Abstract
Studies on new arthropod models such as the beetle Tribolium castaneum are shifting our knowledge of embryonic patterning and morphogenesis beyond the Drosophila paradigm. In contrast to Drosophila, Tribolium embryos exhibit the short-germ type of development and become enveloped by extensive extra-embryonic membranes, the amnion and serosa. The genetic basis of these processes has been the focus of active research. Here, we complement genetic approaches with live fluorescence imaging of Tribolium embryos to make the link between gene function and morphogenetic cell behaviors during blastoderm formation and differentiation, germband condensation and elongation, and extra-embryonic development. We first show that transient labeling methods result in strong, homogeneous and persistent expression of fluorescent markers in Tribolium embryos, labeling the chromatin, membrane, cytoskeleton or combinations thereof. We then use co-injection of fluorescent markers with dsRNA for live imaging of embryos with disrupted caudal gene function caused by RNA interference. Using these approaches, we describe and compare cell and tissue dynamics in Tribolium embryos with wild-type and altered fate maps. We find that Tribolium germband condensation is effected by cell contraction and intercalation, with the latter being dependent on the anterior-posterior patterning system. We propose that germband condensation drives initiation of amnion folding, whereas expansion of the amniotic fold and closure of the amniotic cavity are likely driven by contraction of an actomyosin cable at the boundary between the amnion and serosa. Our methodology provides a comprehensive framework for testing quantitative models of patterning, growth and morphogenetic mechanisms in Tribolium and other arthropod species.
Keywords: Tribolium castaneum, Arthropods, Fluorescence live imaging, Functional genetics, Embryo morphogenesis, Extra-embryonic development
INTRODUCTION
Fluorescence light microscopy has revolutionized live imaging of developing embryos (Khairy and Keller, 2011; Mavrakis et al., 2010; Megason and Fraser, 2007). Most imaging studies employ transgenic lines expressing fluorescent proteins generated predominantly for the major model organisms. Beyond Drosophila melanogaster, over recent years transgenic systems have been developed for new arthropod species, including other Diptera and Lepidoptera (Fraser, 2012; Holtzman et al., 2010), the beetle Tribolium castaneum (Berghammer et al., 1999; Pavlopoulos et al., 2004), the cricket Gryllus bimaculatus (Nakamura et al., 2010) and the crustacean Parhyale hawaiensis (Pavlopoulos and Averof, 2005). For most of these species, transgenesis is a laborious endeavor and appropriate lines for live imaging are still limited or absent. To address this limitation, we have established a versatile and rapid method for transient fluorescence labeling in Tribolium and other arthropod embryos.
Transient labeling methods involve in vivo delivery of vital dyes, DNA constructs, synthesized mRNAs or purified proteins that fluorescently label treated embryos. Transient labeling was first applied in established models such as Drosophila, zebrafish or mouse (Köster and Fraser, 2004; Minden et al., 1989; Zernicka-Goetz et al., 1997), and is still the method of choice in other model and non-model species, including crustaceans, annelids, echinoderms, ascidians and vertebrates (Corbo et al., 1997; Damle et al., 2006; Köster and Fraser, 2004; Price et al., 2010; Teddy and Kulesa, 2004; Zernicka-Goetz et al., 1996; Zhang and Weisblat, 2005). Labeling agents can also be delivered together with DNA/RNA constructs for gene overexpression or knockdown (Link and Megason, 2008).
Here, we demonstrate transient labeling methods for strong, homogeneous and persistent expression of fluorescent markers in embryos of the red flour beetle Tribolium castaneum. Tribolium is a well-established arthropod model representing an ideal system with which to study the diversity and evolution of developmental mechanisms (Brown et al., 2009; Schröder et al., 2008). Tribolium is supported by an extensive repertoire of genetic and genomic resources (Beeman et al., 1989; Berghammer et al., 1999; Posnien et al., 2009; Richards et al., 2008; Trauner et al., 2009), but is still lagging behind in imaging resources. Complementing developmental genetic studies with 3D time-lapse imaging of intact developing embryos is necessary to make the link between identified gene networks and the dynamic cellular contexts in which these networks operate. The potential of this approach is evident in two reports that demonstrate the involvement of vertebrate-like molecular oscillators in Tribolium segmentation (El-Sherif et al., 2012; Sarrazin et al., 2012). Both studies used the single Tribolium transgenic line specifically designed for live imaging that ubiquitously expresses a nuclear green fluorescent protein (nGFP).
In this study, we have extended imaging resources in Tribolium by testing several fusion constructs of fluorescent proteins injected in the form of mRNA to label different cell components. Furthermore, we demonstrate co-labeling with two fluorescent markers, the use of photoconvertible fluorescent proteins for cell marking, and co-injection of fluorescent markers together with dsRNA for live imaging of embryos with disrupted gene function by RNAi. Using these approaches, we have compared various aspects of blastoderm formation, extra-embryonic development, germband condensation and elongation between wild-type Tribolium embryos and embryos in which caudal (Tc-cad) gene function has been knocked down.
MATERIALS AND METHODS
Templates for synthesis of capped mRNAs
Plasmid pT7-DsRed-NLS is a modification of plasmid pSP6-DsRed-NLS (Price et al., 2010), in which the SP6 promoter is replaced with the T7 promoter for higher-yield transcription, and the coding sequence of interest is flanked by the β-globin 5′ and 3′ UTRs. The PCR-amplified coding sequences for D. melanogaster histone H2B and the monomeric Ruby Red Fluorescent Protein (Müller-Taubenberger et al., 2006) were cloned into pT7-DsRed-NLS, replacing DsRed-NLS with H2B-RFP to generate plasmid pT7-H2B-RFP (supplementary material Table S1). Similarly, the LifeAct (provided by Jianying Yang, Max Planck Institute of Immunobiology and Epigenetics, Freiburg, Germany) and EGFP sequences, or the Actin Binding Peptide-tandem Eos Fluorescent Protein fusion (provided by Christian Specht, Ecole Normale Superieure, Paris, France) were cloned into the same vector, generating plasmids pT7-LifeAct-EGFP and pT7-ABP-tdEosFP, respectively. The pCS2-GAP43-YFP construct was kindly provided by Mette Handberg-Thorsager (European Molecular Biology Laboratory, Heidelberg, Germany). For mRNA synthesis, pT7-H2B-RFP, pT7-LifeAct-EGFP and pT7-ABP-tdEosFP were linearized with EcoRI, and pCS2-GAP43-YFP with NsiI. Capped mRNAs were synthesized using Ambion’s T7 or SP6 mMESSAGE mMACHINE kits for pT7-based and pCS2-based constructs, respectively, as previously described (Pavlopoulos et al., 2004).
Embryo microinjection and live imaging
Beetles were reared according to The Beetle Book (http://wwwuser.gwdg.de/~gbucher1/tribolium-castaneum-beetle-book1.pdf) at 32°C. Eggs were dechorionated in 0.5% bleach and lined up on a microscope slide for upright microscopy or on a glass-bottomed Petri dish (MatTek) that had a window cut in the side of the dish to allow injection for inverted microscopy. Gaps were left between neighboring eggs for efficient gas exchange. Eggs were covered with Voltalef 10S Halocarbon oil. The needle was inserted into the egg at the anterior pole and the tip of the needle was moved into the center of the egg for injection. Capped mRNAs were injected at 0.5-3 μg/μl in injection buffer (5 mM KCl, 10 mM NaH2PO4). To monitor the kinetics of fluorescence labeling, we injected H2B-RFP and/or GAP43-YFP mRNAs into eggs 4-6 hours after egg lay (AEL) at 32°C, when nuclei migrate towards the egg surface and the cellularized blastoderm begins to form. In all other applications, injections were performed in eggs 2-3 hours AEL at 32°C to achieve maximum diffusion and homogeneous labeling. Following injection at room temperature, eggs were transferred back to 32°C and incubated in a dark and humid environment. Under these conditions, Tribolium embryos developed normally, but hatching rates were decreased, probably due to egg immersion in halocarbon oil. Microinjections caused a developmental delay of 30-60 minutes, as judged by the time of initiation of the posterior amniotic fold between injected and non-injected embryos. Live imaging was carried out on an inverted Leica SP5 confocal at 32°C. Image stacks of about 40 focal planes were taken with a 20×/0.7NA multi-immersion objective or a 40×/1.3NA oil-immersion objective at 2.5- or 5-minute intervals. Processing of confocal stacks was carried out with Fiji (Schindelin et al., 2012) and cell tracking with MTrackJ (Meijering et al., 2012).
RESULTS
Labeling of Tribolium embryos with fluorescent nuclear, membrane and actin markers
We first injected Tribolium eggs with mRNA encoding the H2B-RFP nuclear fluorescent marker at 3 μg/μl. The strength and homogeneity of the fluorescent signal increased over time (supplementary material Fig. S1; Table S2). Three hours post injection, strong, uniform and ubiquitous expression was visible in 61% of eggs (Fig. 1A; supplementary material Fig. S1; Table S2). We also injected H2B-RFP mRNA at 1 μg/μl, and found that fluorescence was weaker than at higher concentration (supplementary material Fig. S1). This effect was also observed with all other constructs tested. Notably, H2B-RFP labeled chromatin throughout the cell cycle, allowing us to follow mitotic divisions unambiguously (Fig. 1B-B′′).
Fig. 1.
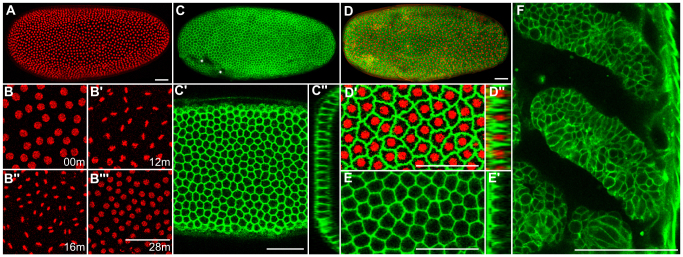
Fluorescence labeling of Tribolium embryos with nuclear, membrane and actin markers. (A) Average intensity projection of a uniform blastoderm labeled with H2B-RFP. (B-B′′) Time series of H2B-RFP-labeled chromatin (B) before, (B′,B′) during and (B′′) after the synchronous 12th mitotic division. Single optical slices are shown timed against B. (C-C′) Uniform blastoderm labeled with GAP43-YFP. (C) Average intensity projection of the entire embryo, (C′) single optical slice and (C′) transverse section that shows labeling of the entire membrane surface. Asterisks in C denote blemishes on the vitelline membrane that locally impair fluorescent signal. (D-D′) Embryo at the uniform blastoderm stage with H2B-RFP-labeled nuclei and GAP43-YFP-labeled membranes. (D) Average intensity projection, (D′) optical slice and (D′) transverse section. (E,E′) Uniform blastoderm labeled with LA-GFP. (E) Single optical slice and (E′) transverse section showing cortical actin fluorescence around the apical surface. (F) Single optical slice through two developing thoracic limbs and the pleuropod of an embryo 48 hours after injection with GAP43-YFP mRNA. Anterior is towards the left in most panels, except in cross-sections C′,D′,E′, where apical is towards the left, and in F, where the ventral midline is towards the left and anterior is towards the top. Scale bars: 50 μm.
To image cell outlines, we used the GAP43-YFP fusion protein that becomes anchored to the plasma membrane (Godinho et al., 2005; Mavrakis et al., 2009). GAP43-YFP specifically labeled cell membranes, marking both the apical and basolateral domains of Tribolium blastoderm cells (Fig. 1C-C′; supplementary material Table S2). By injecting a mixture of the mRNAs encoding H2B-RFP (at 2 μg/μl) and GAP43-YFP (at 1 μg/μl), we were able to label both the nuclei and the cell borders in the same embryo (Fig. 1D-D′).
We also imaged filamentous actin (F-actin) cytoskeleton by fusing EGFP to the LifeAct motif (LA-GFP; Riedl et al., 2008). After injecting Tribolium eggs with LA-GFP mRNA at 3 μg/μl, we observed a weak diffuse fluorescence 3 hours after injection, followed by uniform expression 6 hours after injection in 70% of eggs (supplementary material Table S2). Upon blastoderm formation, LA-GFP specifically labeled the cortical actin layer producing the polygonal pattern of blastoderm cells also observed with GAP43-YFP (Fig. 1E,E′).
Tribolium embryogenesis takes about 3 days at 32°C. Most treated eggs exhibited a strong, uniform, ubiquitous and persistent fluorescence even 48 hours after injection (Fig. 1F). We concluded that transient methods are well suited to fluorescently label different cell compartments in Tribolium embryos and image them live for most of embryogenesis.
Marking clones of cells with a photoconvertible fluorescent protein in Tribolium
We next tested a photoconvertible fluorescent protein to differentially label specific populations of cells. We employed a construct that combines an actin-binding peptide with the tandem Eos fluorescent protein (ABP-tdEosFP) and can be converted from green to red upon 405 nm illumination (Izeddin et al., 2011). One hour after injection, the cortical actin was labeled near the injection site. Over the next hours, fluorescence spread to all blastoderm cells, but expression levels varied within each embryo (supplementary material Table S2). Despite this heterogeneity, we generated clones of cells differentially labeled to their neighbors by photoconverting ABP-tdEosFP in the blastoderm (Fig. 2A-A′). ABP-tdEosFP also allowed us to examine the perdurance of injected mRNA. We photoconverted ABP-tdEosFP in a developing limb-bud of an embryo 26 hours after injection (Fig. 2B-B′). The same limb had produced new unconverted ABP-tdEosFP protein 22 hours later (Fig. 2C), suggesting that the mRNA is stable enough to drive synthesis of the fluorescent protein for more than 26 hours after injection. The photoconverted ABP-tdEosFP was also stably labeling the growing limb (Fig. 2C′,C′).
Fig. 2.
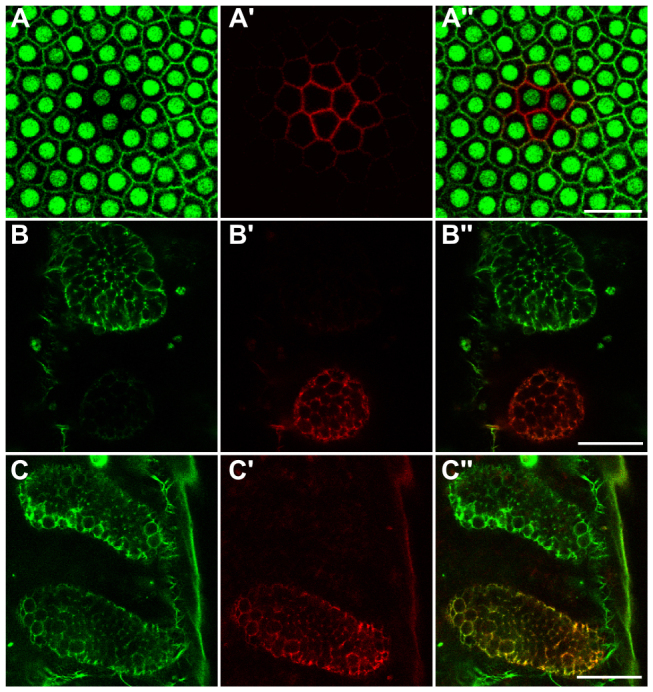
Fluorescence labeling of Tribolium embryos with photoconvertible ABP-tdEosFP. (A-A′) Optical sections through the blastoderm of a transgenic embryo ubiquitously expressing nuclear GFP (Sarrazin et al., 2012), which was injected with ABP-tdEosFP mRNA and photoconverted in the central region. (A) nGFP and unconverted ABP-tdEosFP fluorescence detected at similar levels in the green channel, (A′) photoconverted ABP-tdEosFP fluorescence detected in the red channel and (A′) overlay of the two channels. (B-B′) Optical sections through developing limb-buds of an embryo 26 hours after injection with ABP-tdEosFP mRNA. Most of the protein in the bottom limb-bud has been photoconverted. (B) Green channel showing unconverted ABP-tdEosFP, (B′) red channel showing converted ABP-tdEosFP, and (B′) overlay of the two channels. (C-C′) Optical sections through the same limbs shown in B-B′ 22 hours later. Both green unconverted and red converted ABP-tdEosFP are detected in the bottom photoconverted limb. Anterior is towards the top in all panels, and ventral midline is towards the left in B-C′. Scale bars: 25 μm.
Timing of Tribolium blastoderm cellularization
The preblastoderm stages of Tribolium development are similar to most insects (Handel et al., 2000). After synchronous rounds of division, nuclei migrate to the egg cortex, where further synchronous nuclear divisions occur. Membranes then segregate nuclei into separate cells forming a uniform blastoderm. In agreement with previous descriptions (Handel et al., 2000), after the 12th nuclear division, GAP43-YFP-labeled membranes extended to the basal end of cortical nuclei, but their leading edges appeared very loosely organized (Fig. 3A′,A′). Cells remained open to the yolk for almost the entire interphase of the 13th cycle, which lasted 120-140 minutes at 32°C. The leading edges of membranes started constricting about halfway through the 13th interphase (Fig. 3B′,B′). Over the following hour, the basal membranes continued constricting (Fig. 3C′,C′) and eventually closed cells off (Fig. 3D′,D′). Basal closure was accompanied by a progressive refinement and stretching of the subapical contours of blastoderm cells (Fig. 3A-D). All blastoderm cells completed cellularization in near synchrony irrespective of their fate. The first divisions of the 13th cell cycle occurred 5-30 minutes after completion of cell closure.
Fig. 3.
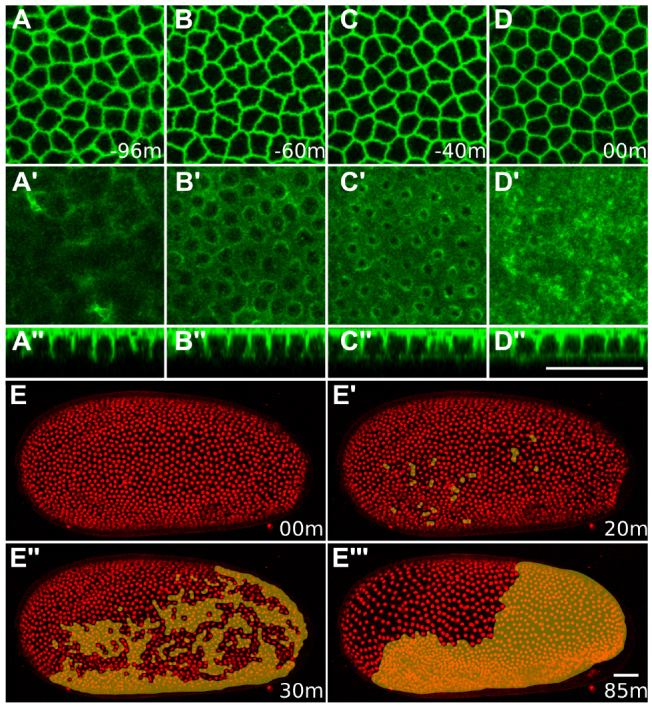
Tribolium blastoderm cellularization and differentiation. (A-D′) Time series of blastoderm cellularization during 13th interphase of a GAP43-YFP-labeled embryo timed against basal cell closure shown in D-D′. (A-D) Subapical optical sections showing progressive refinement of the polygonal packing geometry during blastoderm cellularization. (A′-D′) Basal optical sections showing progressive membrane constriction and eventual basal cell closure. (A′-D′) Cross-sections showing the lateral extension of constricting basal membranes during cellularization. (E-E′′) Time series of the 13th round of cell divisions during blastoderm differentiation. All panels show average intensity projections of an H2B-RFP-labeled embryo (lateral views, anterior towards the left and dorsal towards the top) at different stages timed against E. Divided nuclei in the embryonic rudiment are indicated with an orange mask. (E) Labeled nuclei have uniform size and distribution before the onset of 13th divisions. (E′,E′) Asynchronous nuclear divisions are detected throughout the embryonic rudiment, but not in the serosa primordium. (E′′) Divided nuclei in the embryonic rudiment become smaller and more tightly packed compared with non-dividing serosa nuclei that increase their spacing. Scale bars: 50 μm.
Formation of the Tribolium differentiated blastoderm
Cells have a uniform shape and distribution in the Tribolium uniform blastoderm (Fig. 1A,C,D). The majority of blastoderm cells condense ventrally to form the embryonic rudiment, while the remaining anterior blastoderm cells spread around the entire egg surface to form the extra-embryonic serosa membrane. The embryonic rudiment gives rise to the embryo proper (germband) and the amnion that extends from the margin of the embryo and covers its ventral side (Roth, 2004).
As previously shown (Handel et al., 2005; Handel et al., 2000), asynchronous 13th divisions occurred exclusively in the presumptive embryonic rudiment, but not in the serosa (Fig. 3E-E′′; supplementary material Movie 1). The time taken from first to last division varied between 80 and 110 minutes across embryos. From this stage onwards, known as the differentiated blastoderm (Anderson, 1972a), the distinction between embryonic and extra-embryonic cells became progressively more pronounced.
Cell and tissue dynamics in wild-type and Tc-cadRNAi Tribolium embryos
During germband condensation and gastrulation, the prospective amnion and serosa undergo extensive rearrangements to envelope the germband ventrally. We first followed cell behaviors and tissue movements in wild-type GAP43-YFP-labeled Tribolium embryos from 8 to 21 hours AEL at 32°C. We then compared wild-type with genetically perturbed embryos by RNAi (Brown et al., 1999; Posnien et al., 2009) to make the link between gene function and morphogenesis. We combined transient labeling with embryonic RNAi against the Tribolium caudal gene (Tc-cad) by co-injection of GAP43-YFP mRNA (1 μg/μl) with Tc-cad dsRNA (0.8 μg/μl) that targets both Tc-cad isoforms (A and B). Tc-cad encodes a homeodomain transcription factor with a crucial role in posterior embryonic patterning (Copf et al., 2004; Schoppmeier et al., 2009; Schulz et al., 1998). Knockdown of maternal and zygotic Tc-cad expression by parental RNAi results in severely truncated embryos retaining only their most anterior pregnathal structures (Copf et al., 2004; Schoppmeier et al., 2009). In our experiments, embryonic Tc-cad RNAi caused a moderate change in embryo fate map and resulted in abnormally short and wide germbands enveloped by amnion and serosa (see below).
By tracking cell and tissue dynamics in wild-type Tribolium embryos, we defined six distinct stages spanning the uniform blastoderm through to germband elongation stages (Table 1; relation to other Tribolium staging systems shown in supplementary material Table S3). At each stage, we also assessed the effects of Tc-cad knockdown. Our interpretations were based on screening more than 20 embryos imaged from different views, and on more detailed pairwise comparison between three wild-type and three Tc-cadRNAi embryos described below.
Table 1.
Overview of staging system for Tribolium embryogenesis
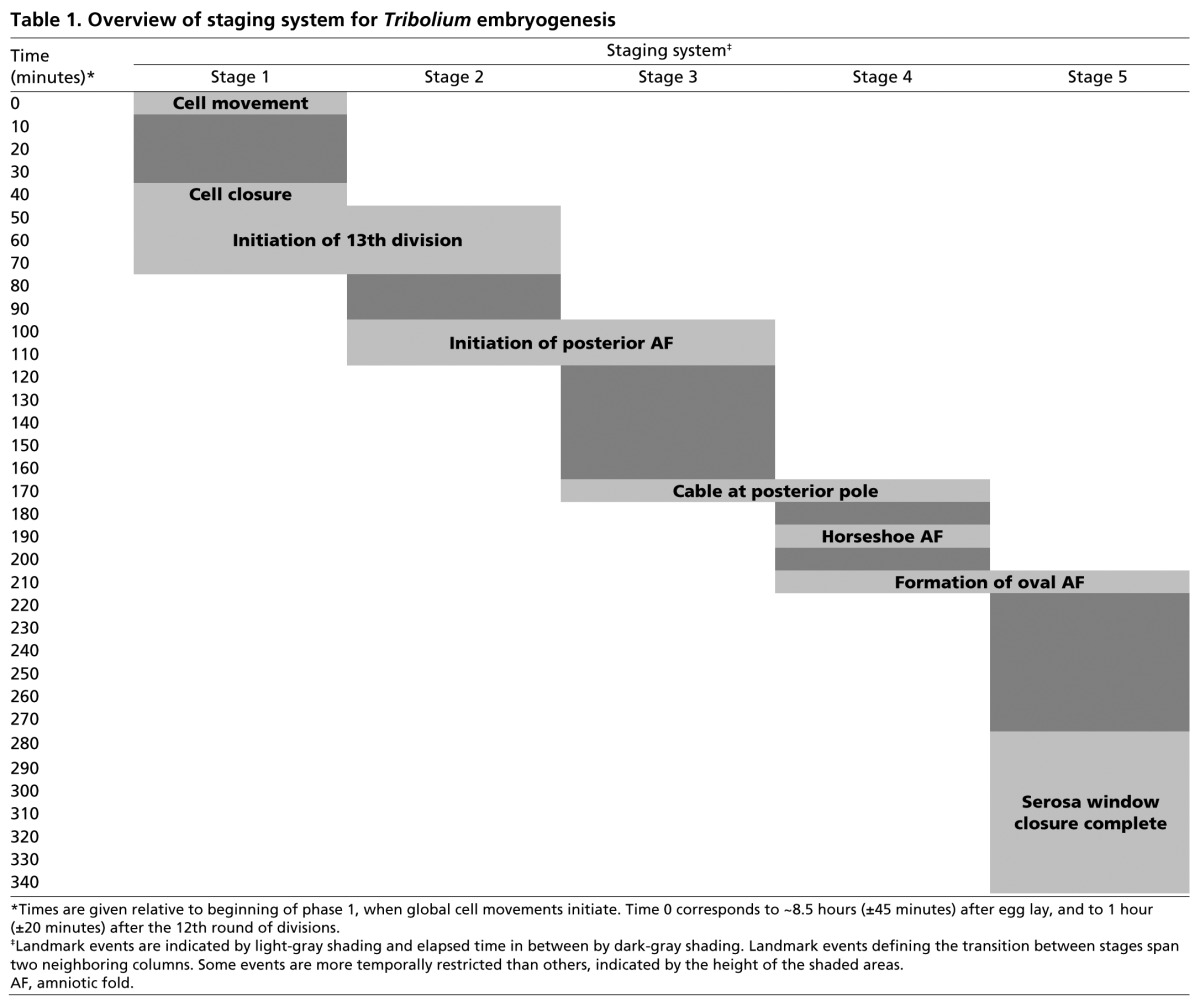
Stage 1: cellularization of uniform blastoderm
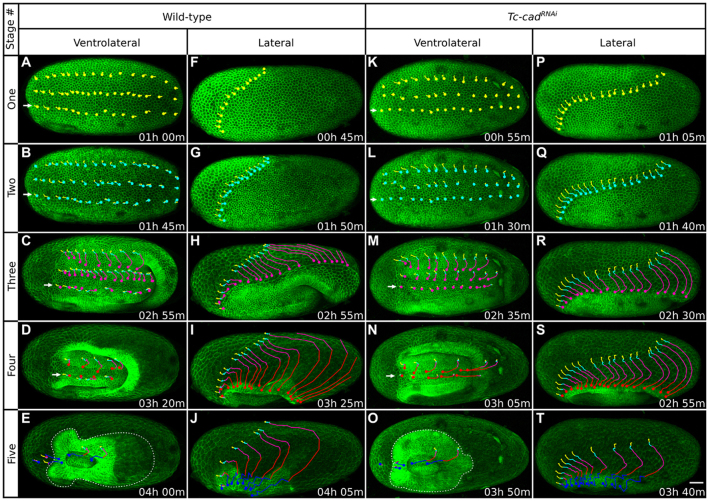
This stage began with the onset of global cell movements in the uniform blastoderm and encompassed blastoderm cellularization during 13th interphase. Wild-type and Tc-cadRNAi blastoderms exhibited very similar numbers of cuboidal cells (Fig. 4A,F,K,P; Fig. 6A,E; supplementary material Fig. S2A-A′,E-E′; Fig. S3A-A′′,C-C′′). We counted on average 268 cells (σ=32 cells) per 30,000 μm2 in three wild-type embryos and 257 cells (σ=32 cells) in three Tc-cadRNAi embryos. In wild-type embryos, we observed minor flows of cells from the two poles towards the middle of the egg, as well as from the dorsal and lateral sides towards the ventral midline (Fig. 4A,F). After Tc-cad RNAi, the pattern of early cell movements in the anterior half was similar to wild-type, but cells in the posterior half moved primarily ventrally and only very little anteriorly (Fig. 4K,P).
Fig. 4.
Patterns of cell movement during Tribolium embryogenesis in wild-type and Tc-cadRNAi embryos. Each column shows a different GAP43-YFP-labeled embryo at representative time-points spanning, from top to bottom, stages 1 to 5. (A-E) Ventrolateral views of a wild-type embryo. (F-J) Lateral views of a wild-type embryo. (K-O) Ventrolateral views of a Tc-cadRNAi embryo. (P-T) Lateral views of a Tc-cadRNAi embryo. In each embryo, cells were tracked from beginning of stage 1, and all panels are timed against this starting point. Cell tracks are color-coded according to stages: stage 1 in yellow, stage 2 in cyan, stage 3 in magenta, stage 4 in red and stage 5 in blue. The corresponding position of cells is indicated with dots, and tracks are displayed up to that time-point. Ventrolateral views show rows of tracked cells across the anterior-posterior axis; at stages 1 to 4, the bottom row (marked with arrows) is very close to the ventral midline; at stage 5, the dotted line outlines the germbands. Lateral views show tracks of leading serosa cells. Please refer to the text for description of cell and tissue movements observed. The fluorescent signal is locally impaired in some embryo regions by autofluorescent and dark blemishes on the vitelline membrane. Average intensity projections, anterior towards the left. Scale bar: 50 μm.
Fig. 6.
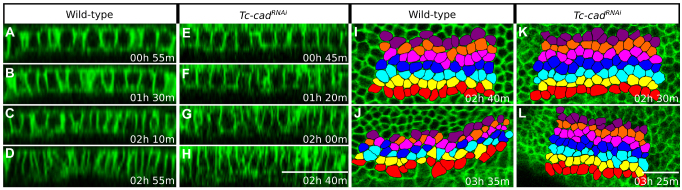
Cell behaviors during Tribolium germband condensation in wild-type and Tc-cadRNAi embryos. (A-H) Progressive contraction of embryonic cells observed both in (A-D) wild-type and (E-H) Tc-cadRNAi embryo at four time-points spanning, from top to bottom, stages 1 to 3. Panels show transverse sections of GAP43-YFP-labeled wild-type and Tc-cadRNAi embryos, also shown in Fig. 4A-E and Fig. 4K-O, respectively. Panels are timed against the onset of stage 1 and apical is towards the top. (I-L) Cell intercalation in wild-type embryo (I,J) and absence thereof in Tc-cadRNAi embryo (K,L). Panels show ventrolateral views of GAP43-YFP-labeled embryos at two time-points during stages 3 (top) and 4 (bottom). All panels are arranged with anterior towards the left and show average intensity projections of 6 μm substacks covering the anterior left side of the condensing germbands. Abutting rows of ectodermal cells, highlighted with different colors, were tracked after their 13th division. (I,J) Cells in each row become separated by ventral or dorsal neighboring cells in the wild type. This cell intercalation narrows the highlighted cluster of cells in the dorsal-ventral axis and lengthens it in the anterior-posterior axis. Many cells also appear to elongate along the anterior-posterior axis. (K,L) Rows of cells remain contiguous as no cell intercalation takes place after Tc-cad RNAi. Under both wild-type and RNAi conditions, the highlighted clusters of cells shrink between the two time-points as a result of cell contraction to about 75% and 73% of their original size, respectively. Scale bars: 50 μm.
Initial cell movements occurred in the absence of cell divisions and were accompanied in some embryos by retraction of the posterior pole from the vitelline membrane (Fig. 7A-B; supplementary material Fig. S2A,A′). Blastoderm cells at the posterior pole then invaginated, forming the primitive pit that is considered the posterior midgut rudiment (Handel et al., 2000). The extent and timing of posterior pole retraction from the vitelline membrane varied significantly, as observed in time-lapses of 10 GAP43-YFP-labeled and 10 nGFP transgenic embryos (compare supplementary material Fig. S2B,B′ and Fig. S3B,B′).
Fig. 7.
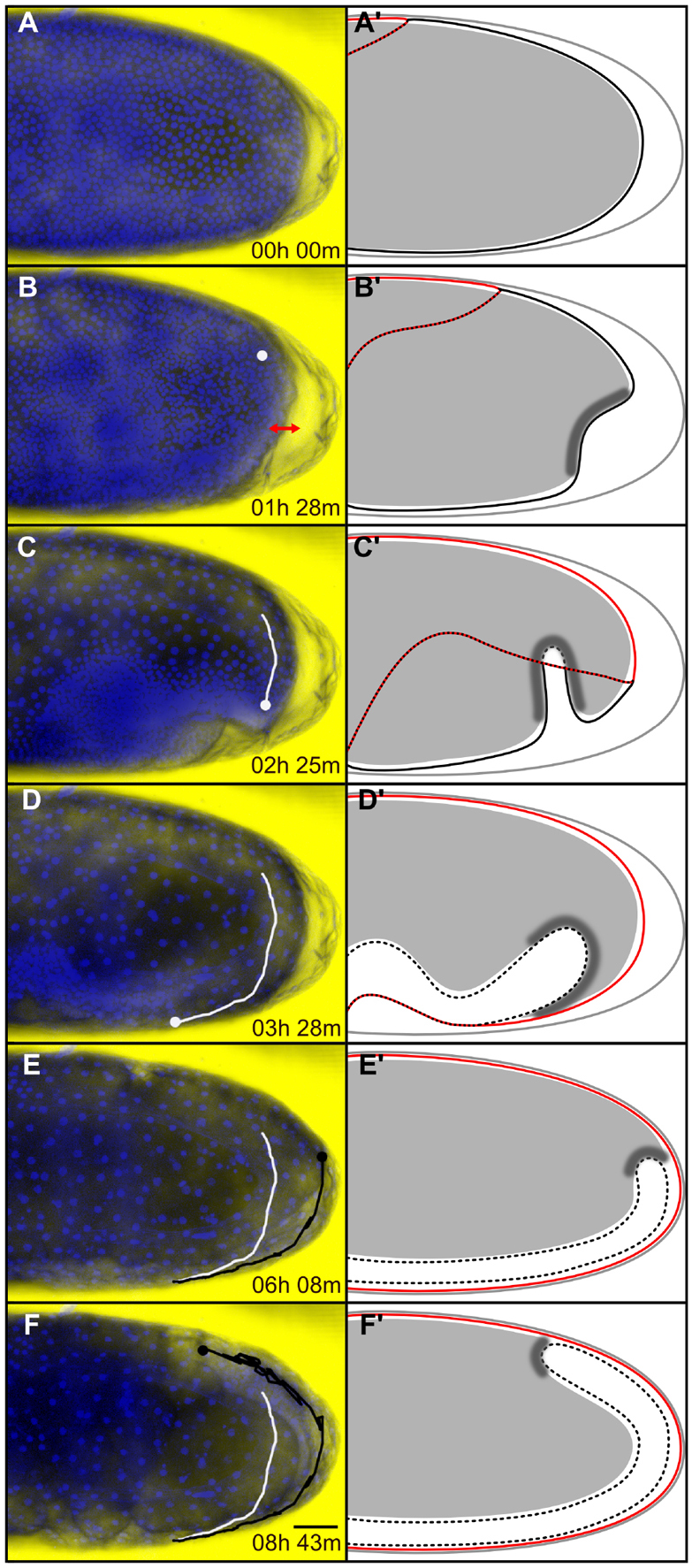
Posterior yolksac dynamics relative to Tribolium germband condensation and elongation. Time series of posterior yolksac deformation in (A-F) an H2B-RFP-labeled embryo and (A′-F′) corresponding schematic representations. (A-F) Average intensity projections of the posterior half of the same embryo timed against the first time-point shown in A. At each time-point, the DIC image of the embryo was overlaid with the corresponding H2B-RFP signal shown in blue. In B-F, the dot indicates the position of the leading edge of the posterior yolk-fold. The white track marks the early ventral extension of the yolk-fold and the black track marks its dorsal retraction up to the corresponding time-point. In A′-F′, the vitelline membrane is shown with a gray line, the serosa outline with a red line, the amnion/serosa boundary with a dotted red/black line, the condensing and internalized elongating embryonic rudiment with solid and broken black lines, respectively, the yolksac with filled gray, and the presumptive posterior yolksac-germband attachment by darker shading. (A,A′) Uniform blastoderm stage. (B,B′) Differentiated blastoderm stage showing blastoderm and yolk depression at the posterior pole (double-headed red arrow). (C,C′) The posterior dorsal wedge of yolk extends ventrally during posterior amniotic fold formation. There is concurrent amnion involution during yolk-fold extension. (D,D′) The yolk-fold completes its ventral extension when posterior amnion involution stops and serosa window forms. (E,E′) Yolk-fold retraction is tightly coupled to posterior germband extension. The yolk levels out posteriorly when the translucent germband reaches the posterior pole. (F,F′) Yolksac displacement continues dorsally as the translucent germband extends around the posterior pole. Lateral views, anterior towards the left and dorsal towards the top. Scale bar: 50 μm.
Stage 2: blastoderm differentiation
This stage began with the onset of the 13th divisions of embryonic cells and their morphological differentiation from serosa cells (Fig. 4G). Embryonic cells started to contract, becoming columnar, whereas serosa cells started to flatten, becoming squamous (Fig. 6B; supplementary material Fig. S2B-B′; Fig. S3B-B′′). Cell differentiation was also evident in Tc-cadRNAi embryos (Fig. 4Q; Fig. 6F; supplementary material Fig. S2F-F′; Fig. S3D-D′′), but the extent of embryonic cell divisions, and the pattern of cell shape change, revealed an altered fate map. The domain of non-dividing and flattening serosa cells expanded considerably along the dorsal side from 55% egg length (EL from posterior pole) in wild-type (σ=2.2 percentage points; four embryos measured) to 22% EL after Tc-cad RNAi (σ=1.7 percentage points; four embryos measured). Thus, after Tc-cad knockdown, the serosa primordium occupied most of the dorsal half of the blastoderm, while the embryonic primordium was restricted to the ventral half of the blastoderm (Fig. 4G,Q).
During this stage, wild-type blastoderm cells moved in concert posteriorly for one-to-two cell diameters (Fig. 4B,G). Dorsal and lateral cells also continued their movement towards the ventral midline. The invaginated primitive pit reached its maximal displacement from the vitelline membrane (Fig. 7B; supplementary material Fig. S2B′). Tracked cells in Tc-cadRNAi embryos continued slight movements along the same trajectories initiated during stage 1 (Fig. 4L,Q). We conclude that the relative position and size of the expansive serosa rudiment and the contractile embryonic rudiment influence the motion patterns detected during stages 1 and 2.
Stage 3: initiation of germband condensation and amnion folding
During this stage, cells in wild-type embryonic rudiments advanced in almost straight trajectories towards the ventral midline (Fig. 4C). By tracking abutting rows of prospective ectodermal cells, we found that this conspicuous ventral movement was effected, in addition to cell contraction (Fig. 6C,D; supplementary material Fig. S2C-D′), by intercalation of cells between their dorsal and ventral neighbors (Fig. 6I,J). In contrast to wild-type embryos, we found no evidence for cell intercalation after Tc-cad RNAi (Fig. 6K,L) and lateral ectodermal cells adopted anterior-ventral trajectories (Fig. 4M). The residual germband condensation was effected entirely by cell contraction (Fig. 6G,H; supplementary material Fig. S2G-H′).
Condensation of the embryonic primordium coincided with the posterior and ventral movement of prospective amnion and serosa cells, and formation of the posterior amniotic fold (Fig. 4C,H). In wild-type embryos, this epithelial rearrangement was accompanied by the progressive submersion of the posterior germband into the yolk, coupled with the flow of superficial prospective amnion cells inwards to cover the inner side of the growing fold (Fig. 4C,H; Fig. 7C; supplementary material Movies 2, 3, 7). At the end of this stage, the amnion/serosa boundary passed around the posterior egg pole and started approaching the rim of the posterior amniotic fold (Fig. 4H). In Tc-cadRNAi embryos, the amnion/serosa boundary moved around the posterior pole shortly after the serosa started to expand (Fig. 4R; supplementary material Movie 5). Amnion involution was also limited, resulting in a much shallower posterior amniotic fold compared with wild type (Fig. 4M,R; supplementary material Movies 4, 5).
Stage 4: progression of germband condensation and amnion folding
During this stage, the posterior amniotic fold extended laterally and anteriorly, progressively covering the germband in a posterior-to-anterior direction (Fig. 4D,I; supplementary material Movies 2, 3). At the same time, the amnion raised slightly around the lateral sides of the condensing head lobes, initiating what is known as the anterior amniotic fold. The advancing lateral edges of the posterior fold were continuous on either side of the germband with the lateral edges of the anterior fold forming a horseshoe-shaped amnion cover (Fig. 4D,I). Soon thereafter, the amnion folded over the head lobes anteriorly, resulting in an oval amniotic fold surrounding the open serosa window (Fig. 4E,J).
Throughout stages 3 to 5, the amnion/serosa boundary was demarcated by a conspicuous membrane thickening in GAP43-YFP-labeled embryos (Fig. 4H,I; supplementary material Movies 2, 3). This thickening also exhibited a higher concentration of F-actin in LA-GFP-labeled embryos (data not shown), and was reminiscent of intercellular actomyosin cables producing contractile forces in other developmental systems (Behrndt et al., 2012; Kiehart et al., 2000; Martin and Lewis, 1992; Panfilio and Roth, 2010). During stage 3, this cable first expanded as it moved around the posterior pole (Fig. 4H), and then started shortening as it converged towards the rim of the posterior amniotic fold. During stage 4, the progressive constriction of the cable coincided precisely with lateral amnion folding and formation of the horseshoe amniotic fold (Fig. 4D,I), and then with amnion folding over the anterior margin of the head and formation of the serosa window (Fig. 4E,J).
This association between cable constriction and amniotic fold progression was also detected after Tc-cad knockdown (Fig. 4N,O; supplementary material Movies 4, 5). The cable, which was conspicuous throughout the epibolic movement of the serosa in wild-type embryos, was visible only in Tc-cadRNAi embryos from the time of formation of the horseshoe amniotic fold, and during formation and closure of the serosa window (Fig. 4S,T). The condensing germband also appeared conspicuously straight-sided and not constricted behind the head lobes, as in controls (Fig. 4N,O).
During stages 3 to 5, our studies confirmed previous descriptions of Tribolium gastrulation that mesoderm internalization progresses from posterior to anterior and extends along the entire length of the germband (supplementary material Fig. S4; compare with Handel et al., 2005). This progression was not as pronounced after Tc-cad knockdown due to decreased germband length (supplementary material Fig. S5). In our recordings, the fluorescence signal emitted from mesodermal cells deteriorated upon cell internalization and completely disappeared once the germband was covered by the amnion and serosa. This limitation prevented us from describing cell and tissue dynamics during later stages of gastrulation.
In wild-type embryos, the lateral ectodermal cells stopped their ventral movement once they were overtaken by the anterior progression of the amniotic fold, they turned anteriorly and came to a standstill shortly afterwards (Fig. 4D). The lateral ectodermal cells in Tc-cadRNAi embryos behaved differently; they advanced anteriorly ahead of the shallow amniotic fold, instead of becoming submerged by it (Fig. 4N). Cells appeared to stop their anterior movement only when the serosa window formed (Fig. 4O). At the end of this stage, the germband in Tc-cadRNAi embryos was much shorter and wider compared with controls (Fig. 4E,O). Our data suggest that the reduced size of the embryonic rudiment, the absence of germband convergent extension by cell intercalation and the prolonged anterior movement of cells all contribute to germband deformation.
During these extensive cell movements, the most anterior-ventral serosa cells were relatively static both in wild-type (Fig. 4E-I) and in Tc-cadRNAi embryos (Fig. 4O-S), reflecting either a zero net balance of forces in these epithelial regions, or attachment to other egg parts.
Stage 5: serosa window closure
In wild-type and Tc-cadRNAi embryos, serosa expansion appeared as a smooth and continuous process until the amnion/serosa cable lined the circumference of the formed serosa window (Fig. 4F-I; Fig. 4P-S). During stage 5, cells at the edge of this window advanced in a zig-zag rather than a smooth manner to close it (Fig. 4J,T). Tc-cadRNAi embryos formed a larger serosa window than wild-type embryos (supplementary material Movies 2, 4). In agreement with expansion of the serosa rudiment, the serosa epithelium in Tc-cadRNAi embryos comprised a larger number of smaller cells compared with controls (Fig. 4J,T); we counted on average 97 serosa cells (σ=9 cells) per 30,000 μm2 in 3 Tc-cadRNAi embryos, versus 76 cells (σ=3 cells) in three wild-type controls.
Immediately after window closure and separation of the serosa from the amnion, the wild-type germband occupied 79% EL (σ=4.1 percentage points; four embryos measured). The anterior end of the germband was positioned at 94% EL (σ=3.2 percentage points) and the posterior end at 15% EL (σ=1.8 percentage points) (Fig. 5A). The truncated Tc-cadRNAi germband occupied 51% EL (σ=2.1 percentage points; four embryos measured) (Fig. 5D). The anterior end was at about the same position as in wild type (93% EL; σ=4.3 percentage points), but the posterior end was positioned much more anteriorly (42% EL; σ=5.3 percentage points).
Fig. 5.
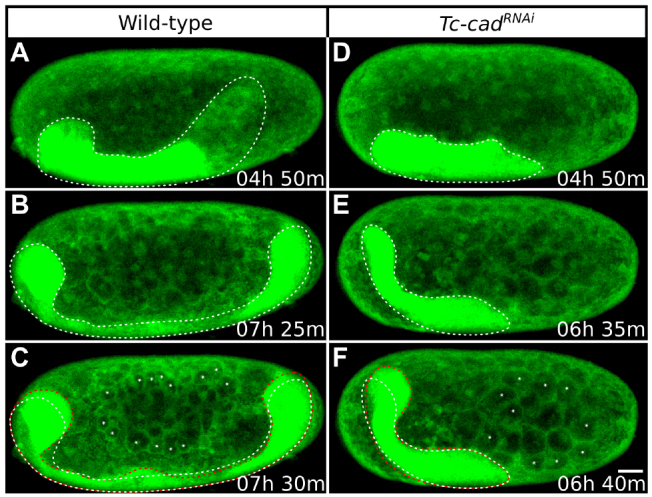
Patterns of Tribolium germband elongation in wild-type and Tc-cadRNAi embryos. Each column shows a single GAP43-YFP-labeled embryo at progressive time-points during stage 6. (A-C) Lateral views of the wild-type embryo also shown in Fig. 4F-J. (D-F) Lateral views of the Tc-cadRNAi embryo also shown in Fig. 4P-T. All images are average intensity projections with uniformly enhanced brightness/contrast to show the germband (outlined with dotted lines) and the membrane-bound yolk spheres. Panels are timed against the onset of stage 1. (A,D) Size and position of germbands immediately after serosa window closure. The germband is truncated posteriorly after Tc-cad RNAi. (B,E) The control germband grows considerably and extends round both poles. The Tc-cadRNAi germband fails to grow, but is displaced anteriorly with the head region curved around the anterior pole. (C,F) At the following time-point, both ends of the control embryo and the anterior end of the Tc-cadRNAi embryos move towards the dorsal side (previous position indicated with white dotted line and new position with red dotted line). This movement is accompanied by the appearance of extra membrane-bound yolk spheres (asterisks) compared with the previous time-point. Anterior is towards the left and dorsal towards the top. Scale bar: 50 μm.
Stage 6: germband elongation
The relative position, size and shape of serosa cells remained unchanged throughout germband elongation (supplementary material Movies 2-5). The head region extended anteriorly and curled dorsally at the anterior pole in both wild-type and Tc-cadRNAi embryos (Fig. 5; supplementary material Movie 6). The wild-type germband also extended posteriorly, curling around the posterior pole and extending along the dorsal side of the egg as new segments were added sequentially from the posterior growth zone (Fig. 5). No posterior elongation was detected in Tc-cadRNAi embryos.
In agreement with previous descriptions (Sarrazin et al., 2012), the entire wild-type germband, as well as the anterior part of the truncated Tc-cadRNAi germband, contracted and expanded repeatedly during stage 6 (supplementary material Movie 6). Every contraction/expansion cycle coincided with the appearance of extra membrane-bound yolk spheres (Fig. 5B,C,E,F). Therefore, we interpreted this movement as a passive response of the germband to the concomitant cleavage of the underlying yolk (Anderson, 1972b).
A possible role for the yolk system in Tribolium morphogenesis
The yolk of insect eggs, together with its accompanying reticular cytoplasm, yolk nuclei and membrane (yolk system or yolksac) plays various roles during insect development (Counce, 1961). Handel et al. described a number of possible interactions between the yolk system and the overlying epithelia during Tribolium embryogenesis (Handel et al., 2000). We examined the earliest and most prominent of these interactions: the involvement of the yolk system in posterior amniotic fold formation. We combined differential interference contrast microscopy with fluorescence confocal microscopy to simultaneously monitor movement of the yolk, while imaging tissue dynamics in H2B-RFP-labeled embryos (Fig. 7; supplementary material Movie 7).
During stages 1 and 2, the yolk contracted globally in concert with the surrounding blastoderm epithelium. A more pronounced yolk depression was observed locally at the posterior pole during primitive pit invagination (Fig. 7A-B′). During stages 3 and 4, we replicated previous observations (Brown et al., 1994; Handel et al., 2000) that a posterior dorsal wedge of yolk initiates movement around the posterior pole, bends ventrally and advances anteriorly, exhibiting strikingly similar dynamics to the posterior amniotic fold (Fig. 7C,C′). This yolk-fold came to a standstill at the end of stage 4, around the time of serosa window formation (Fig. 7D,D′). The yolk-fold stayed in the same position during serosa closure (stage 5), and then retracted posteriorly in tight temporal association with the extending posterior end of the growing germband (stage 6). This coupled movement did not stop when the yolk-fold returned close to its starting point at the posterior pole (Fig. 7E,E′). Instead, it continued dorsally as the posterior germband curled around the posterior pole and extended along the dorsal side of the egg, displacing the abutting yolk further anteriorly (Fig. 7F,F′; supplementary material Movie 7). These observations point to a tight physical connection between the posterior end of the yolksac and the posterior end of the germband during condensation and extension stages that could play a key role during Tribolium epithelial rearrangement (see Discussion). This interpretation is further supported by the Tc-cadRNAi effects: the shallowness of the posterior amniotic fold, the lack of posterior germband submersion into the yolk, and the reduced bouncing of the posterior germband may be related to abnormal patterning of posterior germband structures and improper attachment to the yolksac.
DISCUSSION
Transient fluorescence labeling methods are suitable for superficially cleaving Tribolium embryos
Transient fluorescence labeling methods have found extensive use in embryos with holoblastic (or discoidal meroblastic) cleavage, but only limited application in labeling superficially cleaving insect embryos. In this article, we have shown that transient methods are well suited for imaging Tribolium embryos at a high spatial and temporal resolution. The Tribolium blastoderm cellularizes during interphase of the 13th cycle, about 8.5-10 hours AEL at 32°C. Therefore, material injected at early preblastoderm stages can readily diffuse throughout the entire egg volume to produce a homogeneous and strong fluorescence in most injected embryos until blastoderm cellularization. It is not clear whether this diffusion is principally of injected mRNA, of synthesized protein, or of both. Injected mRNAs encoding H2B-RFP, GAP43-YFP and LA-GFP offer a versatile palette to label the chromatin, the cell membrane, the actin cytoskeleton or combinations thereof. Beyond Tribolium, this labeling methodology will be applicable to other insect and arthropod species for embryonic live imaging.
A model for cell and tissue behaviors that drive Tribolium embryo morphogenesis
With our labeling approach, we have generated the first detailed description of normal and genetically perturbed Tribolium development that is sufficiently precise to allow the formulation of a model for the underlying cell and tissue dynamics.
All formed cells at the uniform blastoderm stage (stage 1) start out with the same cuboidal shape. During blastoderm differentiation (stage 2), embryonic cells start to contract and become columnar, whereas serosa cells start to spread and become squamous. Embryonic and extra-embryonic rudiments are by this stage already asymmetrically positioned, with an oblique dorsal-ventral border between them. The expansive serosa cells occupy an anterior-dorsal domain, whereas the contractile embryonic cells cover most of the posterior and ventral surface of the blastoderm. The relative position of the primordia and the differential properties of their constituent cells cause the germband to condense towards the ventral side of the blastoderm.
The most dramatic and complex tissue rearrangement occurs in stages 3 and 4 during posterior amniotic fold formation and advancement. A key observation to explain this rearrangement is that the movement (and standstill) of the yolk-fold is coupled to that of the posterior end of the embryo throughout stages 3 to 6. This coupling suggests the presence of a physical connection between the primitive pit region of the blastoderm and the posterior pole of the yolksac. This suggestion that the basal surfaces of the abutting posterior amnion and germband are tightly linked to the yolksac has important implications for our model. First, it implies that condensation of the embryonic rudiment will be coupled with posterior amniotic fold progression. As the embryonic rudiment condenses ventrally during stage 3, the anchor on the yolksac would force the posterior end of the forming germband to bend into the yolk and the abutting posterior amnion to fold onto the germband. Second, as condensation proceeds, the anchor pulls the posterior germband and amnion deeper into the yolk. This pulling force drives amnion involution from the blastoderm surface into the inner side of the fold to cover its growing surface, and the movement of the amnion/serosa boundary towards the rim of the posterior amniotic fold. Third, the posterior yolk-fold bends ventrally and extends anteriorly dragged by the involuting amnion that is tightly attached to its inner surface. Extension of the yolk-fold ceases during stage 4 together with posterior amnion involution.
Our GAP43-YFP and LA-GFP imaging studies have suggested the presence of a contractile actomyosin cable at the amnion/serosa boundary. During stage 4, contraction of the cable correlates precisely with amniotic fold progression. We propose that the contractile force exerted by the cable from the rim of the posterior amniotic fold drives folding of the more anterior amnion during this stage. Initially, the cable pulls the amnion over the lateral edges of the head lobes and, later, over the anterior margin of the head. During stage 5, contraction of the cable correlates precisely with, and likely drives, closure of the serosa window. A similar purse string mechanism for serosa window contraction has previously been suggested (Handel et al., 2000). Based on the zig-zag movement of the leading serosa cells, it appears that contraction of the cable during window closure occurs in pulses.
Probing cell behaviors during Tribolium germband condensation and elongation
In our model, condensation of the embryonic rudiment plays a central role in remodeling the single-layered blastoderm into the multi-layered enveloped germband. We were able to follow embryonic cell contraction during germband condensation, as well as polarized cell movements that narrow the Tribolium germband in the dorsal-ventral axis and extend it in the anterior-posterior axis. Convergent extension cell movements have also been reported recently during the subsequent stage of Tribolium early germband extension (Sarrazin et al., 2012). These results show that the morphogenetic mechanism of cell intercalation that drives axial elongation in various vertebrates and invertebrates (Irvine and Wieschaus, 1994; Kanayama et al., 2011; Keller, 2002; Steinmetz et al., 2007) is also at work during Tribolium germband condensation and early extension.
Genetic evidence supporting our model
Support for our model is already available from previous studies that can now be revisited in the light of our findings. The Tribolium zerknüllt-1 gene (Tc-zen1) is expressed in the presumptive serosa at the anterior pole of the blastoderm (van der Zee et al., 2005). Tc-zen1 knock-down changes the blastoderm fate map in the opposite direction to Tc-cad knock-down; all blastoderm cells acquire the embryonic fate at the expense of anterior serosa cells (van der Zee et al., 2005). Interestingly, the following observations were made in these Tc-zen1RNAi embryos: first, the germband condensed ventrally and elongated normally. Second, the posterior amniotic fold formed and covered the germband posteriorly, but never expanded anteriorly. Third, the anterior amnion failed to cover the head lobes anteriorly, and the entire head region appeared enlarged. These observations are in accordance with our proposed model that germband condensation is an autonomous process, which drives initiation of posterior amnion folding, but that expansion of the posterior amniotic fold and closure of the amniotic cavity require serosa function.
Linking caudal gene function to embryo morphogenesis
In this study, we also made the link between Tc-cad function and the highly dynamic cellular contexts in which this gene operates. Tc-cad is first required for specification of the Tribolium fate map, then for dorsal-ventral cell intercalation during germband convergent extension and later on for posterior elongation. Previous studies had addressed the early role of Tc-cad as a posterior determinant (Schoppmeier et al., 2009) and its late role in segmentation (Copf et al., 2004).
Our new finding that Tc-cad activity is required for cell intercalation has many similarities to the situation in Drosophila, where normal anterior-posterior patterning is also required for polarized cell intercalation during germband extension (Butler et al., 2009; Irvine and Wieschaus, 1994). In particular, stripe expression of the pair-rule segmentation genes such as even-skipped is necessary for normal germband extension (Irvine and Wieschaus, 1994; Zallen and Wieschaus, 2004). Previous work in Tribolium has shown that Tc-cad regulates stripe expression of the even-skipped (Tc-eve) pair-rule gene and disruption of Tc-cad expression leads to missing or aberrant Tc-eve stripes (Copf et al., 2004; Schoppmeier et al., 2009). Furthermore, Tc-eve knock-down results in severely truncated and deformed Tribolium germbands, similar to those generated by Tc-cad knockdown (Choe et al., 2006). Following this line of argument, it seems plausible that the effect of Tc-cad on germband convergent extension could be channeled through the control of Tc-eve and/or of other Tribolium pair-rule genes.
Supplementary Material
Acknowledgments
We thank Mette Handberg-Thorsager, Jianying Yang, Christian Specht and Michalis Averof for providing plasmids; Gregor Bucher for providing beetle strains; Andrew Peel and Pavel Tomancak for discussions about the project; and Michalis Averof, Stephan Grill, Maurijn van der Zee, Johannes Schinko, Jack Green, Vera Hunnekuhl, Carsten Wolff and three anonymous reviewers for constructive comments on the manuscript.
Footnotes
Funding
This work was supported by the EVONET European Initial Training Network (to M.A.); by the Cambridge Commonwealth Trusts, the C. T. Taylor scholarship and the John Stanley Gardiner Studentship (to M.A.B.); and by an European Molecular Biology Organization short-term fellowship, a Wellcome Trust VIP fellowship and a Marie Curie Intra-European fellowship (to A.P.). Deposited in PMC for immediate release.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
M.A.B. and A.P. conceived and designed the experiments, performed the experiments and analyzed the data. M.A.B., M.A. and A.P. wrote the paper.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.096271/-/DC1
References
- Anderson D. T. (1972a). The development of Hemimetabolous insects. In Developmental Systems. Insects (ed. Counce S. J., Waddington C. H.), Vol. 1, pp. 95–163 New York, NY: Academic Press; [Google Scholar]
- Anderson D. T. (1972b). The development of Holometabolous insects. In Developmental Systems. Insects (ed. Counce S. J., Waddington C. H.), Vol. 1, pp. 165–242 New York, NY: Academic Press; [Google Scholar]
- Beeman R. W., Stuart J. J., Haas M. S., Denell R. E. (1989). Genetic analysis of the homeotic gene complex (HOM-C) in the beetle Tribolium castaneum. Dev. Biol. 133, 196–209 [DOI] [PubMed] [Google Scholar]
- Behrndt M., Salbreux G., Campinho P., Hauschild R., Oswald F., Roensch J., Grill S. W., Heisenberg C. P. (2012). Forces driving epithelial spreading in zebrafish gastrulation. Science 338, 257–260 [DOI] [PubMed] [Google Scholar]
- Berghammer A. J., Klingler M., Wimmer E. A. (1999). A universal marker for transgenic insects. Nature 402, 370–371 [DOI] [PubMed] [Google Scholar]
- Brown S. J., Patel N. H., Denell R. E. (1994). Embryonic expression of the single Tribolium engrailed homolog. Dev. Genet. 15, 7–18 [DOI] [PubMed] [Google Scholar]
- Brown S. J., Mahaffey J. P., Lorenzen M. D., Denell R. E., Mahaffey J. W. (1999). Using RNAi to investigate orthologous homeotic gene function during development of distantly related insects. Evol. Dev. 1, 11–15 [DOI] [PubMed] [Google Scholar]
- Brown S. J., Shippy T. D., Miller S., Bolognesi R., Beeman R. W., Lorenzen M. D., Bucher G., Wimmer E. A., Klingler M. (2009). The red flour beetle, Tribolium castaneum (Coleoptera): a model for studies of development and pest biology. Cold Spring Harb Protoc 2009, pdb.emo126 [DOI] [PubMed] [Google Scholar]
- Butler L. C., Blanchard G. B., Kabla A. J., Lawrence N. J., Welchman D. P., Mahadevan L., Adams R. J., Sanson B. (2009). Cell shape changes indicate a role for extrinsic tensile forces in Drosophila germ-band extension. Nat. Cell Biol. 11, 859–864 [DOI] [PubMed] [Google Scholar]
- Choe C. P., Miller S. C., Brown S. J. (2006). A pair-rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. Proc. Natl. Acad. Sci. USA 103, 6560–6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copf T., Schröder R., Averof M. (2004). Ancestral role of caudal genes in axis elongation and segmentation. Proc. Natl. Acad. Sci. USA 101, 17711–17715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo J. C., Levine M., Zeller R. W. (1997). Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development 124, 589–602 [DOI] [PubMed] [Google Scholar]
- Counce S. J. (1961). The Analysis of Insect Embryogenesis. Annu. Rev. Entomol. 6, 295–312 [Google Scholar]
- Damle S., Hanser B., Davidson E. H., Fraser S. E. (2006). Confocal quantification of cis-regulatory reporter gene expression in living sea urchin. Dev. Biol. 299, 543–550 [DOI] [PubMed] [Google Scholar]
- El-Sherif E., Averof M., Brown S. J. (2012). A segmentation clock operating in blastoderm and germband stages of Tribolium development. Development 139, 4341–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser M. J., Jr (2012). Insect transgenesis: current applications and future prospects. Annu. Rev. Entomol. 57, 267–289 [DOI] [PubMed] [Google Scholar]
- Godinho L., Mumm J. S., Williams P. R., Schroeter E. H., Koerber A., Park S. W., Leach S. D., Wong R. O. (2005). Targeting of amacrine cell neurites to appropriate synaptic laminae in the developing zebrafish retina. Development 132, 5069–5079 [DOI] [PubMed] [Google Scholar]
- Handel K., Grünfelder C. G., Roth S., Sander K. (2000). Tribolium embryogenesis: a SEM study of cell shapes and movements from blastoderm to serosal closure. Dev. Genes Evol. 210, 167–179 [DOI] [PubMed] [Google Scholar]
- Handel K., Basal A., Fan X., Roth S. (2005). Tribolium castaneum twist: gastrulation and mesoderm formation in a short-germ beetle. Dev. Genes Evol. 215, 13–31 [DOI] [PubMed] [Google Scholar]
- Holtzman S., Miller D., Eisman R., Kuwayama H., Niimi T., Kaufman T. (2010). Transgenic tools for members of the genus Drosophila with sequenced genomes. Fly (Austin) 4, 349–362 [DOI] [PubMed] [Google Scholar]
- Irvine K. D., Wieschaus E. (1994). Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development 120, 827–841 [DOI] [PubMed] [Google Scholar]
- Izeddin I., Specht C. G., Lelek M., Darzacq X., Triller A., Zimmer C., Dahan M. (2011). Super-resolution dynamic imaging of dendritic spines using a low-affinity photoconvertible actin probe. PLoS ONE 6, e15611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama M., Akiyama-Oda Y., Nishimura O., Tarui H., Agata K., Oda H. (2011). Travelling and splitting of a wave of hedgehog expression involved in spider-head segmentation. Nat Commun 2, 500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R. (2002). Shaping the vertebrate body plan by polarized embryonic cell movements. Science 298, 1950–1954 [DOI] [PubMed] [Google Scholar]
- Khairy K., Keller P. J. (2011). Reconstructing embryonic development. Genesis 49, 488–513 [DOI] [PubMed] [Google Scholar]
- Kiehart D. P., Galbraith C. G., Edwards K. A., Rickoll W. L., Montague R. A. (2000). Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J. Cell Biol. 149, 471–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster R. W., Fraser S. E. (2004). Time-lapse microscopy of brain development. Methods Cell Biol. 76, 207–235 [DOI] [PubMed] [Google Scholar]
- Link B. A., Megason S. G. (2008). Zebrafish as a model for development. In Sourcebook of Models for Biomedical Research (ed. Conn P. M.), pp. 103–112 Totowa, NJ: Humana Press; [Google Scholar]
- Martin P., Lewis J. (1992). Actin cables and epidermal movement in embryonic wound healing. Nature 360, 179–183 [DOI] [PubMed] [Google Scholar]
- Mavrakis M., Rikhy R., Lippincott-Schwartz J. (2009). Plasma membrane polarity and compartmentalization are established before cellularization in the fly embryo. Dev. Cell 16, 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis M., Pourquié O., Lecuit T. (2010). Lighting up developmental mechanisms: how fluorescence imaging heralded a new era. Development 137, 373–387 [DOI] [PubMed] [Google Scholar]
- Megason S. G., Fraser S. E. (2007). Imaging in systems biology. Cell 130, 784–795 [DOI] [PubMed] [Google Scholar]
- Meijering E., Dzyubachyk O., Smal I. (2012). Methods for cell and particle tracking. Methods Enzymol. 504, 183–200 [DOI] [PubMed] [Google Scholar]
- Minden J. S., Agard D. A., Sedat J. W., Alberts B. M. (1989). Direct cell lineage analysis in Drosophila melanogaster by time-lapse, three-dimensional optical microscopy of living embryos. J. Cell Biol. 109, 505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Taubenberger A., Vos M. J., Böttger A., Lasi M., Lai F. P., Fischer M., Rottner K. (2006). Monomeric red fluorescent protein variants used for imaging studies in different species. Eur. J. Cell Biol. 85, 1119–1129 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Yoshizaki M., Ogawa S., Okamoto H., Shinmyo Y., Bando T., Ohuchi H., Noji S., Mito T. (2010). Imaging of transgenic cricket embryos reveals cell movements consistent with a syncytial patterning mechanism. Curr. Biol. 20, 1641–1647 [DOI] [PubMed] [Google Scholar]
- Panfilio K. A., Roth S. (2010). Epithelial reorganization events during late extraembryonic development in a hemimetabolous insect. Dev. Biol. 340, 100–115 [DOI] [PubMed] [Google Scholar]
- Pavlopoulos A., Averof M. (2005). Establishing genetic transformation for comparative developmental studies in the crustacean Parhyale hawaiensis. Proc. Natl. Acad. Sci. USA 102, 7888–7893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos A., Berghammer A. J., Averof M., Klingler M. (2004). Efficient transformation of the beetle Tribolium castaneum using the Minos transposable element: quantitative and qualitative analysis of genomic integration events. Genetics 167, 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posnien N., Schinko J., Grossmann D., Shippy T. D., Konopova B., Bucher G. (2009). RNAi in the red flour beetle (Tribolium). Cold Spring Harb Protoc 2009, pdb.prot5256 [DOI] [PubMed] [Google Scholar]
- Price A. L., Modrell M. S., Hannibal R. L., Patel N. H. (2010). Mesoderm and ectoderm lineages in the crustacean Parhyale hawaiensis display intra-germ layer compensation. Dev. Biol. 341, 256–266 [DOI] [PubMed] [Google Scholar]
- Richards S., Gibbs R. A., Weinstock G. M., Brown S. J., Denell R., Beeman R. W., Gibbs R., Beeman R. W., Brown S. J., Bucher G., et al. ; Tribolium Genome Sequencing Consortium (2008). The genome of the model beetle and pest Tribolium castaneum. Nature 452, 949–955 [DOI] [PubMed] [Google Scholar]
- Riedl J., Crevenna A. H., Kessenbrock K., Yu J. H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T. A., Werb Z., et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5, 605–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. (2004). Gastrulation in other insects. In Gastrulation: From Cells to Embryo (ed. Stern C. D.), pp. 105–121 New York, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Sarrazin A. F., Peel A. D., Averof M. (2012). A segmentation clock with two-segment periodicity in insects. Science 336, 338–341 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppmeier M., Fischer S., Schmitt-Engel C., Löhr U., Klingler M. (2009). An ancient anterior patterning system promotes caudal repression and head formation in ecdysozoa. Curr. Biol. 19, 1811–1815 [DOI] [PubMed] [Google Scholar]
- Schröder R., Beermann A., Wittkopp N., Lutz R. (2008). From development to biodiversity—Tribolium castaneum, an insect model organism for short germband development. Dev. Genes Evol. 218, 119–126 [DOI] [PubMed] [Google Scholar]
- Schulz C., Schröder R., Hausdorf B., Wolff C., Tautz D. (1998). A caudal homologue in the short germ band beetle Tribolium shows similarities to both, the Drosophila and the vertebrate caudal expression patterns. Dev. Genes Evol. 208, 283–289 [DOI] [PubMed] [Google Scholar]
- Steinmetz P. R., Zelada-Gonzáles F., Burgtorf C., Wittbrodt J., Arendt D. (2007). Polychaete trunk neuroectoderm converges and extends by mediolateral cell intercalation. Proc. Natl. Acad. Sci. USA 104, 2727–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teddy J. M., Kulesa P. M. (2004). In vivo evidence for short- and long-range cell communication in cranial neural crest cells. Development 131, 6141–6151 [DOI] [PubMed] [Google Scholar]
- Trauner J., Schinko J., Lorenzen M. D., Shippy T. D., Wimmer E. A., Beeman R. W., Klingler M., Bucher G., Brown S. J. (2009). Large-scale insertional mutagenesis of a coleopteran stored grain pest, the red flour beetle Tribolium castaneum, identifies embryonic lethal mutations and enhancer traps. BMC Biol. 7, 73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zee M., Berns N., Roth S. (2005). Distinct functions of the Tribolium zerknüllt genes in serosa specification and dorsal closure. Curr. Biol. 15, 624–636 [DOI] [PubMed] [Google Scholar]
- Zallen J. A., Wieschaus E. (2004). Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell 6, 343–355 [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M., Pines J., Ryan K., Siemering K. R., Haseloff J., Evans M. J., Gurdon J. B. (1996). An indelible lineage marker for Xenopus using a mutated green fluorescent protein. Development 122, 3719–3724 [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M., Pines J., McLean Hunter S., Dixon J. P., Siemering K. R., Haseloff J., Evans M. J. (1997). Following cell fate in the living mouse embryo. Development 124, 1133–1137 [DOI] [PubMed] [Google Scholar]
- Zhang S. O., Weisblat D. A. (2005). Applications of mRNA injections for analyzing cell lineage and asymmetric cell divisions during segmentation in the leech Helobdella robusta. Development 132, 2103–2113 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.