Abstract
Interrogating specific cellular activities often entails the dissection of posttranslational modifications or functional redundancy conferred by protein families, which demands more sophisticated research tools than simply eliminating a specific gene product by gene targeting or RNAi. We have developed a novel methodology that involves engineering a single SCFβTrCP-based ubiquitin ligase that is capable of not only simultaneously targeting the entire family of ErbB receptor tyrosine kinases for ubiquitination and degradation, but also selectively recruiting only activated ErbBs. The engineered SCFβTrCP ubiquitin ligase effectively blocked ErbB signaling and attenuated oncogenicity in breast cancer cells, yet had little effect on the survival and growth of non-cancerous breast epithelial cells. Therefore, engineering ubiquitin ligases offers a simple research tool to dissect the specific traits of tumorigenic protein families, and provides a rapid and feasible means to expand the dimensionality of drug discovery by assessing protein families or posttranslational modifications as potential drug targets.
INTRODUCTION
Normal or oncogenic signaling are often orchestrated by not one, but multiple members of signaling proteins with redundant functions. Efficacy of targeted inhibition of one signaling molecule is frequently compromised as a result of compensatory upregulation of another family member, which accounts in part for the resistance to targeted therapy. Moreover, activation of a given signaling pathway under physiological or pathological conditions further entails posttranslational modifications of signaling molecules, e.g. phosphorylation of receptor tyrosine kinases. Such complexities present challenges of inactivating a disease pathway by targeting one signaling protein, yet offer new avenues or strategies for disease intervention by collectively abrogating the entire family of functionally overlapping proteins, or by ablation of a specific posttranslational event that is preferentially dysregulated in a diseased state. However, functional assessments of these new biological space in drug discovery require more sophisticated tools than simple elimination of a given target protein by established methodology, such as RNAi or gene targeting. In this regard, chemical inhibitors can potentially fulfill the desired functionality, but they are often not available at this early discovery stage, and are developed after a specific drug target is validated. As such, efforts to explore the value of cellular protein families as emerging drug targets are largely hampered by the lack of effective experimental tools in pre-clinical and clinical models.
One example of such cases is the epidermal growth factor receptor (EGFR/ErbB) family, which consists of EGFR (EGFR/HER-1 or ErbB1), ErbB-2 (Neu or HER-2), ErbB3 (HER-3), and ErbB4 (HER-4). It has been established that the ErbB RTKs are frequently hyper-activated in cancers and contribute to tumor survival, growth, progression, angiogenesis, and metastasis (1–7). Overexpression of ErbB2 is found in nearly 20% of breast cancer patients as well as other malignancies, and is correlated with shorter relapse-free survival and poor prognosis. Individual ErbB RTKs have been intensely pursued as therapeutic targets through the attenuation of their signaling activities (8–11). Numerous attempts have been made to inhibit EGFR and ErbB2 signaling, such as inactivating monoclonal antibodies (mAbs) directed against the extracellular domain of the receptors and chemical inhibitors that block intracellular tyrosine kinase activity. Currently, novel therapeutic strategies targeting multiple ErbB RTKs are being intensely pursued as more effective ways to block the ErbB signaling that is often necessary to sustain tumor growth and survival. Moreover, in order to reduce the potential cytotoxicity to normal cells that results from ErbB eradication, it is preferable to selectively target activated, tyrosine-phosphorylated ErbB RTKs instead of knocking out all ErbB expression. However, such specific and multifaceted manipulation of ErbB activity is technically challenging using conventional methods and tools.
Ubiquitin-dependent proteolysis constitutes the major pathway that eukaryotic cells employ to degrade cellular proteins, and involves a cascade of enzymatic reactions catalyzed by the E1 ubiquitin-activating enzyme, the E2 ubiquitin-conjugating enzyme, and the E3 ubiquitin ligase. Of these enzymes, the E3 ubiquitin ligase is exclusively responsible for defining substrate specificity through direct binding to the target protein. The multimeric SCFβTrCP ubiquitin ligase, which consists of Skp1, CUL1, the F-box-containing βTrCP substrate receptor and the Rbx1/Roc1/Hrt1 adaptor, selectively targets a variety of key regulators of the cell cycle, cell signaling and transcription for ubiquitination and degradation (12). Among the SCF subunits, the F-box protein βTrCP is exclusively responsible for substrate recognition and recruitment. Engineering F-box proteins has been established as a powerful strategy to direct the SCF machinery to target a variety of cellular proteins for degradation in cultured eukaryotic cells and animal models (reviewed in (13) and references therein). More specifically, the SCF ubiquitination machinery can be harnessed to degrade non-SCF targets by attaching an F-box protein to a peptide or motif that is able to bind to the protein of interest (13–21).
In this study, we fused the phosphotyrosine-binding domain (PTB) from the Shc signaling protein to the C-terminus of βTrCP to selectively capture all activated (tyrosine-phosphorylated) EGFR/ErbB family proteins and recruit them to the SCF machinery for degradation. Our data demonstrated the efficacy of the engineered F-TrCP-Shc E3 ligase in selectively inducing ubiquitin-mediated degradation of all activated ErbB family proteins. We also showed that F-TrCP-Shc suppressed tumor growth through increased apoptosis and G1 arrest, sensitized ErbB2-overexpressing breast cancer cells to cisplatin-mediated killing, and inhibited tumorigenicity in nude mice.
RESULTS
The engineered F-TrCP-Shc ubiquitin ligase interacts with ErbB2 and induces its degradation in a dose-dependent manner
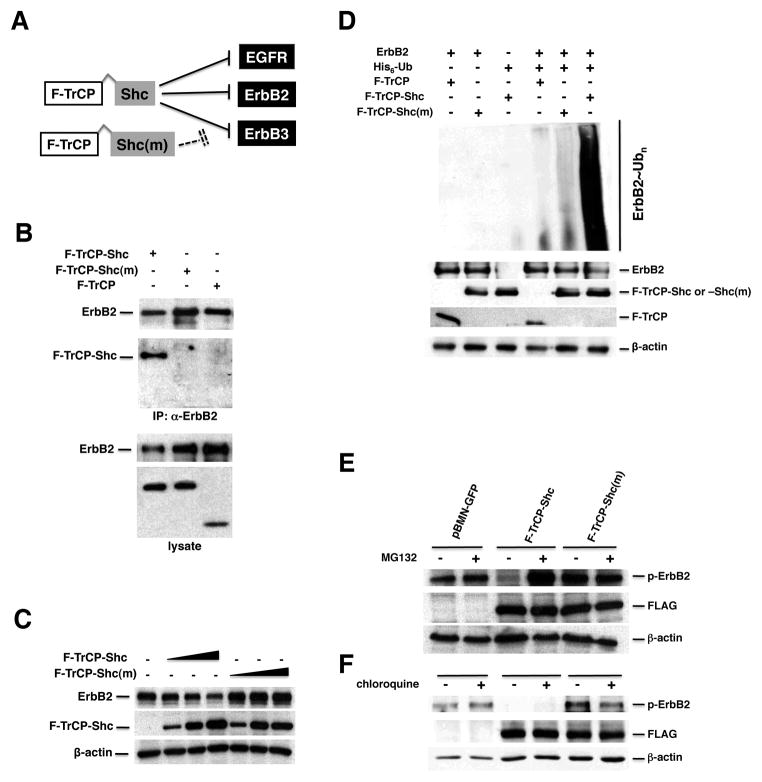
To specifically recruit activated (oncogenic) ErbB proteins to the SCF ubiquitination machinery, we first engineered a chimeric F-TrCP-Shc ubiquitin ligase with the phospho-tyrosine binding (PTB) domain of the Shc adaptor protein fused in-frame to the carboxyl terminus of βTrCP and a FLAG tag at the N-terminus (Fig. 1A). The PTB domain of Shc specifically binds to the tyrosine phosphorylated/activated intracellular domain of EGFR (pY1114, pY1086 and pY1148), ErbB2 (pY1248), ErbB3 (pY1328) and ErbB4 (pY1284) (22–25). In order to verify the specificity of the F-TrCP-Shc/ErbB interaction, we created a phospho-ErbB binding-defective construct, F-TrCP-Shc(m), that possesses the F198V point mutation in the PTB domain but is otherwise identical to F-TrCP-Shc (Fig. 1A) (26). To test the binding of the engineered F-TrCP-Shc ubiquitin ligase to ErbB2, FLAG-tagged chimeric constructs were transiently transfected into 293T cells and co-immunoprecipitation experiments were performed. As shown in Fig. 1B, F-TrCP-Shc, but not F-TrCP-Shc(m) effectively binds to endogenous ErbB2. To investigate whether F-TrCP-Shc can reduce endogenous ErbB2 levels through ubiquitin-dependent proteolysis, 293T cells were transfected with increasing doses of F-TrCP-Shc or F-TrCP-Shc(m). Indeed, the ectopic expression of F-TrCP-Shc induced robust poly-ubiquitination and dose-dependent downregulation of endogenous ErbB2, whereas the binding-deficient F-TrCP-Shc(m) did not (Fig. 1C–D).
Figure 1. The engineered F-TrCP-Shc E3 ligase recruits and induces the dose-dependent degradation of ErbB2.
A. Schematic diagram of the engineered F-TrCP-Shc ubiquitin ligase to target the ErbB RTKs. B. F-TrCP-Shc binds to ErbB2. 293T cells were transiently transfected with the indicated plasmids. ErbB2 was immunoprecipitated with the anti-ErbB2 antibody and immunoblotted with the ErbB2, FLAG and β-actin antibodies. C. Degradation of endogenous ErbB2 by the engineered F-TrCP-Shc. 293T cells were transiently transfected with increasing doses of F-TrCP-Shc or F-TrCP-Shc(m), and levels of endogenous ErbB2, F-TrCP fusions and β-actin were determined by Western blotting. The experiment was repeated three times. D. F-TrCP-Shc promotes ubiquitination of ErbB2. 293T cells were transfected with the indicated plasmids for in vivo ubiquitination assays. E–F. F-TrCP-Shc-induced ErbB2 degradation is proteasome-dependent. SKBR3 and BT474 cells expressing F-TrCP-Shc, F-TrCP-Shc(m) or pBMN-GFP were treated with 50 μM MG132 or 100 μM choloroquine. The levels of endogenous ErbB2, F-TrCP fusions and β-actin were determined by Western blotting.
Ubiquitinated ErbB receptors can be degraded by either the 26S proteasome or trafficked to the lysosomes for destruction (27, 28). As seen in Fig. 1E and 1F, treatment of F-TrCP-Shc-expressing cells with the proteasome inhibitor MG132 blocked ErbB2 degradation, while the lysosomal acidification inhibitor chloroquine had no discernable effect, indicating that the engineered F-TrCP-Shc construct preferentially directs ErbB2 towards proteasomal degradation. Therefore, the PTB domain of Shc, upon fusion to F-TrCP, effectively binds ErbB2 and recruits it to the SCF machinery for ubiquitination and proteasome-dependent degradation.
F-TrCP-Shc promotes degradation of all ErbB family proteins and inhibits ErbB signaling
The compensatory increase in the expression levels of other ErbB family members has been reported to contribute to the acquired resistance of breast cancer cells to ErbB2 inhibitory drugs in patients (29). This finding raises the question of whether the simultaneous attenuation of all ErbB proteins could potentially improve the therapeutic efficacy and outcome in cancer patients with ErbB over-activation. Thus, we selected the Shc PTB domain for fusion with βTrCP since Shc functions as a mediator of ErbB signaling and interacts with all members of the ErbB family upon RTK activation. F-TrCP-Shc is therefore able to degrade all activated ErbB RTKs, which creates a unique opportunity to examine the physiological consequences of fully attenuated ErbB signaling. Indeed, F-TrCP-Shc but not F-TrCP-Shc(m) stimulates ubiquitination of EGFR and ErbB3 (Fig. S1).
To achieve sustained degradation of ErbB RTKs, BT474 and SKBR3 breast cancer cells were infected with recombinant retroviruses expressing either F-TrCP-Shc, F-TrCP-Shc(m) or the pBMN-GFP viral vector. Infected cells with chromosomally-integrated chimeric F-TrCP transgenes were isolated by FACS based on the expression of GFP from the viral vector. The endogenous expression levels of ErbB RTKs (overall protein levels including the tyrosine phosphorylated/activated subpopulations) in BT474 and SKBR3 cells were determined by Western blotting (Fig. 2A–B, lane 1). We note that three ErbB proteins, EGFR, ErbB2 and ErbB3, are expressed in BT474 and SKBR3 cells and the activated subpopulations of these ErbB RTKs can be detected using their respective phospho-specific antibodies (Fig. 2A–B, lane 1). However, ErbB4 expression cannot be detected in BT474 or SKBR3 cells (data not shown), as documented previously (29). As shown in Fig. 2A and 2B, the constitutive expression of F-TrCP-Shc selectively degraded tyrosine-phosphorylated ErbB2 in both SKBR3 and BT474 cells, while the overall ErbB2 expression levels, consisting predominantly of unmodified and inactive ErbB2, were largely unaffected. Furthermore, F-TrCP-Shc also selectively depleted the activated pools of EGFR and ErbB3 without compromising their overall levels of expression (Fig. 2A–B, lane 2). In contrast, F-TrCP-Shc(m), which is unable to interact with ErbB RTKs, had no effect on their steady-state levels (Fig. 2A–B, lane 3). The mRNA levels of EGFR, ErbB2, and ErbB3 in these cell lines were not altered significantly by the ectopic expression of F-TrCP-Shc or F-TrCP-Shc(m) (Fig. S2). Taken together with the rescue of F-TrCP-Shc-induced ErbB2 degradation by the proteasome inhibitor MG132 (Fig. 1E), we conclude that the engineered F-TrCP-Shc ubiquitin ligase operates at the post-translational level to specifically ubiquitinate and degrade the entire family of activated ErbB RTKs.
Figure 2. The engineered F-TrCP-Shc E3 ligase selectively targets the activated ErbB family members for degradation and depletes ErbB2 RTK on the cell surface.
SKBR3 (A) and BT474 (B) cells infected with the indicated recombinant retroviruses were lysed and 50 μg of each cell lysate was analyzed by SDS-PAGE and Western blotting with the indicated antibodies. C. SKBR3 and BT474 cells were plated in 6-well plates for 24 hours, trypsinized and washed in FACS buffer. The live cells were stained with anti-ErbB2 antibody (Santa Cruz) against the extracellular domain of ErbB2. Results are the mean for three replicate experiments. D. Dose-dependent degradation of endogenous ErbB2 upon infection of adenoviral F-TrCP-Shc was determined by Western blotting with the indicated antibodies. E. Depletion of ErbB2 on the cell membrane by adenoviral F-TrCP-Shc. ErbB2 (red) was detected by immunofluorescent staining with the anti-ErbB2 antibody (R&D Systems). Infected cells were marked by GFP expression from the adenoviral vector. The nuclei were visualized by DAPI staining (blue). White arrow, a low F-TrCP-Shc expressing SKBR3 cell.
Ubiquitination of membrane receptors accelerates endocytosis, resulting in an overall reduction in cell surface levels (28, 30, 31). In order to determine whether F-TrCP-Shc induced receptor endocytosis, we employed a flow cytometry-based approach to measure the levels of transmembrane ErbB2 using the anti-ErbB2 antibody against the extracellular domain of the receptor. As shown in Fig. 2C, enforced retroviral expression of F-TrCP-Shc effectively diminished ErbB2 from the cell surface of both SKBR3 and BT474 cells. In comparison, expression of the pBMN-GFP control or F-TrCP-Shc(m) had minimal, if any, influence in the cell surface pool of ErbB2 (Fig. 2C). This result was further corroborated by adenoviral-driven high-level expression of F-TrCP-Shc that essentially eliminated the presence of ErbB2 on the cell membrane of SKBR3 cells (Fig. 2D). Moreover, the efficiency of membrane ErbB2 depletion correlated with the levels of F-TrCP-Shc expression, as SKBR3 cells with low levels of chimeric E3 ligase expression (indicated by low GFP expression) retained residual ErbB2 expression on the cell surface (Fig. 2D, white arrow). Taken together, the targeted ubiquitination of ErbB2 resulted in the effective depletion of membrane ErbB2 through increased ubiquitination.
Suppression of MAPK and PI3K signaling upon targeted degradation of ErbB RTKs
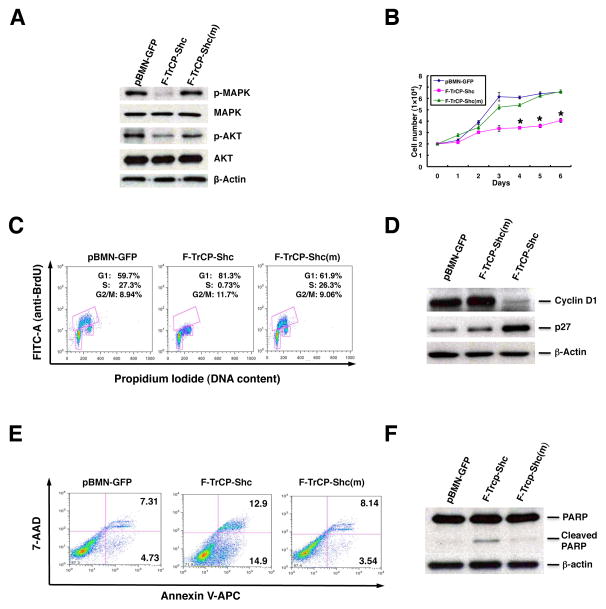
The downstream pathways activated by ErbB RTKs include the MAP kinase and AKT signaling cascades, both of which play critical roles in tumorigenesis (recently reviewed in (32) and references therein). Therefore, we next chose to examine whether targeting the ErbB family of proteins with our E3 ligase had an effect on these crucial oncogenic signaling pathways. As seen in Fig. 3A and 4A, the attenuation of ErbB signaling by targeted proteolysis was accompanied by the diminished phosphorylation (and activation) of AKT and MAPK. Thus, F-TrCP-Shc inhibited both signaling pathways in BT474 and SKBR3 cells, while the overall levels of AKT and MAPK were not affected (Fig. 3A and 4A). In contrast, the constitutive expression of the binding-deficient F-TrCP-Shc(m) did not alter phospho-AKT or phospho-MAPK levels and signaling, as compared with the control pBMN-GFP-infected BT474 and SKBR3 cells (Fig. 3A and 4A). Therefore, the targeted depletion of ErbB RTKs effectively attenuates the downstream MAPK and PI3K/AKT signaling cascade.
Figure 3. Targeted degradation of ErbB RTKs by F-TrCP-Shc led to the attenuation of MAPK and PI3K signaling, cell cycle arrest and inhibition of proliferation in SKBR3 breast cancer cells.
A. SKBR3 cells were infected with pBMN-F-TrCP-Shc, pBMN-F-TrCP-Shc(m) or pBMN-GFP. Infected cells were sorted, lysed and probed with antibodies against p-MAPK, MAPK, p-AKT, AKT and β-actin. B. SKBR3 cells were infected with the same retroviruses as in (A) and were plated in 12-well plates for up to 6 days. The number of cells in each group was counted at the indicated time points. Results are the mean for three replicate experiments. * indicates P<0.05 for pBMN-F-TrCP-Shc-infected cells compared to those with pBMN-GFP or pBMN-F-TrCP-Shc(m). C. SKBR3 cells were infected with pBMN-F-TrCP-Shc, pBMN-F-TrCP-Shc(m) or pBMN-GFP, labeled with BrdU and propidium iodide, and subjected to FACS analysis. Results are represented as the mean ± standard deviation (n=3). D. Western blotting was performed to measure the expression of G1 cell cycle markers cyclin D1 and p27 using the appropriate antibodies. E–F. Apoptotic induction of SKBR3 cells by F-TrCP-Shc, as determined by Annexin V staining-FACS analysis, and PARP1 cleavage by Western blotting.
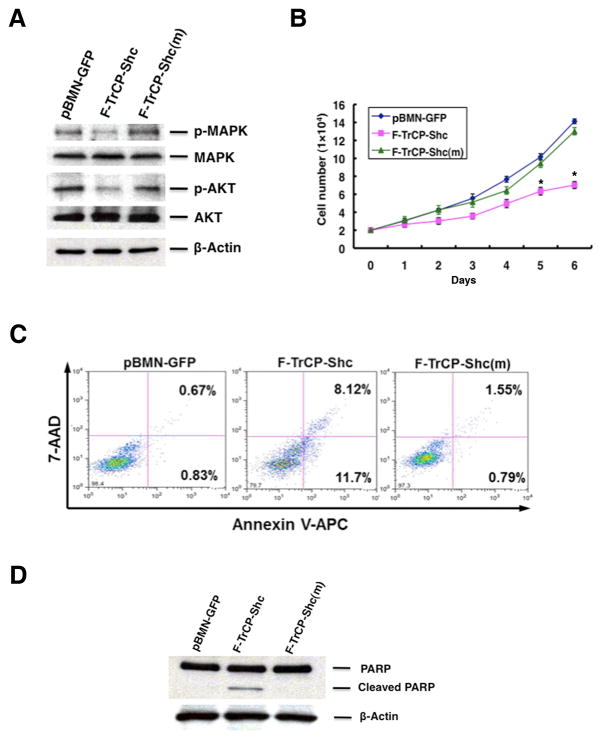
Figure 4. Targeted degradation of ErbB RTKs triggered apoptosis in BT474 cells.
A. BT474 cells were infected with pBMN-F-TrCP-Shc, pBMN-F-TrCP-Shc(m) or pBMN-GFP, sorted and analyzed for inhibition of MAPK and PI3K signaling by Western blotting as in Fig. 4A. B. Retroviral expression of F-TrCP-Shc in BT474 cells inhibited cell growth. Results are the mean for three replicate experiments. * indicates P<0.05 for pBMN-F-TrCP-Shc-infected cells compared to those with pBMN-GFP or pBMN-F-TrCP-Shc(m). C–D. Apoptotic induction of BT474 cells by F-TrCP-Shc, as determined by Annexin V-FACS analysis and PARP1 cleavage by Western blotting. For FACS analysis, cells were stained with Annexin-V-APC and 7-AAD, and the percentage of early (right bottom) and late apoptotic (right top) populations are indicated.
Cell cycle inhibition and increased apoptosis occur upon depletion of ErbB RTK in breast cancer cells
The ErbB receptor family members stimulate cell proliferation and promote the survival of breast cancer as well as other tumor types. Thus, we next examined whether the targeted degradation of all activated ErbB RTKs inhibits the growth and survival of BT474 and SKBR3 cells. As shown in Fig. 3B and Fig. 4B, the ectopic expression of F-TrCP-Shc led to a marked reduction of the growth rates of BT474 and SKBR3 cells relative to their corresponding F-TrCP-Shc(m) or pBMN-GFP controls. This observation is in accordance with the previous finding that siRNAs targeting ErbB2 suppressed the proliferation of ErbB2-overexpressing breast cancer cells (33, 34).
We further determined the cell cycle profiles of SKBR3 cells infected with pBMN-F-TrCP-Shc, pBMN-F-TrCP-Shc(m) or pBMN-GFP retroviruses by measuring BrdU incorporation via FACS analysis. As shown in Fig. 3C, the enforced expression of F-TrCP-Shc arrested the cell cycle in G1 phase (81.3% G1 cells versus 59.7% and 61.9% in pBMN-GFP- and F-TrCP-Shc(m)-expressing SKBR3 cells, respectively). Strikingly, F-TrCP-Shc induced robust inhibition of DNA synthesis, as indicated by the reduction of BrdU-positive S phase cells (0.73%) compared with those infected with control viruses (27.3% and 26.3% for pBMN-GFP and F-TrCP-Shc(m) respectively) (Fig. 3C). The G1 cell cycle arrest induced by F-TrCP-Shc was further demonstrated by a reduction in cyclin D1 expression and an accumulation of the cyclin-dependent kinase inhibitor p27Kip1 (Fig. 3D). These results revealed that F-TrCP-Shc inhibits cell proliferation via G1 cell cycle arrest in SKBR3 breast cancer cells. Furthermore, the targeted degradation of ErbB RTKs induced approximately 3–4 fold increase in apoptosis, as determined by Annexin V/7-AAD staining and FACS analysis (Fig. 3E), as well as increased cleavage of poly(ADP-ribose) polymerase (PARP) (Fig. 3F). Therefore, the inhibition of cell cycle progression and induction of apoptosis by F-TrCP-Shc both contribute to its growth suppressive effect on ErbB2-overexpressing SKBR3 cells.
In BT474 cells, the enforced expression of F-TrCP-Shc led to a modest inhibition of cell cycle progression, as indicated by the approximately 10–14% reduction of S phase populations compared with controls, and a concomitant increase in G1 and G2/M populations (Fig. S3). However, the most striking effect of F-TrCP-Shc in BT474 cells is the robust induction of apoptosis. As measured by Annexin V and 7-AAD staining and FACS analysis, early apoptosis was increased 14-fold in F-TrCP-Shc-expressing cells compared to pBMN-GFP cells (11.7% versus 0.83%), while F-TrCP-Shc(m) did not induce apoptosis (0.79%) (Fig. 4C). Furthermore, increased PARP cleavage was detected in BT474 cells stably expressing F-TrCP-Shc (Fig. 4D). These results indicate that the downregulation of activated ErbB family proteins by F-TrCP-Shc triggers apoptosis in ErbB2-overexpressing BT474 breast cancer cells.
Targeted depletion of ErbB RTKs had no discernable effect on non-cancerous MCF-10A mammary epithelial cells
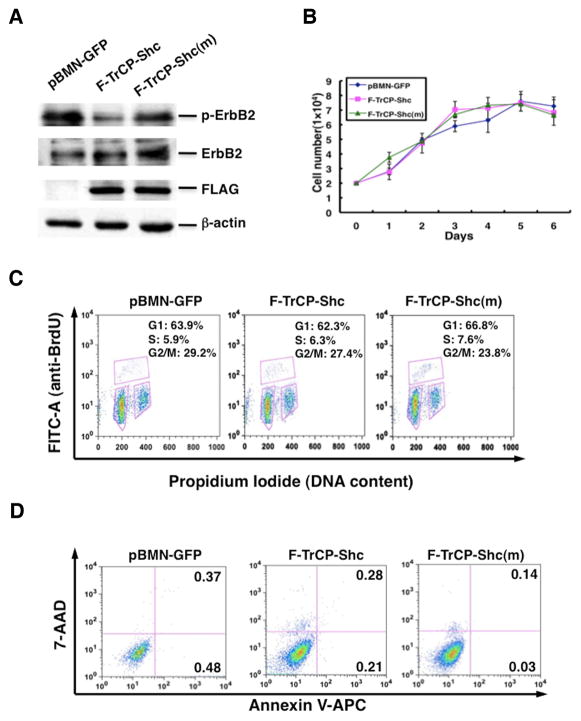
We next sought to determine whether non-cancerous mammary epithelial cells are also subjected to F-TrCP-Shc-mediated growth inhibition, or if ErbB-overexpressing breast cancers are particularly sensitive to the engineered ligase. Non-cancerous MCF-10A mammary epithelial cells were infected with the pBMN-F-TrCP-Shc, pBMN-F-TrCP-Shc(m) or pBMN-GFP retroviruses. Reduction of endogenous ErbB2 levels was observed, similar to that observed in SKBR3 and BR474 cells (Fig. 5A). Strikingly, the depletion of endogenous ErbB RTKs by pBMN-F-TrCP-Shc had little impact on the growth rate of MCF-10A cells, similar to those infected with pBMN-F-TrCP-Shc(m) or pBMN-GFP (Fig. 5B). Cell cycle arrest or apoptotic induction was not observed in MCF-10A cells stably expressing F-TrCP-Shc (Fig. 5C–D). It is noteworthy that while EGF stimulates the proliferation rate of MCF10A (35, 36), ErbB-knockdown by F-TrCP-Shc was carried out under normal culture conditions in the absence of exogenous EGF. This non-exponential growth of MCF10A cells is likely less dependent on EGF signaling. Therefore, the engineered F-TrCP-Shc ligase specifically inhibits the proliferation of ErbB-overexpressing breast cancer cells, but exerts little effect on non-stimulated mammary epithelial cells. These results indicate that the simultaneous family-wide targeting of activated ErbB RTKs is a feasible intervention strategy for breast cancer.
Figure 5. The engineered F-TrCP-Shc E3 ligase had no effect on the non-cancerous MCF-10A mammary epithelial cells.
MCF-10A cells were infected with pBMN-F-TrCP-Shc, pBMN-F-TrCP-Shc(m) or pBMN-GFP, and assayed for (A) ErbB2 degradation by Western blotting, (B) growth rate, (C) cell cycle by BrdU labeling and FACS analysis, and (D) apoptosis by Annexin V/7-AAD staining and FACS analysis as in Figures 3 and 4.
F-TrCP-Shc sensitizes SKBR3 and BT474 cells to cisplatin and inhibits tumorigenicity in vitro and in vivo
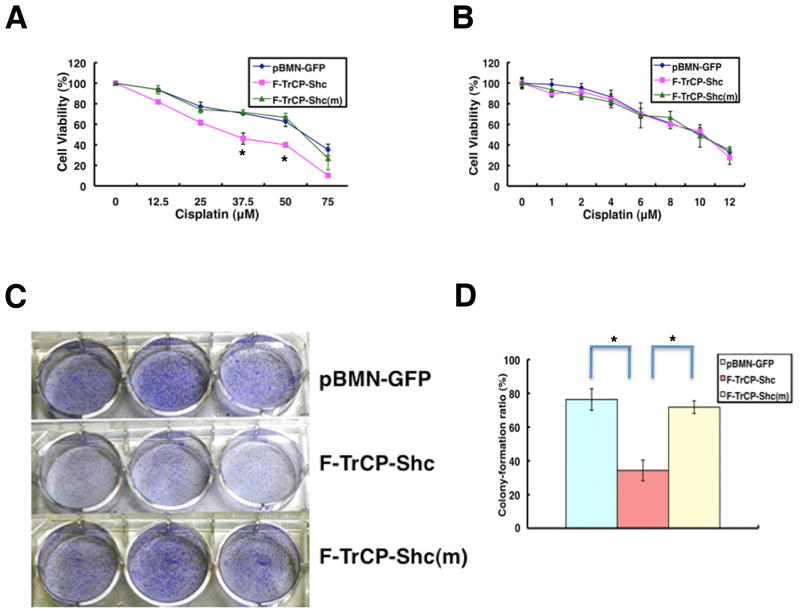
Several retrospective studies have suggested that ErbB2 overexpression is associated with a reduced efficacy of adjuvant chemotherapy (37, 38). Therefore, we hypothesized that F-TrCP-Shc would render breast cancer cells more susceptible to chemotherapy agents. BT474 cells expressing F-TrCP-Shc or F-TrCP-Shc(m) were treated with doses of cisplatin ranging from 12.5 μM to 100 μM, and cell viability was measured 48 hours after treatment. As shown in Fig. 6A, F-TrCP-Shc reduced the viability of BT474 cells upon cisplatin treatment. For example, at 37.5 μM, the number of viable cells is approximately 30% lower than control BT474 cells infected with pBMN-GFP or pBMN-F-TrCP-Shc(m) (Fig. 6A). A similar result was observed in BT474 cells (data not shown). In contrast, MCF-10A cells expressing F-TrCP-Shc did not display enhanced cytotoxicity in response to cisplatin compared to those expressing F-TrCP-Shc(m) or pBMN-GFP (Fig. 6B).
Figure 6.
The engineered F-TrCP-Shc E3 ligase sensitizes BT474 breast cancer cells to cytotoxic killing by cisplatin and decreases their transforming ability.
A–B. BT474 or MCF-10A cells infected with pBMN-F-TrCP-Shc, pBMN-F-TrCP-Shc(m) or pBMN-GFP retroviruses were treated with the indicated amounts of cisplatin for 48 hours, and the cell viability was measured by the XTT assay. Results are represented as the mean ± standard deviation (n=3), for BT474 cells. C–D. Focus formation assay. Retroviral vector infected BT474 cells were seeded in 6-well plates and cultured for 10 days to allow for colony formation. The colonies were fixed, stained and photographed. The colony numbers were counted and the colony formation ratio was calculated according to the formula: . Results are represented as the mean ± standard deviation (n=3). * indicates P<0.05 for pBMN-F-TrCP-Shc-infected cells compared to those with pBMN-GFP or pBMN-F-TrCP-Shc(m).
Next, we assessed the oncogenic potential of F-TrCP-Shc-expressing breast cancer cells by focus formation assays. Retrovirally-expressed F-TrCP-Shc caused a 50% reduction in focus formation in both BT474 and SKBR3 cells compared to control pBMN-GFP- and F-TrCP-Shc(m)-infected cells (Fig. 6C–D and data not shown).
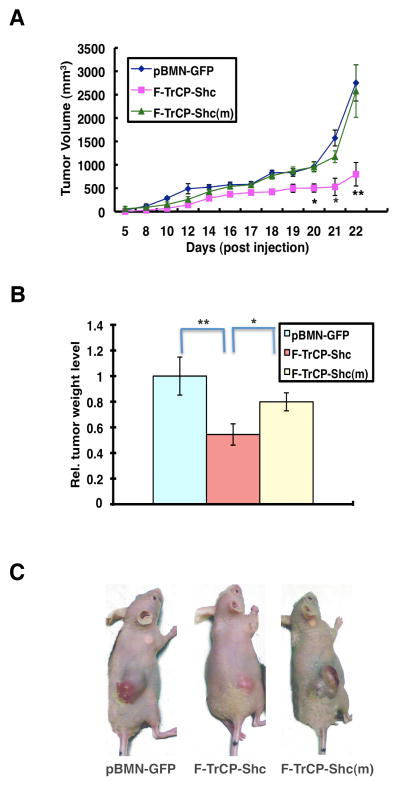
To assess whether the targeted degradation of ErbB RTKs by F-TrCP-Shc inhibits the tumorigenicity of BT474 cells, we subcutaneously injected 7×106 GFP+ BT474 cells expressing either pBMN-F-TrCP-Shc, pBMN-F-TrCP-Shc(m) or pBMN-GFP into nude mice. Expretssion of F-TrCP-Shc dramatically attenuated xenograft tumor growth compared to expression of F-TrCP-Shc(m) or pBMN-GFP (Fig. 7). At week 3, the average tumor weight of F-TrCP-Shc-expressing BT474 xenografts was 50% less than pBMN-GFP- and F-TrCP-Shc(m)-expressing BT474 xenografts (P<0.01 and P<0.05, respectively) (Fig. 7B). Collectively, these data indicate that the targeted degradation of activated ErbB family proteins significantly reduced the oncogenic potential of ErbB2-overexpressing breast cancer cells, and sensitized those cells to the cisplatin chemotherapeutic agent.
Figure 7. The engineered F-TrCP-Shc E3 ligase suppressed BT474 xenograft tumor growth in nude mice.
A. Nude mice were injected subcutaneously in the right flank with 5×106 BT474 cells infected with pBMN-F-TrCP-Shc, pBMN-F-TrCP-Shc(m) or pBMN-GFP retroviruses. Tumor volumes were measured over a 4-week period and calculated by the formula: . B. Tumor weight (g) was recorded at the end of the experiment. Columns, mean value of tumor weight; bars, SD. C. Representative images of tumor-bearing mice. * P<0.05 or **P<0.01 indicate tumors expressing pBMN-F-TrCP-Shc compared to those infected with pBMN-GFP or pBMN-F-TrCP-Shc(m).
DISCUSSION
Given the extensive crosstalk among the ErbB RTKs, it is plausible that blocking signaling from more than one family member may be necessary to effectively eradicate tumors and limit the occurrence of drug resistance. Currently, there are no effective tools or reagents available that can simultaneously abrogate the functions of all ErbB RTKs, and no studies have evaluated whether broad ErbB RTK inactivation preferentially kills tumor versus normal cells. Moreover, while the complete eradication of ErbB RTK expression is highly toxic to all cells, selectively targeting the active, tyrosine-phosphorylated forms of ErbB RTKs may be an effective strategy to preferentially kill tumor cells. However, more refined tools are required to achieve this delicate selection.
Targeting proteins for ubiquitination and subsequent proteasomal degradation is the primary mode of regulating the abundance of cellular proteins on the post-transcriptional level. Here, we showed that fusion of the Shc PTB domain to the βTrCP ubiquitin ligase allows for the selective recruitment of the activated subpopulations of the ErbB family of RTKs to the SCF machinery for ubiquitination and proteasome-dependent degradation, and effectively attenuates MAPK and PI3K signaling. Simultaneous degradation of all activated ErbB RTKs exerts dramatic cytotoxic effects on ErbB2-overexpressing breast cancer cells by inducing robust growth inhibition and apoptosis, and enhancing sensitivity to the chemotherapy drug cisplatin (Figs. 4–5). Our findings are in sharp contrast to ErbB tyrosine kinase inhibitors that primarily induce cytostatic attenuation of tumor growth (29).
Major inroads for targeted therapy against ErbB2-positive breast cancers were achieved with the development of trastuzumab, a monoclonal antibody that targets the extracellular domain of ErbB2. However, a majority of patients with advanced disease, as well as a proportion of those receiving treatment for operable disease, experience metastatic progression within one year of treatment because of the development of resistance to trastuzumab (39–41). One such mechanism of resistance is the compensatory upregulation of other ErbB RTKs, e.g. ErbB3, which effectively restores ErbB signaling through the PI3K/Akt axis of signaling and promotes the survival of tumor cells despite the continued presence of trastuzumab (29) (42). Moreover, ErbB3 lacks a kinase domain that renders it refractory to small-molecule inhibitors. Our F-TrCP-Shc construct addresses this particular concern by successfully targeting activated ErbB3 for proteasomal degradation (Fig. 2A–B).
Our engineered F-TrCP-Shc construct has the unique capability of selectively recruiting the active forms of all ErbB RTKs for degradation (Fig. 2). This unique property allows for the specific targeting of the oncogenic forms of ErbB RTKs while leaving the native forms intact. Strikingly, F-TrCP-Shc induced apoptosis, increased cisplatin sensitivity and reduced the tumorigenicity of ErbB2-overexpressing breast cancer cells, but demonstrated negligible toxicity in non-malignant mammary epithelial cells (Figs. 3–5). A somewhat surprising yet encouraging observation is that other cellular proteins that can interact with the Shc PTB domain and are theoretically also subjected to ubiquitin-dependent degradation by F-TrCP-Shc, such as c-Met, were not degraded in ErbB2-overexpressing BT474 cells (Fig. S4). This finding suggests that the engineered ubiquitin ligase preferentially engages with the active forms of Shc-binding partners (i.e., the ErbB RTKs). Collectively, our studies suggest that the perceived adverse effects upon the eradication of all ErbB RTKs can be mitigated by designing agents that target the activated subpopulations of ErbBs, thereby achieving the optimal tumoricidal outcome while minimizing the therapy-associated cytotoxic side-effects to normal cells.
The goal of the present study is to test the engineered ubiquitin ligase strategy for simulating the consequences of pharmacological eradication of the entire family of activated ErbB RTKs in ErbB-overexpressing tumor cells, and further assess the impact on non-malignant mammary epithelial cells. Compared with gene knockout approaches to assess the outcomes of ErbB RTK ablation, the engineered F-TrCP-Shc ubiquitin ligase is anticipated to more closely recapitulate the actions of pharmaceutical agents. It reduces but does not completely eliminate its intended targets, and therefore may not elicit the same severity of consequences as those that could arise from gene knockout. Moreover, the unique properties of the protein knockout system in interrogating complex protein functions, including post-translational modifications, subcellular distributions and functional redundancy, offer a versatile and powerful investigative tool for the rapid assessment of specific traits in tumorigenesis, maintenance and resistance to therapy, and inform new and improved therapeutic modalities against human malignancy. Future studies should directly compare the therapeutic effects of eradicating a single or multiple ErbB family members in pre-clinical models, and evaluate the potential of pan-ErbB suppression as a feasible strategy for the development of the next generation of targeted therapies against ErbB-positive tumors. The engineered ubiquitin ligase system developed here should be widely applicable to the assessment of protein families or specific posttranslational modifications as potential new targets for therapeutic intervention.
EXPERIMENTAL PROCEDURES
Cell lines, antibodies and reagents
Human mammary carcinoma cell lines SKBR3 and BT474, and non-cancerous MCF-10A mammary epithelial cells were obtained from ATCC. Antibodies against β-actin, EGFR (1005), ErbB2(C-18), cyclin D1 (M-20), and p27 (C-19) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA), Akt, p-Akt (Ser473), p38MAPK, p-MAPK, p-EGFR (Y1148), p-ErbB2 (Y1248), ErbB3, and p-ErbB3 (pY1328) from Cell Signaling (Andover, MA, USA). Anti-ErbB2 antibodies used for immunofluorescence and FACS analysis were purchased from R&D Systems (Minneapolis, MN, USA) and Santa Cruz, respectively. SYBR Green universal master mix and Multiscript RT were purchased from Applied Biosystems (Foster City, CA, USA). Cisplatin from Enzo Life Sciences (Plymouth Meeting, PA, USA), and the proteasome inhibitor MG132 and lysosomal inhibitor chloroquine from Calbiochem (Billerica, MA, USA).
Plasmid construction, transfection and retroviral infection
The PTB domain (1–209 aa) and mutant PTB(F198V) from Shc were fused to carboxyl terminus of FLAG-tagged βTrCP, and the resulting F-TrCP-Shc and F-TrCP-Shc(F198V) were subcloned into the BamHI/NotI sites of the retroviral pBMN-GFP vector to generate recombinant retroviruses (43) for infection of SKBR3, BT474 and MCF10A cells, as detailed in (44). Fluorescence-activated cell sorting (FACS) analysis was carried out to isolate infected cells, which was based on the expression of GFP from the recombinant retroviruses. Immunoprecipitation and Western blot analysis were carried out as described (45). F-TrCP-Shc was also subcloned into the pAdTrack-CMV adenoviral vector to generate recombinant adenoviruses for infection of SKBR3 cells (46).
Flow cytometry analysis of cell membrane-localized ErbB2
Flow cytometric analysis of cell membrane-localized ErbB2 was performed on live SKBR3 cells. Briefly, SKBR3 cells infected with pBMN-F-TrCP-Shc, pBMN-F-TrCP-Shc(m) or pBMN-GFP retroviruses were trypsinized, washed in ice-cold FACS buffer (2% FBS, 2% BSA in PBS containing 0.02% sodium azide) and serially stained with primary (anti-ErbB2) and secondary APC-conjugated anti-goat IgG antibodies (R&D Systems, Minneapolis, MN, USA) on ice in the same buffer. Stained cells were analyzed using the BD™ LSR II flow cytometer (BD Biosciences, San Jose, CA, USA).
Cell cycle, cell proliferation, apoptosis, and focus formation assays
For cell cycle analysis, cells were incubated with 10 mM BrdU for 30 min at 37°C, harvested and fixed overnight in 70% ethanol, and analyzed by flow cytometry following incubation with FITC-conjugated anti-BrdU antibodies (BD Pharmingen™ FITC-BrdU Flow kit, BD Biosciences, San Jose, CA, USA) and counterstaining with PBS containing 10 mg/ml propidium iodide. Cell proliferation rate was determined by plating 2×104 cells and counting at the indicated time points (Figs. 3B, 4B, and 5D) using an automated cell counter (Invitrogen, Grand Island, NY, USA). Each experiment was carried out in triplicate and repeated twice. The results are expressed as the mean ± SD for each set of experiments. Apoptosis was measured by Annexin V-APC and 7-AAD staining and flow cytometry using the Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences, San Jose, CA, USA). To measure focus formation, 3.0×103 SKBR3 cells or 6.0×103 BT474 cells were seeded in 6-well plates for 10 days, stained with 0.2% crystal violet, and foci were counted. Images of the colonies were obtained using a NIKON digital camera.
In vitro drug sensitivity assay
BT474 or SKBR3 cells were seeded in 96-well plates (5,000 cells/well) for 24 hr, treated with the specified concentrations of cisplatin (Fig. 6A–B) for 48 hr, and cell viability detected by 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5- [(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) using the Cell Prolifereation Kit II (Roche, Indianapolis, IN, USA).
BT474 tumor xenografts
A single 17 β-estradiol pellet (Innovative Research of America, Sarasota, FL, USA) was implanted subcutaneously into each 6-wk-old female athymic nude mouse (Harlan Laboratories, Indianapolis, IN, USA) 1 day before cell injection. 5×106 BT474 cells were suspended in Matrigel basement membrane matrix (BD Biosciences), and injected subcutaneously into the right flank. Tumor xenografts were measured with calipers three times/week and tumor volume was determined using the formula: . he results are presented as mean ± SEM.
Statistical analysis
Statistical analysis was performed with the SPSS15.0 software package for Windows by using the Student’s t-test for independent groups. Statistical significance was based on a value of P≤0.05. Data are expressed as mean ± SD.
Supplementary Material
Acknowledgments
We thank Bruce Mayer (University of Connecticut) for Shc plasmids, Alexandra Grassian for helpful discussions, and Jeffrey Hannah and Jennifer Lee for critical reading of the manuscript. This work is supported by National Institutes of Health Grant CA92792, the Irma T. Hirschl Career Scientist Award and the Speakers Fund from the New York Academy of Medicine to P.Z., and the Natural Science Foundation of China (No. 30901754) and Innovation Funding for Graduates of Tianjin Medical University, third Phase of the 211 Project for Higher Education (No. 2010GSI01).
References
- 1.Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10578–82. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hynes NE, Stern DF. The biology of erbB-2/neu/HER-2 and its role in cancer. Biochim Biophys Acta. 1994 Dec 30;1198(2–3):165–84. doi: 10.1016/0304-419x(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 3.Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988 Jul 1;54(1):105–15. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 4.Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000 Jul 3;19(13):3159–67. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001 Feb;2(2):127–37. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 6.Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol. 2009 Apr;21(2):177–84. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 7.Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N Engl J Med. 2005 Jul 14;353(2):172–87. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- 8.Zhou P, Fernandes N, Dodge IL, Reddi AL, Rao N, Safran H, et al. ErbB2 degradation mediated by the co-chaperone protein CHIP. J Biol Chem. 2003 Apr 18;278(16):13829–37. doi: 10.1074/jbc.M209640200. [DOI] [PubMed] [Google Scholar]
- 9.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005 May;5(5):341–54. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 10.Rabindran SK. Antitumor activity of HER-2 inhibitors. Cancer Lett. 2005 Sep 8;227(1):9–23. doi: 10.1016/j.canlet.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Klapper LN, Waterman H, Sela M, Yarden Y. Tumor-inhibitory antibodies to HER-2/ErbB-2 may act by recruiting c-Cbl and enhancing ubiquitination of HER-2. Cancer Res. 2000 Jul 1;60(13):3384–8. [PubMed] [Google Scholar]
- 12.Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12(4):220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou P. Targeted protein degradation. Curr Opin Chem Biol. 2005;9(1):51–5. doi: 10.1016/j.cbpa.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Zhou P, Bogacki R, McReynolds L, Howley PM. Harnessing the ubiquitination machinery to target the degradation of specific cellular proteins [In Process Citation] Mol Cell. 2000;6(3):751–6. doi: 10.1016/s1097-2765(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Zheng N, Zhou P. Exploring the functional complexity of cellular proteins by protein knockout. Proc Natl Acad Sci U S A. 2003;100(24):14127–32. doi: 10.1073/pnas.2233012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong F, Zhang J, Pao W, Zhou P, Varmus H. A protein knockdown strategy to study the function of beta-catenin in tumorigenesis. BMC Mol Biol. 2003;4(1):10. doi: 10.1186/1471-2199-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su Y, Ishikawa S, Kojima M, Liu B. Eradication of pathogenic beta-catenin by Skp1/Cullin/F box ubiquitination machinery. Proc Natl Acad Sci U S A. 2003 Oct 28;100(22):12729–34. doi: 10.1073/pnas.2133261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen JC, Scott D, Miller J, Zhang J, Zhou P, Larson JE. Transient in utero knockout (TIUKO) of C-MYC affects late lung and intestinal development in the mouse. BMC Dev Biol. 2004;4(1):4. doi: 10.1186/1471-213X-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Stevens J, Matsunami N, White RL. Targeted degradation of beta-catenin by chimeric F-box fusion proteins. Biochem Biophys Res Commun. 2004 Jan 23;313(4):1023–9. doi: 10.1016/j.bbrc.2003.12.035. [DOI] [PubMed] [Google Scholar]
- 20.Chen W, Lee J, Cho SY, Fine HA. Proteasome-mediated destruction of the cyclin a/cyclin-dependent kinase 2 complex suppresses tumor cell growth in vitro and in vivo. Cancer Res. 2004 Jun 1;64(11):3949–57. doi: 10.1158/0008-5472.CAN-03-3906. [DOI] [PubMed] [Google Scholar]
- 21.Hannah J, Zhou P. Maximizing target protein ablation by integration of RNAi and protein knockout. Cell Res. 2011 Jul;21(7):1152–4. doi: 10.1038/cr.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suenaga A, Hatakeyama M, Kiyatkin AB, Radhakrishnan R, Taiji M, Kholodenko BN. Molecular dynamics simulations reveal that Tyr-317 phosphorylation reduces Shc binding affinity for phosphotyrosyl residues of epidermal growth factor receptor. Biophys J. 2009 Mar 18;96(6):2278–88. doi: 10.1016/j.bpj.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006 Jan 12;439(7073):168–74. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 24.Sithanandam G, Anderson LM. The ERBB3 receptor in cancer and cancer gene therapy. Cancer Gene Ther. 2008 Jul;15(7):413–48. doi: 10.1038/cgt.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol. 2005;1:2005–0008. doi: 10.1038/msb4100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yajnik V, Blaikie P, Bork P, Margolis B. Identification of residues within the SHC phosphotyrosine binding/phosphotyrosine interaction domain crucial for phosphopeptide interaction. J Biol Chem. 1996 Jan 26;271(4):1813–6. doi: 10.1074/jbc.271.4.1813. [DOI] [PubMed] [Google Scholar]
- 27.Carraway KL., 3rd E3 ubiquitin ligases in ErbB receptor quantity control. Semin Cell Dev Biol. 2010 Dec;21(9):936–43. doi: 10.1016/j.semcdb.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longva KE, Blystad FD, Stang E, Larsen AM, Johannessen LE, Madshus IH. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J Cell Biol. 2002 Mar 4;156(5):843–54. doi: 10.1083/jcb.200106056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007 Jan 25;445(7126):437–41. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panigada M, Porcellini S, Barbier E, Hoeflinger S, Cazenave PA, Gu H, et al. Constitutive endocytosis and degradation of the pre-T cell receptor. J Exp Med. 2002 Jun 17;195(12):1585–97. doi: 10.1084/jem.20020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2008 Oct 15;314(17):3093–106. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castaneda CA, Cortes-Funes H, Gomez HL, Ciruelos EM. The phosphatidyl inositol 3-kinase/AKT signaling pathway in breast cancer. Cancer Metastasis Rev. 2010 Dec;29(4):751–9. doi: 10.1007/s10555-010-9261-0. [DOI] [PubMed] [Google Scholar]
- 33.Yang G, Cai KQ, Thompson-Lanza JA, Bast RC, Jr, Liu J. Inhibition of breast and ovarian tumor growth through multiple signaling pathways by using retrovirus-mediated small interfering RNA against Her-2/neu gene expression. J Biol Chem. 2004 Feb 6;279(6):4339–45. doi: 10.1074/jbc.M311153200. [DOI] [PubMed] [Google Scholar]
- 34.Faltus T, Yuan J, Zimmer B, Kramer A, Loibl S, Kaufmann M, et al. Silencing of the HER2/neu gene by siRNA inhibits proliferation and induces apoptosis in HER2/neu-overexpressing breast cancer cells. Neoplasia. 2004 Nov-Dec;6(6):786–95. doi: 10.1593/neo.04313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmieri D, Bouadis A, Ronchetti R, Merino MJ, Steeg PS. Rab11a differentially modulates epidermal growth factor-induced proliferation and motility in immortal breast cells. Breast Cancer Res Treat. 2006 Nov;100(2):127–37. doi: 10.1007/s10549-006-9244-6. [DOI] [PubMed] [Google Scholar]
- 36.Worster DT, Schmelzle T, Solimini NL, Lightcap ES, Millard B, Mills GB, et al. Akt and ERK control the proliferative response of mammary epithelial cells to the growth factors IGF-1 and EGF through the cell cycle inhibitor p57Kip2. Sci Signal. 2012 Mar 6;5(214):ra19. doi: 10.1126/scisignal.2001986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross JS, Fletcher JA. The HER-2/neu oncogene in breast cancer: prognostic factor, predictive factor, and target for therapy. Stem Cells. 1998;16(6):413–28. doi: 10.1002/stem.160413. [DOI] [PubMed] [Google Scholar]
- 38.Colomer R, Montero S, Lluch A, Ojeda B, Barnadas A, Casado A, et al. Circulating HER2 extracellular domain and resistance to chemotherapy in advanced breast cancer. Clin Cancer Res. 2000 Jun;6(6):2356–62. [PubMed] [Google Scholar]
- 39.Burstein HJ, Kuter I, Campos SM, Gelman RS, Tribou L, Parker LM, et al. Clinical activity of trastuzumab and vinorelbine in women with HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2001 May 15;19(10):2722–30. doi: 10.1200/JCO.2001.19.10.2722. [DOI] [PubMed] [Google Scholar]
- 40.Marty M, Cognetti F, Maraninchi D, Snyder R, Mauriac L, Tubiana-Hulin M, et al. Randomized phase II trial of the efficacy and safety of trastuzumab combined with docetaxel in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer administered as first-line treatment: the M77001 study group. J Clin Oncol. 2005 Jul 1;23(19):4265–74. doi: 10.1200/JCO.2005.04.173. [DOI] [PubMed] [Google Scholar]
- 41.Gasparini G, Gion M, Mariani L, Papaldo P, Crivellari D, Filippelli G, et al. Randomized Phase II Trial of weekly paclitaxel alone versus trastuzumab plus weekly paclitaxel as first-line therapy of patients with Her-2 positive advanced breast cancer. Breast Cancer Res Treat. 2007 Mar;101(3):355–65. doi: 10.1007/s10549-006-9306-9. [DOI] [PubMed] [Google Scholar]
- 42.Garrett JT, Olivares MG, Rinehart C, Granja-Ingram ND, Sanchez V, Chakrabarty A, et al. Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. Proc Natl Acad Sci U S A. 2011 Mar 22;108(12):5021–6. doi: 10.1073/pnas.1016140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Persons DA, Mehaffey MG, Kaleko M, Nienhuis AW, Vanin EF. An improved method for generating retroviral producer clones for vectors lacking a selectable marker gene. Blood Cells Mol Dis. 1998 Jun;24(2):167–82. doi: 10.1006/bcmd.1998.0184. [DOI] [PubMed] [Google Scholar]
- 44.Chen X, Zhang J, Lee J, Lin PS, Ford JM, Zheng N, et al. A kinase-independent function of c-Abl in promoting proteolytic destruction of damaged DNA binding proteins. Mol Cell. 2006 May 19;22(4):489–99. doi: 10.1016/j.molcel.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 45.Chen X, Zhang Y, Douglas L, Zhou P. UV-damaged DNA-binding Proteins Are Targets of CUL-4A-mediated Ubiquitination and Degradation. J Biol Chem. 2001;276(51):48175–82. doi: 10.1074/jbc.M106808200. [DOI] [PubMed] [Google Scholar]
- 46.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95(5):2509–14. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.