Abstract
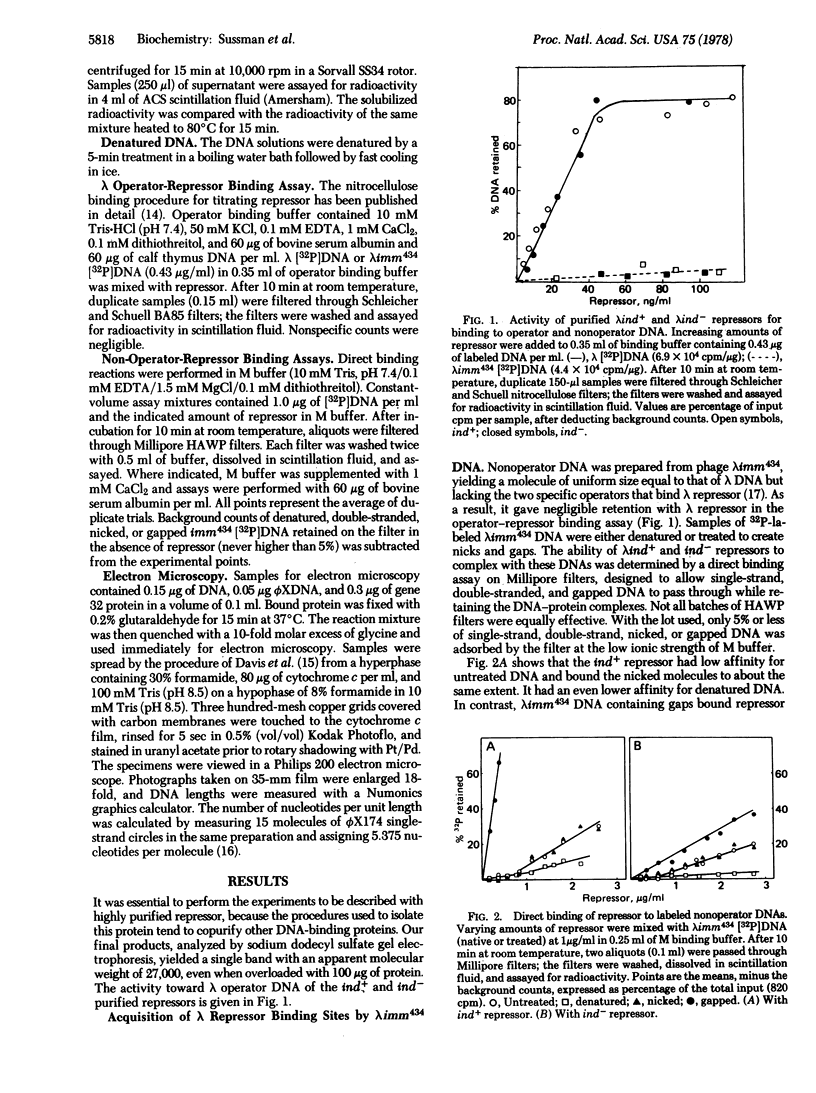
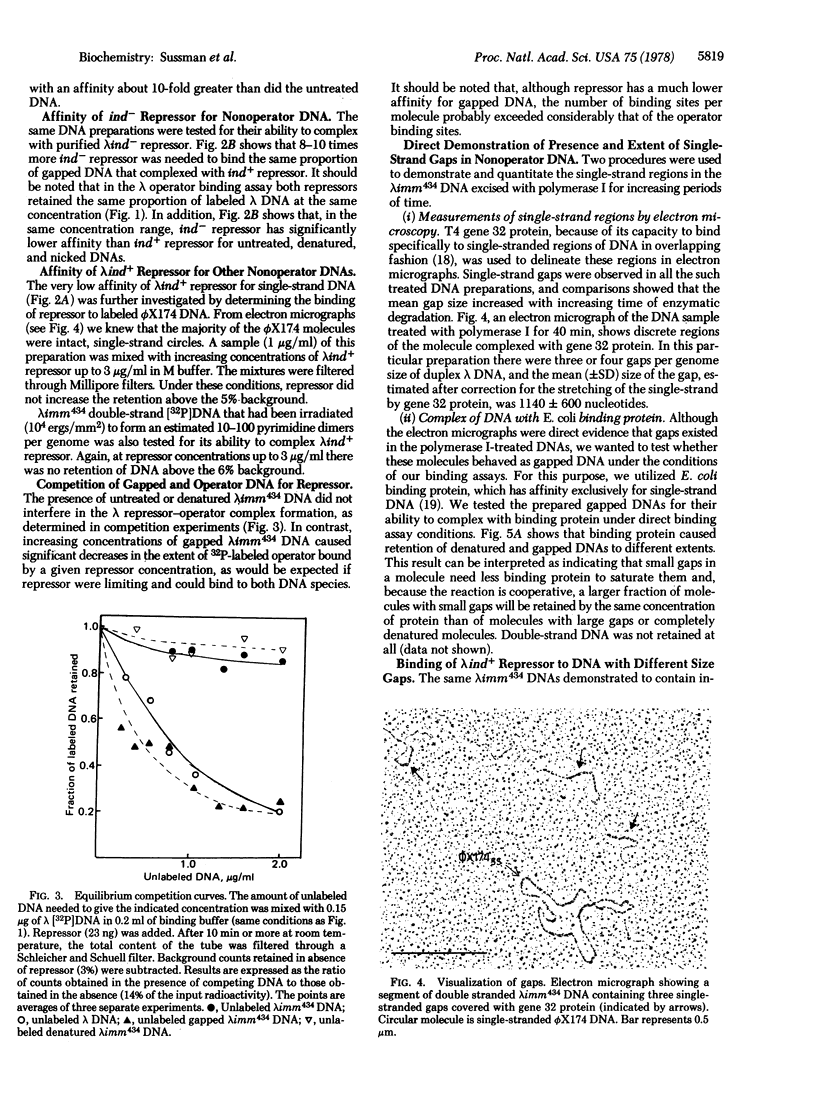
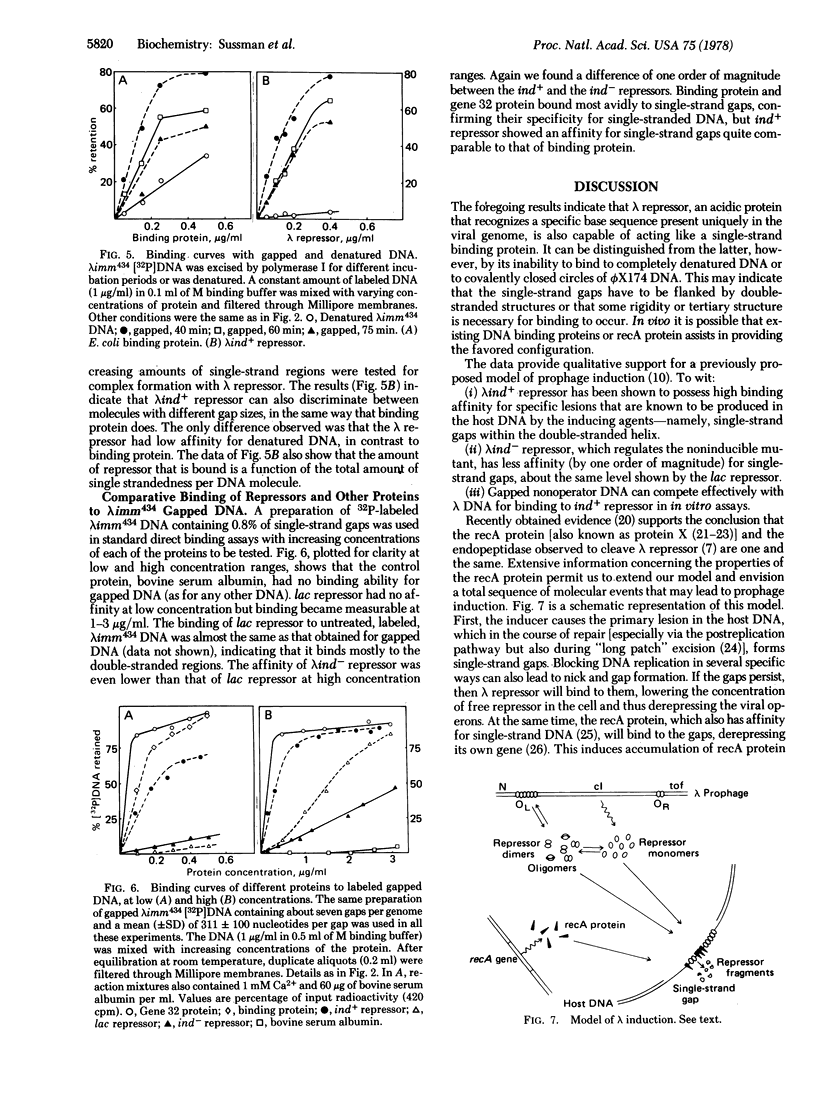
In direct binding assays, purified lambdaind+ repressor displayed high affinity for nonoperator DNA containing single-strand gaps. Its affinity for this same DNA but completely double-stranded, nicked, or denatured was considerably lower. In contrast, purified lambdaind- repressor had 1/10th the affinity for the gapped DNA, a level comparable to that of purified lac repressor. In the presence of limiting amounts of ind+ repressor, nonoperator DNA containing gaps could be shown to compete effectively with lambda DNA for binding of repressor. A previous model of lambda induction [Sussman, R. & Ben-Zeev, H. (1975) Proc. Natl. Acad. Sci. USA 72, 1973--1976], based on the assumption that this phenomenon involves the binding of repressor to lesions in the host DNA, is reevaluated in the light of the data reported here.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M., Frey L. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970 Sep 26;227(5265):1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- Backman K., Ptashne M., Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluch J., Sussman R. Correlation between UV dose requirement for lambda bacteriophage induction and lambda repressor concentration. J Virol. 1978 Jun;26(3):595–602. doi: 10.1128/jvi.26.3.595-602.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delius H., Mantell N. J., Alberts B. Characterization by electron microscopy of the complex formed between T4 bacteriophage gene 32-protein and DNA. J Mol Biol. 1972 Jun 28;67(3):341–350. doi: 10.1016/0022-2836(72)90454-8. [DOI] [PubMed] [Google Scholar]
- Emmerson P. T., West S. C. Identification of protein X of Escherichia coli as the recA+/tif+ gene product. Mol Gen Genet. 1977 Sep 21;155(1):77–85. doi: 10.1007/BF00268563. [DOI] [PubMed] [Google Scholar]
- Grossman L., Braun A., Feldberg R., Mahler I. Enzymatic repair of DNA. Annu Rev Biochem. 1975;44:19–43. doi: 10.1146/annurev.bi.44.070175.000315. [DOI] [PubMed] [Google Scholar]
- Gudas L. J., Mount D. W. Identification of the recA (tif) gene product of Escherichia coli. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5280–5284. doi: 10.1073/pnas.74.12.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudas L. J., Pardee A. B. DNA synthesis inhibition and the induction of protein X in Escherichia coli. J Mol Biol. 1976 Mar 15;101(4):459–477. doi: 10.1016/0022-2836(76)90240-0. [DOI] [PubMed] [Google Scholar]
- JACOB F., CAMPBELL A. Sur le système de répression assurant l'immunité chez les bactéries lysogenes. C R Hebd Seances Acad Sci. 1959 Jun 1;248(22):3219–3221. [PubMed] [Google Scholar]
- KAISER A. D., JACOB F. Recombination between related temperate bacteriophages and the genetic control of immunity and prophage localization. Virology. 1957 Dec;4(3):509–521. doi: 10.1016/0042-6822(57)90083-1. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- LWOFF A., SIMINOVITCH L., KJELDGAARD N. Induction de la production de bacteriophages chez une bactérie lysogène. Ann Inst Pasteur (Paris) 1950 Dec;79(6):815–859. [PubMed] [Google Scholar]
- McEntee K., Hesse J. E., Epstein W. Identification and radiochemical purification of the recA protein of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3979–3983. doi: 10.1073/pnas.73.11.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noack D., Klaus S. Inactivation kinetics of lambda phage repressors in a mutant of E. coli temperature sensitive in DNA replication. Mol Gen Genet. 1972;115(3):216–224. doi: 10.1007/BF00268885. [DOI] [PubMed] [Google Scholar]
- Pollard E. C. Induced radioresistance in four strains of Escherichia coli, two with lambda lysogens. Biophys J. 1978 Jun;22(3):431–438. doi: 10.1016/S0006-3495(78)85497-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. W., Roberts C. W., Mount D. W. Inactivation and proteolytic cleavage of phage lambda repressor in vitro in an ATP-dependent reaction. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2283–2287. doi: 10.1073/pnas.74.6.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. W., Roberts C. W. Proteolytic cleavage of bacteriophage lambda repressor in induction. Proc Natl Acad Sci U S A. 1975 Jan;72(1):147–151. doi: 10.1073/pnas.72.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Anderegg R. Primary structure of the lambda repressor. Biochemistry. 1978 Mar 21;17(6):1092–1100. doi: 10.1021/bi00599a024. [DOI] [PubMed] [Google Scholar]
- Schuster H., Beyersmann D., Mikolajczyk M., Schlicht M. Prophage induction by high temperature in thermosensitive dna mutants lysogenic for bacteriophage lambda. J Virol. 1973 Jun;11(6):879–885. doi: 10.1128/jvi.11.6.879-885.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick S. G., Levine A., Bailone A. Induction of recA+-protein synthesis in Escherichia coli. Mol Gen Genet. 1978 Apr 17;160(3):267–276. doi: 10.1007/BF00332970. [DOI] [PubMed] [Google Scholar]
- Shinagawa H., Ito T. Inactivation of DNA-binding activity of repressor in extracts of lambda-lysogen treated with mitomycin C. Mol Gen Genet. 1973 Nov 2;126(2):103–110. doi: 10.1007/BF00330987. [DOI] [PubMed] [Google Scholar]
- Shinagawa H., Mizuuchi K., Emmerson P. T. Induction of prophage lambda by gamma-rays, mitomycin C and tif; repressor cleavage studied by immunoprecipitation. Mol Gen Genet. 1977 Sep 21;155(1):87–91. doi: 10.1007/BF00268564. [DOI] [PubMed] [Google Scholar]
- Sigal N., Delius H., Kornberg T., Gefter M. L., Alberts B. A DNA-unwinding protein isolated from Escherichia coli: its interaction with DNA and with DNA polymerases. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3537–3541. doi: 10.1073/pnas.69.12.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Oishi M. Early events and mechanisms in the induction of bacterial SOS functions: analysis of the phage repressor inactivation process in vivo. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1657–1661. doi: 10.1073/pnas.75.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg R. A., Ptashne M. In vitro repression of RNA synthesis by purified lambda phage repressor. Nat New Biol. 1971 Mar 17;230(11):76–80. doi: 10.1038/newbio230076a0. [DOI] [PubMed] [Google Scholar]
- Sussman R., Zeev H. B. Proposed mechanism of bacteriophage lambda induction: acquisition of binding sites for lambda repressor by DNA of the host. Proc Natl Acad Sci U S A. 1975 May;72(5):1973–1976. doi: 10.1073/pnas.72.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin E. M. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976 Dec;40(4):869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]



