldrich, St Louis, MO). Polyclonal rabbit anti-mouse Bcl-10 antibody (2 μg) (Santa Cruz sc-5611) and protein A/G PLUS-agarose (20 μl) (Santa Cruz sc-2003) were added to each 1 ml sample (100–500 μg/ml total protein) followed by rotation at 4° for 2 hr. Beads were washed three times with RIPA buffer, boiled in SDS loading buffer, resolved on 10% SDS–PAGE gels and Western blotted. Primary antibodies used were rabbit mAb to CARD11 (1D12) and polyclonal rabbit anti-mouse Bcl-10 antibody. For improved Western blotting detection, blots were incubated with 1 : 5000 dilution of Protein A-horseradish peroxidase (GE Healthcare, Amersham, UK) prepared in blocking buffer, in place of secondary antibody.
Statistical analysis
All results are given as mean values with SD. A non-parametric unpaired t-test (two-tailed) was applied: ***P < 0.001, **P = 0.001–0.01, *P = 0.01–0.05, ns = not significant. All statistical calculations were performed using Prism5 software (GraphPad, San Diego, CA).
Results
Dominant ENU-induced mutations vulpo and zerda impair nTreg cell development
N-Ethyl-N-nitrosourea-induced germline G1 mutants (n = 766) were screened for dominant mutations resulting in abnormal nTreg cell development, defined as a CD25+ FOXP3+ CD4+ cell population outwith 2 SD of the mean. Chronic antigenic exposure represents one of the main sources of iTreg cells25 and therefore, it was expected that these non-immunized, 6-week-old mice housed under clean, specific pathogen-free conditions had a negligible iTreg cell contribution. Two ENU mutant mice, Vulpo and Zerda, with > 90% reduction in the frequency of FOXP3+ CD25+ CD4+ cells in peripheral blood leucocytes (PBL) compared with the mean value for the 766 animals screened (P < 0.001), were identified (Fig. 1a,b). These dominant mutations were confirmed to be heritable in the germline. G2 phenodeviant mutants were inter-crossed and the G3 offspring were screened and inter-crossed to establish homozygous strains.
Figure 1.
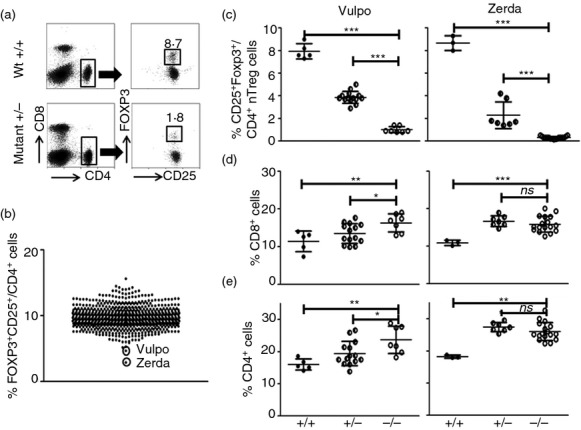
Identification of Vulpo and Zerda mutants with impaired natural regulatory T (nTreg) cell development. (a) A representative dot plot defining the percentage of FOXP3+ CD25+ nTreg cells in the CD4+ T lymphocyte population from peripheral blood leucocytes (PBL) in one wild-type (Wt) mouse (top panel) and one heterozygous N-ethyl-N-nitrosourea (ENU) mutant (Vulpo) mouse (Mutant ±) (bottom panel). Forward scatter plot allowed the exclusion of non-singlet cells and gates were applied (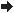 ) to the lymphocyte (not shown) and CD4+ populations (left column) to define the FOXP3+ CD25+/CD4+ population (right column). (b) Two ENU mutant mice, Vulpo and Zerda, with reduced FOXP3+ CD25+/CD4+ nTreg cell frequencies in PBL (values outwith 2SD of the mean). Each symbol represents data from an individual mouse (n = 766). The graph is the cumulative result of at least 20 independent experiments. (c) Percentage of FOXP3+ CD25+/CD4+ nTreg cells in PBL. (d) Percentage of CD8+ CD4− cells in PBL. (e) Percentage of CD8− CD4+ cells in PBL. (c–e) Each symbol represents data from an individual mouse. n = 5 for Wt (+/+), 14 for heterozygotes (±) and eight for homozygous (−/−) Vulpo mutants (left column). n = 3 for Wt, seven for heterozygotes and 17 for homozygous Zerda mutants (right column). Error bars represent mean ±1 SD. A non-parametric unpaired t-test (two-tailed) was applied: ***P < 0.001, **P < 0.01, *P < 0.05, ns = not significant. Graphs are representative of at least two independent experiments.
) to the lymphocyte (not shown) and CD4+ populations (left column) to define the FOXP3+ CD25+/CD4+ population (right column). (b) Two ENU mutant mice, Vulpo and Zerda, with reduced FOXP3+ CD25+/CD4+ nTreg cell frequencies in PBL (values outwith 2SD of the mean). Each symbol represents data from an individual mouse (n = 766). The graph is the cumulative result of at least 20 independent experiments. (c) Percentage of FOXP3+ CD25+/CD4+ nTreg cells in PBL. (d) Percentage of CD8+ CD4− cells in PBL. (e) Percentage of CD8− CD4+ cells in PBL. (c–e) Each symbol represents data from an individual mouse. n = 5 for Wt (+/+), 14 for heterozygotes (±) and eight for homozygous (−/−) Vulpo mutants (left column). n = 3 for Wt, seven for heterozygotes and 17 for homozygous Zerda mutants (right column). Error bars represent mean ±1 SD. A non-parametric unpaired t-test (two-tailed) was applied: ***P < 0.001, **P < 0.01, *P < 0.05, ns = not significant. Graphs are representative of at least two independent experiments.
The level of nTreg cells (CD25+ Foxp3+/CD4+ cells) in the PBL of homozygous mutant Vulpo mice was only 12.8% of Wt (87.2% reduction) (P < 0.0001) (Fig. 1c). The level of nTreg cells in the PBL of homozygous mutant Zerda mice was only 3.4% of that in Wt (96.6% reduction) (P < 0.0001) (Fig. 1c). Organ dissection supported these findings (Fig. 2e): we observed a 57.5% decrease in absolute number of nTreg cells in the thymus of Vulpo mutants (mean 4.2 × 105 ± 1 SD (8.9 × 104) cells, n = 5) and a 92.2% decrease in absolute number of nTreg cells in the thymus of Zerda mutants (mean 7.8 × 104 ± 1SD (4.9 × 103) cells, n = 6) compared with Wt (mean 1 × 106 ± 1SD (8.7 × 104) cells, n = 6). The absolute number of nTreg cells in the thymus of Zerda mutants was significantly less than for Vulpo mutants (P < 0.0001) (Fig. 2e). The Treg cell lineage remained virtually absent in older Vulpo and Zerda mice (1 year) indicating that these were persistent defects and not compensated over time (data not shown).
Figure 2.
The mutations vulpo and zerda affect all stages of thymocyte maturation. Thymus dissection was performed in wild-type (Wt), Vulpo and Zerda mutants. Graphs show absolute numbers for (a) CD4− CD8− (DN), (b) CD4+ CD8+ (DP), (c) CD4− CD8+, (d) CD4+ CD8− and (e) CD25+ Foxp3+ CD4+ CD8− thymocytes. Each symbol represents data from an individual homozygous mouse thymus: Wt (n = 6), Vulpo (n = 5) and Zerda (n = 6). A non-parametric unpaired t-test was applied to compare data sets. ***P < 0.001, *P < 0.05, ns = not significant. Graphs are representative of two independent experiments.
We also observed a generalized increase in Tconv cells in the PBL of both mutants versus Wt mice. We recorded a 42.1% increase in the frequency of CD8+ T cells in the PBL of homozygous mutant Vulpo mice compared with Wt (P = 0.0078) and a 50% increase for homozygous mutant Zerda mice compared with Wt (P < 0.0001) (Fig. 1d). We recorded a 47.9% increase in the frequency of CD4+ T cells in the PBL of homozygous mutant Vulpo mice compared with Wt (P = 0.0037) and a 75.4% increase for homozygous mutant Zerda mice compared with Wt (P < 0.0001) (Fig. 1e). Organ dissection of spleen (data not shown) and thymus (Fig. 2a–d) supported these findings: we observed a significant increase in absolute cell number in Vulpo and Zerda mutants at every stage of thymocyte maturation compared with Wt mice. In contrast to the difference observed in frequency of mature nTreg cells between the two mutants, we observed no significant difference between Vulpo and Zerda mutants at all stages of thymocyte maturation (Fig. 2a–d).
Both vulpo and zerda mutations showed incomplete dominance, as evidenced by heterozygous (±) phenotypes intermediate to the Wt (+/+) and homozygous (−/−) mutant phenotypes (Fig. 1c–e). This semi-dominant effect is seen on nTreg cell development and not just FOXP3 expression because no decrease in mean fluorescence intensity was observed during flow cytometric analysis (data not shown).
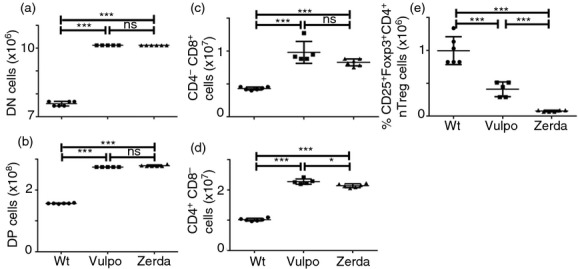
In the two-step model of nTreg cell development1,15,16 (Fig. 3a), vulpo and zerda mutations affected both Treg precursor populations (Fig. 3b,c). Both mutations resulted in > 70% reduction in frequency of proximal CD4+ CD8+ FOXP3+ thymocytes compared with Wt (P < 0.01) (Fig. 3b) and > 40% reduction in frequency of more distal CD4+ CD25+ GITR+ FOXP3− thymocytes compared with Wt (P < 0.05) (Fig. 3c). No signifcant difference was observed between the two mutants. The vulpo and zerda mutations therefore block the initial TCR/CD28-mediated induction of γ-chain cytokine-responsive FOXP3− Treg precursor cells from immature thymocytes.
Figure 3.
The vulpo and zerda mutations affect natural regulatory T (nTreg) precursor cell development in the thymus, as well as inducible (i) Treg cell differentiation in the periphery. (a) A schematic diagram illustrating the putative two-step model of nTreg cell development from immature thymocytes. (b) Thymi from wild-type (Wt), Vulpo and Zerda mice were analysed for the presence of CD25+ FoxP3+ cells among CD4+ CD8+ thymocytes. Numbers in the plots represent the percentage of FoxP3+ cells among the double-positive (DP) thymocytes (i.e. proximal nTreg precursor population). The adjacent graph compiles the mean ± SD of percentage FoxP3+ events among DP thymocytes generated from the indicated numbers of mice. Each symbol represents data from an individual mouse. Thymocytes from Wt (n = 4), Vulpo (n = 3) and Zerda (n = 4) homozygous mutants were analysed. (c) Similarly, thymi from Wt, Vulpo and Zerda mice were analysed for the presence of GITRhi CD25+ FoxP3− cells among CD4+ CD8− thymocytes. This graph compiles the mean ± SD of the percentage of CD4+ CD25+ GITR+ FOXP3− precursor cells recorded. Each symbol represents data from an individual mouse. Thymocytes from Wt (n = 4), Vulpo (n = 3) and Zerda (n = 4) homozygous mutants were analysed. A non-parametric unpaired t-test (two-tailed) was applied: ***P < 0.001, **P < 0.01, *P < 0.05. Each graph is representative of two independent experiments. (d) FACS-sorted, naive CD4+ splenocytes were stimulated with plate-bound anti-CD3 mAb (0–10 μg/ml) and anti-CD28 mAb (1 μg/ml) for 3 days in the presence of interleukin-2 (IL-2) and transforming growth factor-β (TGF-β). Percentage of FOXP3+ iTreg cells was then measured for Wt, Vulpo or Zerda by intracellular flow cytometry. Shown are mean values ± 1 SD from three replicates in two independent experiments. A parametric unpaired t-test was applied to compare data sets: Wt versus Vulpo P = 0.04; Wt versus Zerda P = 0.02; Vulpo versus Zerda P = 0.3, not significant.
Dominant ENU-induced mutations vulpo and zerda impair iTreg cell development
We next examined the effects of vulpo and zerda mutations on iTreg differentiation in vitro. Purified splenic CD4+ T cells were sorted as naive CD4+ CD25− CD44lo cells then stimulated with anti-CD3 and anti-CD28 for 3 days in the presence of recombinant IL-2 and recombinant TGF-β1 and stained for FOXP3 expression.26 The up-regulation of FOXP3 in Wt naive CD4+ T cells correlated positively with anti-CD3 dose up to 0.5 μg/ml but was reduced in Vulpo (P < 0.05) and Zerda (P < 0.05) cells (Fig. 3d). At higher TCR stimulation (doses of anti-CD3 > 0.5 μg/ml), we observed progressive impairment of iTreg cell differentiation in Wt cells, as previously reported,27 as well as Vulpo and Zerda cells (Fig. 3d). We conclude that vulpo and zerda mutations impede iTreg differentiation. No significant difference was observed between Vulpo and Zerda iTreg differentiation (P = 0.3).
vulpo and zerda are point mutations in the carma1 gene
Given the similar nature of both mutant phenotypes, initial genotyping work was performed on Vulpo. Using a meiotic positional cloning strategy,22 the vulpo mutation was mapped to a 20 mega-base pair region on chromosome 5, bounded by markers 128944450 and 149044358, with an LOD score of 7.2 (P < 0.001) (Fig. 4a). A search of the corresponding published gene list (Ensembl.org) revealed 317 candidate genes including caspase recruitment domain family, member 11 (card11) – more commonly known as carma1 – which maps to chromosome 5, base pairs 141348953–141476550. Using previously identified carma1 primers, DNA prepared from both Vulpo and Zerda mutants was amplified by PCR and sequenced. The resulting sequences were compared with the reference carma1 sequence and point mutations were identified in exon 4 of the carma1 gene for both vulpo (C→G at base 92061) and zerda (T →A at base 92108) mutations. These mutations translate into the substitution of Wt hydrophobic isoleucine for mutant hydrophobic methionine at aa 79 (I79M) for vulpo and the substitution of Wt hydrophobic leucine for positively charged glutamine at aa 95 (L95Q) for zerda (Fig. 4b). Both mutations affect the CARD (aa 15–105) of the CARMA1 protein (Fig. 4c): an aa sequence conserved across mouse and human species.17 Western blotting revealed Wt levels of CARMA1 protein expression in the thymi of both vulpo and zerda mutant mice and so we conclude that neither CARD mutation affected CARMA1 stability (Fig. 4d). As newly identified alleles of a known gene, homozygous mutants are hereafter referred to as Carma1vul/vul and Carma1zer/zer.
Figure 4.
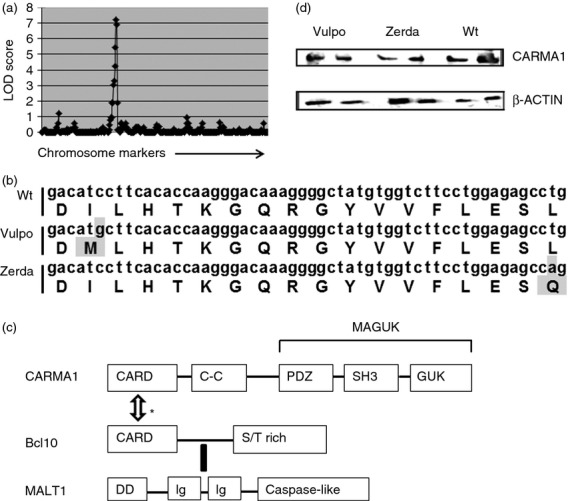
vulpo and zerda are point mutations in the carma1 gene. (a) The vulpo mutation was mapped to a 20 mega-base pair region on chromosome 5, bound by markers 128944450 and 149044358, with a LOD score of 7.2 (P < 0.001). (b) Point mutations in exon 4 of the carma1 gene from vulpo (C→G at base 92061) and zerda (T→A at base 92108) mutant mice, and corresponding amino acid substitutions at position 79 (I79M) for vulpo and position 95 (L95Q) for zerda. (c) The structures of CARMA1, Bcl-10 and MALT1 proteins. Upon T-cell receptor activation, CARMA1 recruits the preformed Bcl-10-MALT1 complex to the immunological synapse. * Based on extrapolated findings from non-regulatory T (Treg) cell studies in vitro, this is thought to be via a CARMA1-CARD–Bcl-10-CARD interaction.17,18 MALT1 contains two immunoglobulin-like domains that mediate binding to Bcl-10 by means of a motif comprising a short stretch of amino acids following the CARD.43 The C-terminal caspase-like domain of MALT1 is then thought to interact directly with the coiled-coil (C-C) domain of CARMA1.28 Functional domains: CARD, caspase recruitment domain; C-C, coiled-coil domain; MAGUK, membrane-associated guanylate kinase (GuK) -like domain; S/T-rich, Ser/Thr-rich domain; DD, death domain; Ig, immunoglobulin-like domain. The vulpo and zerda mutations are located in the CARD (amino acids 15–105) of the CARMA1 protein. (d) Western blot analysis of protein expression in homozygous Vulpo, Zerda and Wt thymus lysates using anti-CARMA mAb (top panel) and anti-ACTIN mAb (lower panel). Two independent samples are shown for each phenotype. This figure is representative of three independent experiments.
Carma1vul/vul and Carma1zer/zer mice do not develop spontaneous autoimmunity
Given the significantly reduced nTreg and increased Tconv cell populations in the periphery (Fig. 1c–e), it was initially surprising that Carma1vul/vul and Carma1zer/zer mice remained healthy at 1 year and showed no macroscopic signs of spontaneous autoimmune disease upon examination, e.g. weight loss, red eyes, dermatitis, spontaneous abortion, or splenomegaly. Both mutants demonstrated Wt levels of circulating anti-ssDNA IgG antibodies, 90% less than MRL/Mp.lpr/lpr positive-control mice at 1 year (P < 0.005) (Fig. 5a).
Figure 5.
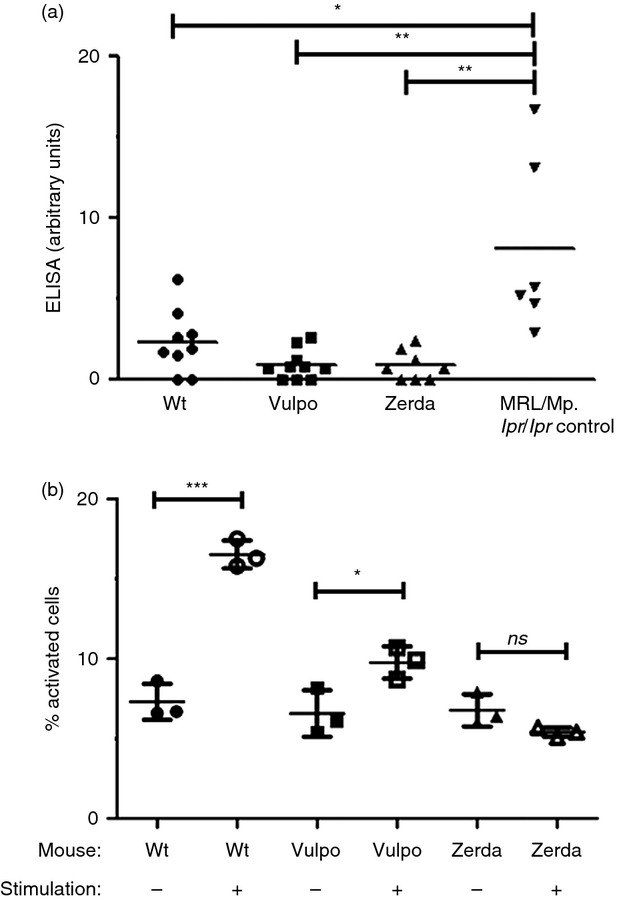
Vulpo and Zerda mutant mice do not develop autoimmunity and harbour non-reactive conventional T (Tconv) cells. (a) ELISA for serum anti-ssDNA IgG auto-antibodies in 1-year-old wild-type (Wt) (n = 8), Vulpo (n = 10), Zerda (n = 8) and MRL/Mp.lpr/lpr positive control (n = 6) female mice. The antibody levels are expressed in arbitrary ELISA units related to a standard positive sample, which was assigned a value of 10 arbitrary units. Serum samples were screened at 1/80 and positive samples were titrated to end-point. Each symbol represents data from one mouse. Horizontal bars indicate the mean. Data were analysed using a non-parametric unpaired t-test (two-tailed): **P < 0.01, *P < 0.05. This graph is representative of two independent experiments. (b) Percentage of activated (CD69+ CD44+ CD25+ CD62Llow) CD4+ T cells before (−) and after (+) stimulation with plate-bound anti-CD3 and anti-CD28 for 2 day in Wt, Vulpo and Zerda mutant mice. Data were analysed using a non-parametric unpaired t-test (two-tailed): ***P < 0.001, *P < 0.05, ns = not significant. This graph is representative of four independent experiments in which CD4+ T-cell activation was analysed in triplicate with samples pooled from five mice per genotype. Black error bars represent mean ± 1SD. Similar results are achieved upon stimulation of mutant peripheral CD8+ T cells (data not shown).
Impaired TCR signalling in Carma1vul/vul and Carma1zer/zer mice
In an attempt to explain this lack of autoimmunity, we measured the activation of Tconv cells from Carma1vul/vul and Carma1zer/zer mice. The percentage of Carma1vul/vul CD4+ T cells expressing an activated phenotype (CD69+ CD44+ CD25+ CD62Llow), increased 1.5-fold in response to anti-CD3/CD28 stimulation (P = 0.03), compared with a 2.3-fold increase in the percentage of activated Wt CD4+ T cells (P = 0.00004) (Fig. 5b). The activation of CD4+ T cells from Carma1zer/zer mutants was even more impaired, as shown by the complete absence of activation following TCR stimulation (P < 0.0001) (Fig. 5b). A similar effect was observed in mutant CD8+ T cells from both Carma1vul/vul and Carma1zer/zer mice (data not shown). We suggest that Carma1vul/vul and Carma1zer/zer mice, like CARMA1 knockout (KO) mice, harbour a generalized TCR/CD28 activation defect in their peripheral Tconv cells, which would explain their lack of autoimmunity despite minimal nTregs. We suggest that this reduced peripheral Tconv cell activation may drive the increased cell production observed at every stage of thymocyte maturation in both mutants (Fig. 2a–d). The blockade in mature nTreg development was more pronounced in Carma1zer/zer compared with Carma1vul/vul mutants (Fig. 2e) and we also observe a corresponding increase in the degree of impairment of TCR activation in Tconv cells from Carma1zer/zer compared with Carma1vul/vul mutants (Fig. 5b).
vulpo and zerda mutations disrupt the CARMA1/Bcl-10 association upon TCR activation
Upon TCR activation, CARMA1 recruits the preformed Bcl-10-MALT1 complex to the immunological synapse and, based on extrapolated findings from non-Treg cell studies in vitro, CARMA1 is thought to bind with Bcl-10 via the CARD17,18 (Fig. 4c). The C-terminal caspase-like domain of MALT1 then interacts directly with the coiled-coil (C-C) domain of CARMA128 (Fig. 4c). No other proteins are known to interact with the CARD of CARMA1. The tri-molecular, CARMA1-Bcl-10-MALT1 complex (CBM), triggers IĸB kinase (IKK)/NF-κB activation in response to TCR/CD28 co-ligation.29 We used immunoprecipitation to examine the effects of vulpo and zerda CARD mutations on CARMA1 function after TCR/CD28 activation. Using PMA/ionomycin-activated splenic CD4+ T cells, we quantified the amount of CARMA1 protein co-immunoprecipitating with anti-Bcl-10 antibody by Western blot, in Carma1vul/vul and Carma1zer/zer versus Wt (Fig. 6). As expected, Bcl-10 interacts with CARMA1 in Wt but not Carma1vul/vul and Carma1zer/zer activated CD4+ T cells (Fig. 6). We conclude that the mutations in the CARD of both CARMA1-vulpo and CARMA1-zerda proteins prevent association with Bcl-10 after TCR stimulation.
Figure 6.
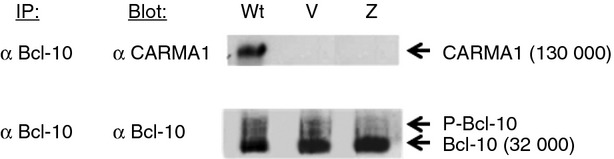
Impaired interaction of mutant CARMA1 proteins with Bcl-10. Bcl-10 was immunoprecipitated from PMA/ionomycin-activated CD4+ T cells then Western blotted and probed with antibodies specific for CARMA1 (top panel) or Bcl-10 (lower panel). Lanes for wild-type (Wt), Vulpo (V) and Zerda (Z) cells are shown. CARMA1 runs at 130 000 molecular weight. Bcl-10 runs at 32 000. Two additional, fainter bands are seen above Bcl-10 in each lane. Bcl-10 is known to be mono- and bi-phosphorylated upon lymphocyte activation.44 These phosphorylated Bcl-10 species (P-Bcl10) migrate slightly slower than Bcl-10 itself and are commonly observed immediately above the 32 000 Bcl-10 band.29
Discussion
The balance of Treg cells and Tconv cells is key to immune homeostasis. This study emphasizes the role of CARMA1 at the fulcrum. We report two new mutant mice, Vulpo and Zerda, with a profound block in the development of nTreg cells in the thymus, as well as impaired iTreg differentiation in the periphery. Despite being independently derived, the mutants harbour different point mutations in the CARD of the crucial scaffold protein, CARMA1. The level of CARMA1 is not affected in Carma1vul/vul and Carma1zer/zer mice but rather the mutant CARMA1 proteins are impaired in their ability to recruit Bcl-10 after TCR stimulation. For the first time, our findings provide genetic and in vivo evidence of a critical role for the CARD of the CARMA1 protein in nTreg development in the thymus, as well as in vitro evidence to augment our understanding of its role in iTreg differentiation in the periphery.
Mice lacking Bcl-10 or CARMA1 are known to be deficient in nTreg cells4,30,31 and CARMA1 is now thought to play a Treg cell-intrinsic role in both the initial TCR/CD28 activation step and the subsequent IL-2 receptor (IL-2R) signalling maturation step of nTreg cell development.4,32 However, before our work, the role for CARD-CARD binding of CARMA1 and Bcl-10 in nTreg cell development was based on results from co-transfection studies in unstimulated, non-Treg cell lines17,18,33–36 and TCR stimulation with non-physiological ligands in murine Tconv cells.29 In 2003, Newton and Dixit generated knock-in mice expressing a CARD-less form of CARMA1 but the impact of this mutation on the Bcl-10–CARMA1 interaction was never reported.24 In addition to CARMA1-KO mice,4,37–39 two other CARMA1 mutant mice have been described: Unmodulated mice, which have an L298Q substitution in the C-C domain,40 and Carma1king/king mice, which have an L525G substitution in the linker domain.30 The CARMA1-king protein is degraded due to instability30 and Unmodulated mice develop intense dermatitis and atopy by 10 weeks of age.40 By definition, not one of these mutants is able to provide in vivo evidence of CARD–CARD binding of CARMA1 and Bcl-10 in nTreg cell development. For the first time, we show that the CARD of CARMA1 is required for TCR-mediated signals triggered by physiological self-MHC ligands in thymic nTreg cell devlopment.
We present two new, in vivo models, Vulpo and Zerda, both expressing Wt levels of mutant CARMA1 protein. These mutants remain healthy at 1 year, do not develop spontaneous autoimmune disease and have allowed us to identify critical residues in the CARD that mediate CARMA1 function in T-cell activation and Treg cell development. Using co-immunoprecipitation studies to examine the CARMA1–Bcl-10 interaction in activated CD4+ T cells from Vulpo and Zerda mutant mice, we demonstrate complete impairment of CARMA1–Bcl-10 binding in both mutants. Given that downstream NF-κB and JNK signalling is completely dependent on CARMA1–Bcl-10 binding in thymic nTreg cell development, we use CARMA1–Bcl-10 binding as an analogue for NF-κB and JNK signalling after TCR stimulation and conclude that Carma1-vulpo and -zerda mutations, like Bcl-10−/− and Carma1−/− mutations,4,30,31 result in complete downstream impairment of NF-κB and JNK signalling in nTreg cell development. In the two-step model of nTreg cell development, we demonstrate a role for the CARD of CARMA1 in the TCR/CD28-mediated induction of γ-chain cytokine-responsive FOXP3− nTreg precursor cells from immature thymocytes.
We observe significant differences in the frequency of mature FOXP3+ nTreg cells (Fig. 2e) and also in Tconv cell activation (Fig. 5b) between our two mutants. In both cases, the zerda mutation results in a more profound phenotype. Given that both mutations result in complete impairment of Bcl-10: CARMA1 binding and presumed downstream NF-κB and JNK signalling during nTreg cell development, we conclude that our findings suggest a Bcl-10-independent role for CARD in TCR signalling during nTreg cell development. Unlike the I79M vulpo mutation, the L95Q zerda mutation results in an amino acid charge change from hydrophobic leucine to positively charged glutamine in the CARD and it is likely that this charge change is responsible for the more severe phenotype observed in Zerda compared with Vulpo mutant mice, through as yet unknown alterations in protein–protein interactions. In addition to its role in the NF-κB and JNK pathway-mediated generation of FOXP3− nTreg precursor cells from immature thymocytes, CARMA1 has recently been shown to play a critical role in the IL-2R signalling pathway-mediated maturation of these precursor cells into mature FOXP3+ nTreg cells.32 How the CARMA1 protein is integrated into this pathway to drive these events is yet to be defined. We suggest that the CARD of CARMA1 facilitates the IL-2R-mediated maturation stage of nTreg cell development, and that it is the effect of vulpo and zerda mutations on this role that accounts for the differences we observe between their phenotypes. Our ENU-mutant CARMA1 mice will allow the determination of how CARD function influences IL-2R signalling in nTreg cell development.
To the best of our knowledge, vulpo and zerda mutations only affect the amino acid sequence of the CARD of CARMA1. As the CARD motifs are protein interaction modules, their surface features dictate their mode of interactions with their binding partners and our results suggest that vulpo and zerda mutations impair CARMA1's interaction with its binding partners. However, we recognize that it is a formal possibility that the mutations that affect CARD function may also alter the conformation of non-CARD domains in CARMA1.
In contrast to the high dependency on TCR-CARMA1-NF-κB activity for thymic development of nTreg cells and their precursors, TCR-CARMA1-NF-κB activity is thought to be dispensable for iTreg cell induction in the periphery.30 In agreement with published findings in CARMA1-KO cells,4,30,31 our CARMA1 mutants retain some differentiation of naive CD4+ T cells into iTreg cells at low-dose TCR stimulation, supporting a role for the CARMA1-independent the TGF-β-TAK1-JNK signalling pathway in iTreg cell differentiation. TGF-β does not play a role in the thymic selection of nTreg cells so we assume that nTreg precursors are dependent on CBM complex formation for TAK1 activation.41 However, as observed in CARMA1-KO cells,26 Carma1vul/vul and Carma1zer/zer CD4+ T cells are less sensitive to increasing concentrations of anti-CD3 and up-regulate FOXP3 less rapidly when compared with Wt naive CD4+ T cells. We suggest that despite adequate CARMA1-independent TGF-β-TAK1-JNK signalling, the increased sensitivity offered by the anti-CD3 dose-dependent induction of TCR-CARMA1-NF-κB activity in Wt CD4+ T cells is absent in Carma1vul/vul and Carma1zer/zer CD4+ T cells, as well as CARMA1-KO cells. We propose that TCR-CARMA1-NF-κB activity is only partially dispensable for iTreg induction at low-dose TCR stimulation in the periphery.
Strong TCR stimulation prevents conversion of naive T cells into iTreg cells and overrules the ability of TGF-β/IL-2 to drive FOXP3 expression,27 although the molecular mechanisms by which FOXP3 expression is inhibited are not well understood. CARMA1-KO CD4+ T cells are partially resistant to this high-dose TCR-mediated inhibition of iTreg differentiation suggesting that NF-κB abrogates FOXP3 induction upon high TCR stimulation.26 In contrast Carma1vul/vul and Carma1zer/zer CD4+ T cells retain sensitivity to this high TCR-induced abrogation of iTreg cell differentiation. We support the view of Molinero et al.26 that high-dose TCR-mediated inhibition of iTreg cell differentiation is at least in part NF-κB dependent, but we propose that this negative regulation is not via the conventional CARD-dependent TCR-CBM-NF-κB axis but via a novel MAGUK region-dependent TCR-PKCδ-CARMA1 signalling pathway.42 Unlike CARMA1-KO cells, the MAGUK region of CARMA1 is preserved in CARMA1-vulpo and CARMA1-zerda proteins and so we predict that PKCδ-mediated inhibition of TCR signalling remains intact in Vulpo and Zerda CARD mutants.
Therapeutic targeting of the conserved canonical NF-κB signalosome is difficult because of its ubiquitous functions and multiple inputs. By contrast, because of its non-redundant role in antigen-receptor activation and its specificity to signals from the antigen-receptors, CARMA1 is a unique and appealing target for the design of therapeutic interventions that promote immune tolerance. For the first time, we provide two healthy in vivo models, Vulpo and Zerda, in which to further study the interactions of the CARMA1 protein with its binding partners and contribute to the goal of expounding the essential pathways required for Treg cell development.
Acknowledgments
This work was supported by a Wellcome Trust Clinical Research Fellowship awarded to EMS and a Medical Research Council grant for the ENU mutagenesis. The authors would like to thank C. Pusey and M. Botto for helpful discussions, and F. Carlucci, D. Heathcote, P. Mueller, S. Alyahya and Central Biomedical Services for technical support.
Glossary
- αα
amino acid
- CARD
caspase recruitment domain
- CARMA1
CARD-MAGUK1
- CBM
CARMA1, Bcl-10, MALT1 complex
- ENU
N-ethyl-N-nitrosourea
- G
generation
- GITR
glucocorticoid-induced tumor necrosis factor receptor
- iTreg
inducible T regulatory cell
- IL-2R
IL-2 receptor
- nTreg
natural T regulatory cell
- Tconv
conventional CD4+ and CD8+ T effector cells
Disclosures
EMS, LW, OC, SR and PAR have no conflicts of interest to disclose and no competing financial interests to declare.
References
- 1.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–87. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Wood KJ, Sakaguchi S. Regulatory T cells in transplantation tolerance. Nat Rev Immunol. 2003;3:199–210. doi: 10.1038/nri1027. [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi T, Sakaguchi S. Regulatory T cells in immune surveillance and treatment of cancer. Semin Cancer Biol. 2006;16:115–23. doi: 10.1016/j.semcancer.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Molinero LL, Yang J, Gajewski T, Abraham C, Farrar MA, Alegre ML. CARMA1 controls an early checkpoint in the thymic development of FoxP3+ regulatory T cells. J Immunol. 2009;182:6736–43. doi: 10.4049/jimmunol.0900498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wildin RS, Ramsdell F, Peake J, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 6.Zheng SG, Gray JD, Ohtsuka K, Yamagiwa S, Horwitz DA. Generation ex vivo of TGF-β-producing regulatory T cells from CD4+CD25– precursors. J Immunol. 2002;169:4183–9. doi: 10.4049/jimmunol.169.8.4183. [DOI] [PubMed] [Google Scholar]
- 7.Isomura I, Palmer S, Grumont RJ, et al. c-Rel is required for the development of thymic Foxp3+ CD4 regulatory T cells. J Exp Med. 2009;206:3001–14. doi: 10.1084/jem.20091411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25– naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–86. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caton AJ, Cozzo C, Larkin J, 3rd, Lerman MA, Boesteanu A, Jordan MS. CD4+ CD25+ regulatory T cell selection. Ann N Y Acad Sci. 2004;1029:101–14. doi: 10.1196/annals.1309.028. [DOI] [PubMed] [Google Scholar]
- 10.Salomon B, Lenschow DJ, Rhee L, Ashourian N, Singh B, Sharpe A, Bluestone JA. B7/CD28 costimulation is essential for the homeostasis of the CD4+CD25+ immunoregulatory T cells that control autoimmune diabetes. Immunity. 2000;12:431–40. doi: 10.1016/s1074-7613(00)80195-8. [DOI] [PubMed] [Google Scholar]
- 11.Tang Q, Henriksen KJ, Boden EK, et al. Cutting edge: CD28 controls peripheral homeostasis of CD4+CD25+ regulatory T cells. J Immunol. 2003;171:3348–52. doi: 10.4049/jimmunol.171.7.3348. [DOI] [PubMed] [Google Scholar]
- 12.Tai X, Cowan M, Feigenbaum L, Singer A. CD28 costimulation of developing thymocytes induces Foxp3 expression and regulatory T cell differentiation independently of interleukin 2. Nat Immunol. 2005;6:152–62. doi: 10.1038/ni1160. [DOI] [PubMed] [Google Scholar]
- 13.D'Alise AM, Auyeung V, Feuerer M, Nishio J, Fontenot J, Benoist C, Mathis D. The defect in T-cell regulation in NOD mice is an effect on the T-cell effectors. Proc Natl Acad Sci U S A. 2008;105:19857–62. doi: 10.1073/pnas.0810713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liston A, Nutsch KM, Farr AG, Lund JM, Rasmussen JP, Koni PA, Rudensky AY. Differentiation of regulatory Foxp3+ T cells in the thymic cortex. Proc Natl Acad Sci U S A. 2008;105:11903–8. doi: 10.1073/pnas.0801506105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–11. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burchill MA, Yang J, Vang KB, et al. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 2008;28:112–21. doi: 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaide O, Martinon F, Micheau O, Bonnet D, Thome M, Tschopp J. Carma1, a CARD-containing binding partner of Bcl10, induces Bcl10 phosphorylation and NF-κB activation. FEBS Lett. 2001;496:121–7. doi: 10.1016/s0014-5793(01)02414-0. [DOI] [PubMed] [Google Scholar]
- 18.Pomerantz JL, Denny EM, Baltimore D. CARD11 mediates factor-specific activation of NF-κB by the T cell receptor complex. EMBO J. 2002;21:5184–94. doi: 10.1093/emboj/cdf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Concepcion D, Seburn KL, Wen G, Frankel WN, Hamilton BA. Mutation rate and predicted phenotypic target sizes in ethylnitrosourea-treated mice. Genetics. 2004;168:953–9. doi: 10.1534/genetics.104.029843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blonska M, Joo D, Zweidler-McKay PA, Zhao Q, Lin X. CARMA1 controls Th2 cell-specific cytokine expression through regulating JunB and GATA3 transcription factors. J Immunol. 2012;188:3160–8. doi: 10.4049/jimmunol.1102943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laird PW, Zijderveld A, Linders K, Rudnicki MA, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi O, Rutschmann S. Dissecting immunity by germline mutagenesis. Immunology. 2012;137:124–30. doi: 10.1111/j.1365-2567.2012.03613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlucci F, Cortes-Hernandez J, Fossati-Jimack L, Bygrave AE, Walport MJ, Vyse TJ, Cook TH, Botto M. Genetic dissection of spontaneous autoimmunity driven by 129-derived chromosome 1 Loci when expressed on C57BL/6 mice. J Immunol. 2007;178:2352–60. doi: 10.4049/jimmunol.178.4.2352. [DOI] [PubMed] [Google Scholar]
- 24.Newton K, Dixit VM. Mice lacking the CARD of CARMA1 exhibit defective B lymphocyte development and impaired proliferation of their B and T lymphocytes. Curr Biol. 2003;13:1247–51. doi: 10.1016/s0960-9822(03)00458-5. [DOI] [PubMed] [Google Scholar]
- 25.Curotto de Lafaille MA, Lafaille JJ. Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity. 2009;30:626–35. doi: 10.1016/j.immuni.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Molinero LL, Miller ML, Evaristo C, Alegre ML. High TCR stimuli prevent induced regulatory T cell differentiation in a NF-κB-dependent manner. J Immunol. 2011;186:4609–17. doi: 10.4049/jimmunol.1002361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner MS, Kane LP, Morel PA. Dominant role of antigen dose in CD4+Foxp3+ regulatory T cell induction and expansion. J Immunol. 2009;183:4895–903. doi: 10.4049/jimmunol.0901459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Che T, You Y, Wang D, Tanner MJ, Dixit VM, Lin X. MALT1/paracaspase is a signaling component downstream of CARMA1 and mediates T cell receptor-induced NF-κB activation. J Biol Chem. 2004;279:15870–6. doi: 10.1074/jbc.M310599200. [DOI] [PubMed] [Google Scholar]
- 29.Wegener E, Oeckinghaus A, Papadopoulou N, et al. Essential role for IκB kinase β in remodeling Carma1-Bcl10-Malt1 complexes upon T cell activation. Mol Cell. 2006;23:13–23. doi: 10.1016/j.molcel.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 30.Barnes MJ, Krebs P, Harris N, et al. Commitment to the regulatory T cell lineage requires CARMA1 in the thymus but not in the periphery. PLoS Biol. 2009;7:e51. doi: 10.1371/journal.pbio.1000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt-Supprian M, Tian J, Grant EP, et al. Differential dependence of CD4+CD25+ regulatory and natural killer-like T cells on signals leading to NF-κB activation. Proc Natl Acad Sci U S A. 2004;101:4566–71. doi: 10.1073/pnas.0400885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee AJ, Wu X, Cheng H, Zhou X, Cheng X, Sun SC. CARMA1 regulation of regulatory T cell development involves modulation of interleukin-2 receptor signaling. J Biol Chem. 2010;285:15696–703. doi: 10.1074/jbc.M109.095190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Guo Y, Huang WJ, et al. Card10 is a novel caspase recruitment domain/membrane-associated guanylate kinase family member that interacts with BCL10 and activates NF-κB. J Biol Chem. 2001;276:21405–9. doi: 10.1074/jbc.M102488200. [DOI] [PubMed] [Google Scholar]
- 34.Bertin J, Wang L, Guo Y, et al. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-κB. J Biol Chem. 2001;276:11877–82. doi: 10.1074/jbc.M010512200. [DOI] [PubMed] [Google Scholar]
- 35.McAllister-Lucas LM, Inohara N, Lucas PC, et al. Bimp1, a MAGUK family member linking protein kinase C activation to Bcl10-mediated NF-κB induction. J Biol Chem. 2001;276:30589–97. doi: 10.1074/jbc.M103824200. [DOI] [PubMed] [Google Scholar]
- 36.Gaide O, Favier B, Legler DF, et al. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-κB activation. Nat Immunol. 2002;3:836–43. doi: 10.1038/ni830. [DOI] [PubMed] [Google Scholar]
- 37.Egawa T, Albrecht B, Favier B, Sunshine MJ, Mirchandani K, O'Brien W, Thome M, Littman DR. Requirement for CARMA1 in antigen receptor-induced NF-κB activation and lymphocyte proliferation. Curr Biol. 2003;13:1252–8. doi: 10.1016/s0960-9822(03)00491-3. [DOI] [PubMed] [Google Scholar]
- 38.Hara H, Wada T, Bakal C, et al. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity. 2003;18:763–75. doi: 10.1016/s1074-7613(03)00148-1. [DOI] [PubMed] [Google Scholar]
- 39.Medoff BD, Sandall BP, Landry A, Nagahama K, Mizoguchi A, Luster AD, Xavier RJ. Differential requirement for CARMA1 in agonist-selected T-cell development. Eur J Immunol. 2009;39:78–84. doi: 10.1002/eji.200838734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jun JE, Wilson LE, Vinuesa CG, et al. Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity. 2003;18:751–62. doi: 10.1016/s1074-7613(03)00141-9. [DOI] [PubMed] [Google Scholar]
- 41.Feuerer M, Hill JA, Mathis D, Benoist C. Foxp3+ regulatory T cells: differentiation, specification, subphenotypes. Nat Immunol. 2009;10:689–95. doi: 10.1038/ni.1760. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Song R, Gao Y, et al. Protein kinase C-δ negatively regulates T cell receptor-induced NF-κB activation by inhibiting the assembly of CARMA1 signalosome. J Biol Chem. 2012;287:20081–7. doi: 10.1074/jbc.M111.335463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thome M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol. 2004;4:348–59. doi: 10.1038/nri1352. [DOI] [PubMed] [Google Scholar]
- 44.Cannons JL, Yu LJ, Hill B, et al. SAP regulates TH2 differentiation and PKC-θ-mediated activation of NF-κB1. Immunity. 2004;21:693–706. doi: 10.1016/j.immuni.2004.09.012. [DOI] [PubMed] [Google Scholar]


