Abstract
Hybridization between species is a genomic instability factor involved in increasing mutation rate and new chromosomal rearrangements. Evidence of a relationship between interspecific hybridization and transposable element mobilization has been reported in different organisms, but most studies are usually performed with particular TEs and do not discuss the real effect of hybridization on the whole genome. We have therefore studied whole genome instability of Drosophila interspecific hybrids, looking for the presence of new AFLP markers in hybrids. A high percentage (27–90%) of the instability markers detected corresponds to TEs belonging to classes I and II. Moreover, three transposable elements (Osvaldo, Helena and Galileo) representative of different families, showed an overall increase of transposition rate in hybrids compared to parental species. This research confirms the hypothesis that hybridization induces genomic instability by transposition bursts and suggests that genomic stress by transposition could contribute to a relaxation of mechanisms controlling TEs in the Drosophila genome.
Introduction
Natural hybridization is a well known phenomenon in organisms living in sympatry and constitutes an important mechanism of speciation [1], [2]. Whereas a large number of natural hybridization examples were reported in plants [3], [4], natural hybrid reports in animals are less frequent likely due to the lack of suitable markers for their detection. The importance of hybridization in evolution of species has been debated for decades and is under major reevaluation [1], [2], [5], [6]. One of the most evaluated consequences of hybridization is that the merging of two different genomes triggers a “genomic shock” leading to genomic modifications including cascades of new gene expressions often accompanied by transposable element (TE) mobilizations [7]. TEs are found in genomes of almost all living organisms [8]. Considered as enigmatic sequences with an uncertain role in the genome, we now know their importance in the building of the genome [9] and in particular its regulation [9]. Their mobility in the genome is usually regulated at a low transposition frequencies, which may greatly increase when different stresses deregulate them [10]. Barbara McClintock in the early 1980s proposed that TEs could be activated by a genomic shock, like hybridization, which could confer an adaptive value to the host. The role of TEs has been ascribed to hybrid incompatibility and speciation [11], [12], intraspecific crosses between different Drosophila strains can trigger the mobilization of different TEs. The best known examples are hybrid dysgenesis mechanisms P-M [13], [14], I-R [15], H-E [16] in D. melanogaster and the dysgenesis of Penelope and other elements in D. virilis [17]. Different elements are sometimes mobilized simultaneously [17] due to an absence of maternal piRNAs impeding the mobilization of paternally inherited families in dysgenic crosses [18] and D. simulans intraspecific crosses involving the retrotransposon tirant [19]. As in Drosophila, most of the organisms have activated mechanisms of epigenetic control to avoid the negative effect produced by high levels of transposition created by genomic stress. During interspecific crosses TE seem to escape to the genomic control allowing their mobilization and the creation of new insertions. Genomic stresses due to hybridization have an impact on TE mobilization and activity in plants, mammals and insects [20]. The early more numerous examples of increase of TE activity in interspecific hybrids have been reported in plants [21]–[24]. For example, in interspecific hybrids of Solanum lycopersicum and Solanum pennellii epigenetic and gene expression changes due to accumulation of transgressive small RNAs in the hybrid genomes were found [25]. In Drosophila, interspecific hybrids have given sometimes different results; while interspecific hybrids between species from the melanogaster subgroup [26] and affinis [27] subgroup do not have an increased transposition rate, two well described examples of TEs activation associated to interspecific hybridization are known. The first one concerns kangaroos, where an induction of TE transposition and centromeric expansion was observed in hybrids of wallabies [28], [29]. The second example was found in Drosophila where D. buzzatii and D. koepferae hybrids underwent an increase of transposition of the Osvaldo retrotransposon of one order of magnitude higher than in the parental species [30], [31]. The scarcity of animal data is probably due to the difficulty of obtaining a large number of hybrid individuals to perform experimental crosses, technical difficulties in transposition detection, or simply because the direct detection of transposition events is difficult.
Previous work on the same species were restricted to the behaviour of a single element [30], [31]. We ignore whether, in response to genomic stresses, TE mobilization is a universal response for all genomic TEs or, on the contrary, it depends on the kind of element. If this were the case, it would explain why no TE mobilization was found in other Drosophila species. We undertook a genome-wide dissection of TE mobilization in Drosophila interspecific hybrids, including a quantitative estimation of transposition rates of some TEs. Knowledge of the impact of hybridization in TE activation is a prerequisite for understanding the implication of TEs in reproductive isolation, speciation and, eventually, in the evolution of hybrid lineages.
Materials and Methods
Drosophila Stocks
D. buzzatii Bu28 stock was originated by the fusion of 4 stocks (LN13, 19, 31 and 33) collected in Bolivia (Los Negros) and D. koepferae Ko2 stock collected in Argentina (San Luis Valley). Both stocks correspond to inbred lines maintained by brother–sister matings for several years and kept thereafter by mass culturing.
Crosses
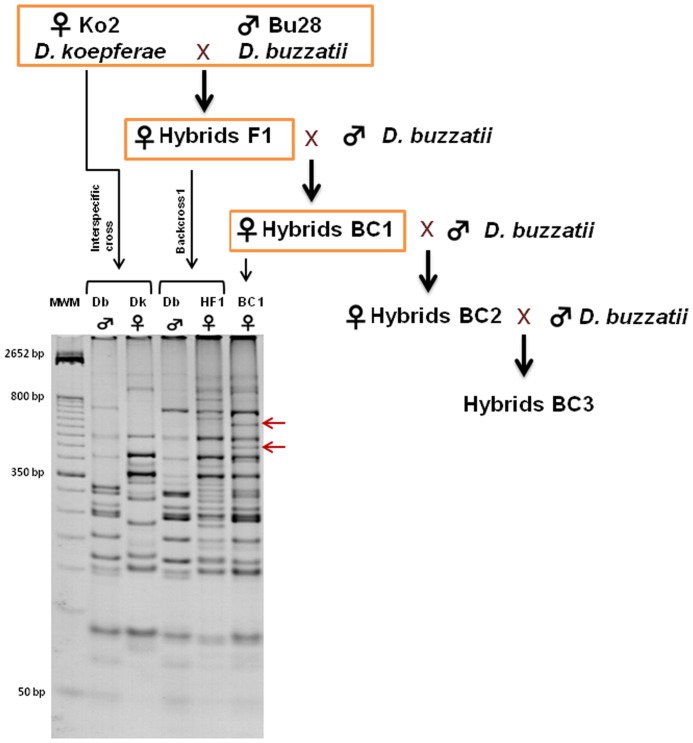
50 interspecific crosses (Figure 1) were established between 2 D. koepferae females and one D. buzzatii male. While the backcross 1 (BC1) was carried out by mass crossing, backcrosses 2 (BC2) and 3 (BC3) were done by individual crosses between a fertile hybrid female and a D. buzzatii male. Due to the presence of high number of sterile or semi-sterile females, especially in the F1, we only obtained the complete progeny (from F1 to BC3) of 3 crosses. Families named 10, 13 and 40, were analyzed by AFLP and transposon display and one additional family (family 1) was added to the transposon display experiments (see transposon display section). A segmental hybrid stock F3–F4 was also analyzed by AFLP; these segmental hybrids carry the F3–F4 region of chromosome 4 from D. koepferae introgressed in the genomic background of D. buzzatii. This cytological region was selected for 60 generations by cytological observations of polytene chromosomes. Simultaneously with the first hybrid crosses, individual intraspecific crosses of D. buzzatii and D. koepferae parental species were also established and kept for 4 generations as controls in experiments on AFLP (families B4 and K9 of D. buzzatii and D. koepferae respectively) and transposon display (families B4, B8 of D. buzzatii and K3, K9 of D. koepferae).
Figure 1. Hybrid crosses and an example of AFLP gel showing bands of parental species and hybrids.
A) Interspecific cross and backcrosses used in experiments B) Selective PCR AFLP band patterns using primers with selective nucleotides GG (EcoRI) and CTG (MseI). The arrows indicate two instability markers detected in hybrids from backcross 1 as an example. MWM, molecular weight marker; Dk, D. koepferae; Db, D. buzzatii; HF1, F1 hybrids; BC1, backcross 1.
DNA Extraction and AFLPs
Individual DNA extractions from adult hybrids and individuals from the parental species involved in each cross,stored at −80°C, were performed as described in the Piñol protocol [32]. AFLP markers were obtained using the Vela protocol [33] where the DNA of each individual was digested by a frequent cutter enzyme (MseI) and an infrequent one (EcoRI) and then ligated to oligonucleotide adapters. Fragments, after linking an adapter to both extremities, are amplified with primers having a supplementary base. A second round of amplification was given with primers where 3 (for MseI) and 2 nucleotides (EcoRI) were added to the 3′ end of the initial primer sequence. The resulting bands were seen on 8% poly-acrylamide gels each containing individual samples from: parental species, parents and the hybrids at the generation being examined (Figure 1). To identify instability, each band present in hybrids was carefully checked for its presence in parents and parental species. Those in hybrids (backcrossed introgression individuals) but absent in parents, parental species and in hybrids of previous generations, were considered new AFLP bands (instability markers) of new genomic rearrangements induced by hybridization that were not exclusively associated to TEs. These selected instability markers were subsequently cloned, sequenced and analyzed. The AFLP technique was chosen because does not need prior knowledge of TEs sequence in species (not yet sequenced) under examination allowing a genome-wide screening from a random sampling of the whole genome.
Transposon Display
Transposon display is an AFLP-based specific technique [34], [35] allowing the simultaneous amplification of the TE insertions from a particular element. Individual transposons are identified by a ligation-mediated nested PCR that starts within the transposon and amplifies part of the flanking sequence up to a specific restriction site. The resulting PCR products are sequenced and the bands separated according to size. From the putative TE related sequences detected by AFLP as new bands, we have studied only those whose sequence known in our species (not yet sequenced) because this technique requires a previous knowledge of the TE sequence.
We estimated the transposition rates of Osvaldo, Helena and Galileo, three elements belonging to different TE classes and being well characterized in D. buzzatii. DNA, from parents and hybrids of each family cross and generation was individually digested with HpaII enzyme (Osvaldo and Galileo elements) and MseI (Helena element), in a total volume of 20 µl (10×enzyme buffer, 1U enzyme, 12 µl H2O and 5 µl of DNA) and incubated one hour at 37°C. A mix of adaptors was prepared with 10 µl of each adaptor (10 µM) and 80 µl H2O. The adaptors used for HpaII were: 5′AACAGCTGGACGATGAGTCCTGAGATACG 3′ and 5′CGCGTATCTCAGGAGTGTA 3′ and for MseI: 5′AAAAGCTGGACGATGAGTCCTGAGA 3′ and 5′TATCTCAGGAGTGTA 3′. The ligation reaction was done in a final volume of 42 µl, including 5×ligase buffer, 2.5 U of T4 DNA ligase, 0.2 µM adaptors mix, 10.7 µl of H2O and 20 µl of digestion product, and was later incubated for 1h at 24°C. External amplification was carried out in a final volume of 25 µl including: 1×PCR buffer, 0.2 µM of dNTPs, 0.4 µM MseI primer, 2 µM external primer, 0.625 U DNA polymerase, 17.9 µl H2O and 2 µl digestion-ligation product. External amplification was programmed as follows: 2 min 94°C, 25 cycles of [1 min 94°C, 1 min 58°C, 1 min 72°C], 4 min 72°C. The product of external amplification was diluted 20 times with water. The external primers used were: Osvaldo 5′AGCCCATTTGCTGACACTTTA3′, Helena 5′TGTTGTTGTCATGGTGCTGA3′, and Galileo 5′CATGGGGCAGAAAGAGAAAG3′. For the internal amplification, 1.5 µl of the external amplification product (diluted 20 times) was used as template by mixing 10×PCR Buffer, 0.2 µM dNTPs, 0.2 µM primer MseI, 0.25 µM internal primer, 1.25 U DNA polymerase and 15.1 µl of H2O. The PCR thermocycler was programmed as follows: 1 min 94°C, 35 cycles of [45 s 94°C, 45 s 58°C, 45 s 72°C], 3 min 72°C. In this last amplification step, specific labelled primers (4, 7, 2′, 4′, 5′, 7′-hexachloro-6-carboxyfluorescein (HEX) fluorochrome) were used: Osvaldo 5′CTCTCTGACCCTTCCAGTCG3′, Helena 5′GAATTCAGCCCTCAGCTCAA3′, and Galileo 5′TTTGGAAAATCGACCGTCAC3′.
Transposon display results were analyzed by capillary electrophoresis, to obtain fragment sizes by Biofidal sequencing services (Lyon, France), and analyzed using the Peak Scanner v4.0 software (Applied Biosystem, CA). Because one enzyme restriction site is inside the element and the other in the flanking sequence, the bands represent the number of insertions. Comparisons between band sizes in hybrid, parents and parental species allowed us to identify those present in hybrids and absent in parents and/or parental species (new bands), which correspond to transposition events.
To verify the reliability of the transposon display technique, several bands per element were sequenced showing that most of them corresponded to the TEs insertions being examinated.
FISH (Fluorescent in situ Hybridization)
Polytene chromosome squashes from salivary glands of third-instar larvae were hybridized with fluorescein labelled probes of Osvaldo, Helena and Galileo. The Osvaldo probe consisted of a 6.4-kb Osvaldo fragment [36], and those of Helena and Galileo consisted of PCR fragments of 440 and 1150 pb respectively containing endonuclease and transposase regions. Prehybridization solutions and posthybridization washes, were done following a protocol by Roche. PCR reactions were carried out in a final volume of 25 µl, including 1× activity buffer (Ecogen), 1.6 mM MgCl2, 0.2 mM of each dNTP (Roche), 0.4 µM primer (SigmaAldrich), 10–20 ng genomic template DNA, and 0.04 units per µl of Taq polymerase (Kapataq from Cultek). Amplifications were run in a MJ Research Inc. thermocycler programmed as follows: 5 min preliminary denaturation at 94°, 30 cycles of 45 s at 94° (denaturation), 45 s at specific PCR annealing temperatures, 1.5 min at 72° (extension) and a final extension for 10 min at 72°. PCR products were gel purified with NucleoSpin kit (Machery-Nagel) and labelled by PCR using Alexa Fluor 488 signal amplification kit (Roche). After hybridization, signal development was done with 2 antibodies: rabbit anti-fluorescein and goat anti-rabbit. Chromosomes were stained with 4′, 6-diamidino-2-phenylindole (DAPI) containing Vectashield mounting media and images taken with a fluorescence microscope.
Transposition Rates
Transposition rates (T) of Osvaldo, Helena and Galileo were estimated at each generation as: T = Ni/N.2.A [30] where Ni is the number of new insertions (those found in progeny, but not in parents or in the parental stock they come from), N the number of analyzed individuals, 2 being the number of parental genomes and A the number of original insertions (sum of parental insertions). The number of new and original insertions was computed from the copies amplified by transposon display for every TE and is provided as supplementary material (Tables S1). The transposition rate was estimated for 3 hybrid generations (BC1, BC2 and BC3) of 4 hybrid families (1, 10, 13, 40), and for the control families (B4, B8, K3, K9) of parental species at the fourth generation (F4). The transposon display technique cannot distinguish completely between transposition events in somatic and germinal line although if a new band is detected in multiple individuals from the same cross, it probably occurred in the parent germline. In contrast, if a new insertion event is present in only one individual of the progeny, it is impossible to distinguish between transpositions occurring in somatic cells or meiotic germline events.
RT-PCR
Males of D. buzzatii and females of D. koepferae parental species were dissected and their testes and ovaries extracted, respectively, in order to separate somatic (carcasses) and germinal (gonads) tissues, and to analyze the transcriptional activity of TEs in each tissue. For this analysis, 10 samples of each tissue were stored at −70°C in a buffer containing beta-mercaptoethanol and RLT buffer (Qiagen) to avoid RNA degradation. RNA extractions were done with RNeasy mini kit (Qiagen) according to the manufacturer’s protocol and treated with DNase I (Ambion) to eliminate DNA contamination. cDNA was synthesized using the Transcriptor First Strand cDNA Syntesis kit (Roche). RT-PCR reactions were carried out in a final volume of 25 µl including 10×PCR buffer, 0.8 mM MgCl2, 0.2 µM dNTPs, 0.8 µM primer 1, 0.8 µM primer 2, 1.2 U DNA polymerase, 10 pg cDNA and 13.9 µl H2O. Amplifications were run in a thermocycler programmed as follows: 5 min 94°C; 30 cycles of 30 sec at 94°C, 45 sec at 59°C and 30 sec at 72°C; and a final extension of 10 min at 72°C. Reverse transcriptase, endonuclease and transposase regions of Osvaldo, Helena and Galileo respectively, were amplified using the following RT-PCR primers: 5′GAGGCACGAACTGGAGAAAT3′ and 5′ACTCCCATTTGACGCCCTTT3′ for Osvaldo, 5′CGACATACTCGCTTCCTGTG3′ and 5′CAATGCAAGAGGGAGTGTGA3′ for Helena, and 5′TTGACACTCAACTTCCGAACC3′ and 5′TTTCAAACCCCTGAATCTCG3′ for Galileo.
Statistical Methods and Sequence Analyses
Most statistical analyses were performed using the statistical software SPSS version 19.0. The Kruskal and Wallis test [37] was done to see whether the differences in stability and transposition between hybrid families were significant. The Mann-Whitney test was used to compare genomic instability induced by transposition between hybrids and parental species.
For identification of AFLP marker sequences, a similarity was searched between the query sequences and TEs sequences of Genbank and Repbase [38] databases using the parameters defined by the CENSOR search tool (http://www.girinst.org/censor). The sequence data have been deposited in GenBank under the accession nos: JX997188; JX997190; JX997191; JX997192; JX997193; JX997194; JX997195: JX997196; JX997197; JX997198; JX997199; JX997200; JX997201; JX997202; JX997203; JX997204; JX997205; JX997206; JX997207; JX997208; JX997209; JX997210; JX997211; JX997212; JX997213; JX997214; JX997216; JX997217.
Results
AFLP band patterns of 40 primer combinations were analyzed in each backcross generation and 28 in segmental hybrids. AFLP instability markers were selected in 3 backcross generations of hybrids (BC1, BC2, BC3) and in segmental hybrids between D. buzzatii and D. koepferae. Instability markers homologous to TE sequences by analysis in the data bases, were considered transposition markers, resulting probably from new insertions of TEs in the hybrid genome.
Instability Markers
The number of different AFLP instability markers segregating in the genome of hybrids of BC1, BC2, BC3 generations was 30, 17, 25, respectively, and 11 in segmental hybrids. Its distribution by family was 19, in family 10; 26 in family 13, and 17, in family 40; the 10 remaining markers corresponding to other families where it was impossible to complete the 3 backcross generations (details Table S2). Parental species D. buzzatii and D. koepferae only showed 6 and 4 AFLP instability markers, respectively. The total number of instability markers segregating in the hybrid genomes was 83 (72 in interspecific hybrids and 11 in segmental hybrids) versus 10 markers in parental genomes. Among all instability markers, some corresponded to non repetitive sequences with homology to different regions of the D. mojavensis genome, and are supplied as supplementary material (Text S1). Comparisons of the total number of instability markers in BC3 show significant differences between hybrid families using a Kruskal & Wallis test (P = 0.01) (Table 1) but there were no significant differences between generations (not shown).
Table 1. Comparison of the number of instability markers in backcross 3 (BC3) between hybrid families.
| Kruskal-Wallis Test | |||
| χ2 | k | P value | |
| Total instability | 8.761 | 2 | 0.010** |
| Instability by transposition | 0.97 | 2 | 0.653 |
Sample size: 12, 12 and 16 flies for families 10, 13 and 40 respectively, χ2: chi square, k: degrees of freedom, **P≤0.01.
Transposition Markers
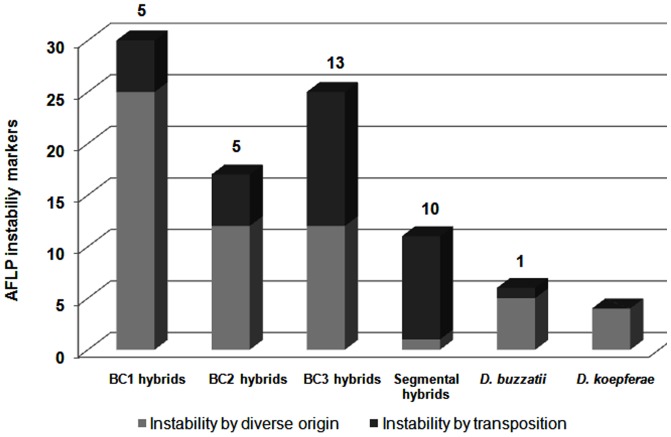
The number of transposition markers was 5, 5 and 13 in the BC1, BC2 and BC3 generations, respectively, and 10 in segmental hybrids (Figure 2). A total of 33 transposition markers were detected in the hybrid genomes whereas only one was found in the parental species (D. buzzatii). All except 3 transposition markers (CGGGG22, CACAT21 and TGCGG21) were longer that 200 bp (Table 2) and showed a wide range of similarity (0.1 to 0.94) with the TE sequences. In a few cases, transposition markers showed homology to internal or coding regions of one, or two TEs. This internal homology probably occurs because these TE regions are the most conserved in the database. The percentage of genomic instability due to transposition (number of transposition markers divided by the number of total markers in each backcross generation, multiplied by 100) was 16.6, 29.4 and 52.0% in the BC1, BC2 and BC3 generations, respectively.
Figure 2. Markers in hybrids and parental species.
Table 2. Sequences of AFLP markers showing homology to TE sequences.
| TEs | AFLP marker | Marker size | Pos | Sim | Homologous region | Hybrid | Genbank reference |
| Baggins1# | TGTCG21 | 377 bp | 0.52 | 0.38 | reverse transcriptase/RNAasa | SH | JX997206 |
| BATUMI* | CACTC22 | 500 bp | 0.60 | 0.60 | polyprotein | SH | JX997209 |
| Bel_1* | GGATT21 | 450 bp | 0.93 | 0.93 | internal region | BC1 | JX997190 |
| BS# | TGTCG22 | 443 bp | 0.71 | 0.62 | internal region | SH | JX997207 |
| CIRCE* | CACTC21 | 529 bp | 0.62 | 0.55 | internal region | SH | JX997212 |
| DIVER* | TGTCG23 | 415 bp | 0.63 | 0.33 | endonuclease/integrase | SH | JX997208 |
| Frogger* | CACAT21 | 153 bp | 0.61 | 0.41 | internal region | SH | not annotated |
| Galileo▪ | TGATT22 | 408 bp | 0.99 | 0.1 | internal region | BC3 | JX997200 |
| Galileo▪ | CGGGG22 | 172 bp | 0.75 | 0.60 | internal region | BC3 | not annotated |
| Gypsy1* | CAGCA21 | 491 bp | 0.83 | 0.70 | internal region | BC1 | JX997198 |
| Gypsy1* | CCCCC22 | 540 bp | 0.83 | 0.83 | internal region | BC1 | JX997192 |
| Gypsy1* | CCGAT22 | 264 bp | 0.72 | 0.64 | internal region | BC3 | JX997217 |
| Gypsy1* | TGATT21 | 554 bp | 0.52 | 0.34 | LTR | BC3 | JX997196 |
| Gypsy3* | CGAGT21 | 707 bp | 0.45 | 0.27 | polyprotein | SH | JX997211 |
| Gypsy8* | TGTCG21 | 377 bp | 0.45 | 0.32 | internal region | SH | JX997206 |
| Gypsy8* | TGATT26 | 505 bp | 0.70 | 0.70 | poliprotein | BC3 | JX997199 |
| Gypsy9* | CCGAT23 | 303 bp | 0.48 | 0.38 | internal region | BC3 | JX997204 |
| Gypsy10* | TGTCG23 | 415 bp | 0.71 | 0.64 | internal region | SH | JX997208 |
| Gypsy12* | CGGCA21 | 1231 bp | 0.60 | 0.47 | internal region | SH | JX997214 |
| Helena# | TGTCG22 | 443 bp | 0.76 | 0.62 | ORF2/endonuclease | SH | JX997207 |
| Helena# | TGTCG27 | 441 bp | 0.80 | 0.76 | reverse transcriptase/RNAasa | BC2 | JX997216 |
| Helena# | TGTCG41 | 441pb | 0.78 | 0.67 | ORF2/endonuclease | DB | not annotated |
| Helitron-2▪ | TGCGG21 | 145 bp | 0.77 | 0.46 | internal region | BC3 | not annotated |
| Helitron-2▪ | CGGCA21 | 1231 bp | 0.62 | 0.51 | 5′region | SH | JX997214 |
| Helitron-1N1▪ | CCGAT22 | 264 bp | 0.92 | 0.92 | internal region | BC3 | JX997217 |
| HETA# | CACTC21 | 592 bp | 0.41 | 0.31 | internal region | SH | JX997212 |
| Homo6▪ | TGATT25 | 442 bp | 0.62 | 0.50 | internal region | BC2 | JX997193 |
| Homo6▪ | TCAGT21 | 347 bp | 0.88 | 0.88 | internal region | BC2 | JX997194 |
| Homo6▪ | TGATT27 | 441 bp | 0.68 | 0.58 | internal region | BC3 | JX997201 |
| Homo6▪ | CCGAT24 | 320 bp | 0.94 | 0.94 | internal region | BC3 | JX997205 |
| LSU-rRNA_HsaØ | CAGCG23 | 448 bp | 0.42 | 0.31 | HSU 13369 locus | BC2 | not annotated |
| MAX* | CACTC22 | 500 bp | 0.66 | 0.50 | polyprotein | SH | JX997209 |
| MINI-ME# | GGCTC21 | 647 bp | 0.58 | 0.46 | internal region | BC1 | JX997188 |
| MINI-ME# | CCCCC21 | 540 bp | 0.71 | 0.71 | internal region | BC1 | JX997191 |
| MINI-ME# | TCAGT22 | 306 bp | 0.71 | 0.62 | internal region | BC2 | JX997195 |
| Osvaldo* | TGATT24 | 530 bp | 0.63 | 0.51 | internal region | BC3 | JX997197 |
| Osvaldo* | TGATT26 | 519 bp | 0.56 | 0.46 | internal region | SH | JX997213 |
| Penelope# | CCGAT21 | 309 bp | 0.91 | 0.13 | internal region | BC3 | JX997203 |
| TART# | TGTCG23 | 415 bp | 0.55 | 0.41 | polyprotein | SH | JX997208 |
| TART# | TCTCG21 | 218 bp | 0.55 | 0.29 | polyprotein | BC3 | JX997202 |
| Transib1▪ | CAGCA22 | 359 bp | 0.73 | 0.60 | transposase | SH | JX997210 |
| TRIM# | TGTCG23 | 415 bp | 0.58 | 0.41 | internal region | SH | JX997208 |
| UHU▪ | TGATT25 | 513 bp | 0.66 | 0.52 | internal region | BC3 | JX997198 |
| ZAM* | TGATT26 | 519 bp | 0.58 | 0.35 | internal region | SH | JX997213 |
Sim: similarity between 2 aligned fragments using the parameters of CENSOR tool referenced in material and methods section, * LTR retrotransposon, # non-LTR retrotransposon, ▪ DNA transposon, Ø pseudogen, SH: segmental hybrid, BC1, BC2 and BC3:hybrids from backcrosses 1,2 and 3 respectively; DB: parental species D. buzzatii.
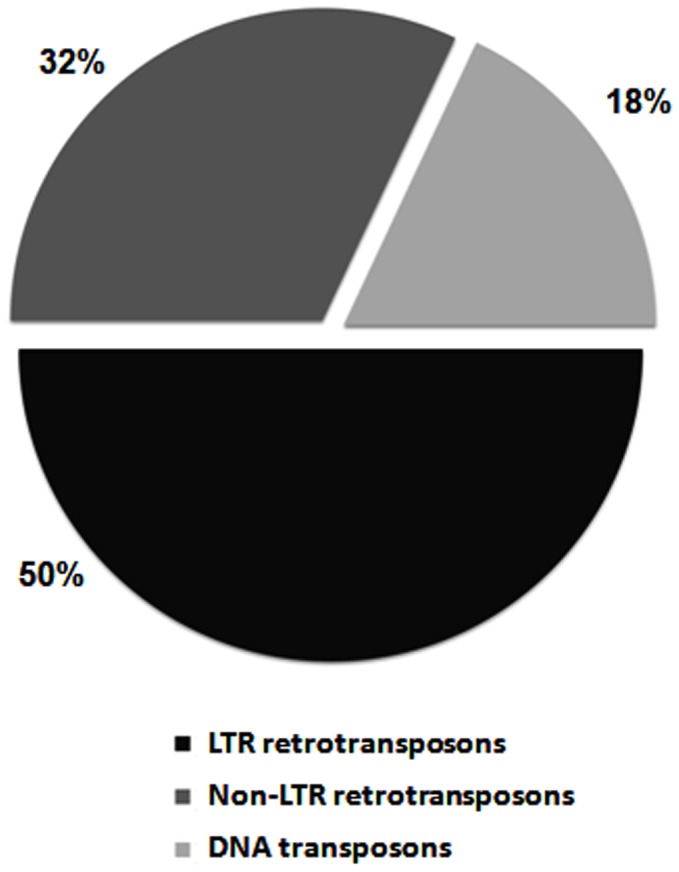
Detailed analysis of the sequenced transposition markers allowed us to detect a pseudogene and 28 different TEs belonging to 14 TE families, including class I and class II elements (Figure 3). These TEs are probably responsible for most of the genomic instability in hybrids (Table 2). Three, 3 and 11 different TEs were detected in BC1, BC2, and BC3 generations, respectively, whereas 19 were identified in segmental hybrids. Interestingly, some specific TEs were detected in more than one generation. The genome of the parental species seems quite stable: only one element (Helena) was mobilized in D. buzzatii and none in D. koepferae.
Figure 3. Percentage of transposition markers in hybrid.
No significant differences were detected between hybrid families for the number of markers associated to TEs (P = 0.65) in BC3 (Table 1). However, significant differences (P = 0.045*) were detected when the number of transposition markers between BC3 hybrids and parental species were compared (Table 3), bolstering the idea that the genome of hybrids induces a significant increment of instability by transposition. The value is only significant in family 40 (P = 0.035*) when analyzing each family separately, despite new TE markers have been detected in all of them.
Table 3. Comparison of the number of transposition markers between BC3 hybrid families and parental species.
| Comparisons | N | Mann-Whitney Test | ||
| Hybrids | Parentals | U | P value | |
| Hybrids vs.Parental species | 40 | 30 | 513.5 | 0.045* |
| Family 10 vs.Parental species | 12 | 30 | 170.5 | 0.286 |
| Family 13 vs.Parental species | 12 | 30 | 155.5 | 0.134 |
| Family 40 vs.Parental species | 16 | 30 | 187.5 | 0.035* |
N: sample size, U: statistic value, *: P≤0.05; BC3: backcross 3.
Transposition Rates
From the set of TEs detected by AFLP, Osvaldo, Helena and Galileo were studied by in situ hybridization (Table 4) and transposon display in parental species, but only by transposon display in hybrids. Although both techniques allow us to assess the number of euchromatic copies in the genome, transposon display has the advantage of amplifying both euchromatic and heterochromatic copies, whereas in situ hybridization only allows one to discern euchromatic copies, in spite of having been traditionally used to estimate total transposition rates in Drosophila [30], [39]. Both techniques were used for parental species analyses to estimate how many of the insertions detected by transposon display corresponded to heterochromatic insertions. Using FISH Osvaldo gave 5 euchromatic copies and a heavy stained centromeric signal in D. buzzatii, and only a centromeric signal in D. koepferae, whereas up to 40 and 39 copies were detected by transposon display in the 2 species, respectively. In the case of Helena, 5 and 12 euchromatic copies were detected by FISH in D. buzzatii and D. kopeferae respectively, but up to 28 and 25 copies by transposon display. Galileo had just one euchromatic copy in each parental species, and up to 43 and 45 copies by transposon display in D. buzzatii and D. koepferae, respectively.
Table 4. Chromosomal insertion sites of Osvaldo, Helena and Galileo in parental species.
| Element/Species | Chromosomes | |||||
| X | 2 | 3 | 4 | 5 | Total | |
| Osvaldo | ||||||
| D. buzzatii | G2, C | E4, D4 | D1, C | C | C | 4 |
| D. koepferae | C | C | C | C | C | 0 |
| Helena | ||||||
| D. buzzatii | A2 | E2 | G1, C | A5, D2 | 5 | |
| D. koepferae | A2, F3, G2 | B2, E2, F1 | D4, G2 | C4.1,C4.2,C5, F2 | 12 | |
| Galileo | ||||||
| D. buzzatii | E3 | 1 | ||||
| D. koepferae | G1 | 1 | ||||
C: centromere.
In view of its advantage the transposon display technique was used to estimate the transposition rates in hybrids and parentals species for each element in each family and backcross generation (Tables S1).
Osvaldo Transposition Rates
The bands of Osvaldo detected by transposon display ranged in size from 171 to 1177 bp, each insertion including 152 bp of a 5′ LTR fragment (determined after the restriction map of this TE) and its flanking genomic region (variable in size depending on the genomic insertion site of each copy). The number of insertions in hybrid genomes ranged between 35 (BC1 family 40) and 54 (BC3 family 10), showing new insertions in all families (e.g. 48 in BC3 of families 1 and 13) except family 40 (BC1). However, only one new insertion was observed in the family B8 in the parental species D. buzzatii (Table 5). The basal transposition rates of Osvaldo estimated for D. buzzatii and D. koepferae ranged from 0 to 3.3×10−3. The transposition rate of Osvaldo increased by one order of magnitude (10−2) in hybrid families compared to parental species; this was observed in the 3 backcross generations, although not in all hybrid families (Table 6). The number of new insertions in the 3 hybrid backcrosses was significantly higher in hybrids than the parental species (Table 7), with differences between hybrid families in BC1 and BC3 (Table 8).
Table 5. Number of Osvaldo, Helena and Galileo insertions detected by transposon display.
| HYBRIDS | BC1 | BC2 | BC3 | ||||||
| Totalinsertions | Newinsertions | Ni | Totalinsertions | Newinsertions | Ni | Totalinsertions | Newinsertions | Ni | |
| Osvaldo | |||||||||
| Family 1 | 40 | 3 | 9 | 40 | 3 | 7 | 48 | 7 | 53 |
| Family 10 | 38 | 1 | 1 | 48 | 9 | 30 | 54 | 11 | 60 |
| Family 13 | 38 | 1 | 1 | 38 | 1 | 2 | 48 | 11 | 22 |
| Family 40 | 34 | 0 | 0 | 40 | 5 | 10 | 50 | 11 | 46 |
| Helena | |||||||||
| Family 1 | 18 | 0 | 0 | 23 | 3 | 11 | 22 | 2 | 14 |
| Family 10 | 18 | 0 | 2 | 22 | 2 | 5 | 25 | 1 | 4 |
| Family 13 | 17 | 2 | 2 | 22 | 0 | 0 | 24 | 1 | 6 |
| Family 40 | 21 | 0 | 0 | 24 | 1 | 2 | 25 | 1 | 3 |
| Galileo | |||||||||
| Family 1 | 25 | 1 | 5 | 30 | 0 | 0 | 32 | 2 | 2 |
| Family 10 | 35 | 0 | 0 | 39 | 0 | 0 | 40 | 0 | 0 |
| Family 13 | 48 | 0 | 0 | 47 | 0 | 0 | 49 | 1 | 4 |
| Family 40 | 52 | 2 | 7 | 55 | 1 | 5 | 56 | 0 | 0 |
| PARENTALS | Osvaldo | Helena | Galileo | ||||||
| Total insertions | New insertions | Ni | Total insertions | New insertions | Ni | Total insertions | New insertions | Ni | |
| D. buzzatii | |||||||||
| Family B4 | 40 | 0 | 0 | 27 | 1 | 6 | 41 | 0 | 0 |
| Family B8 | 35 | 1 | 3 | 28 | 0 | 0 | 43 | 0 | 0 |
| D. koepferae | |||||||||
| Family K3 | 39 | 0 | 0 | 25 | 0 | 0 | 42 | 0 | 0 |
| Family K9 | 34 | 0 | 0 | 22 | 0 | 0 | 45 | 0 | 0 |
Ni: insertions copy number; BC1, BC2 and BC3 correspond to backcrosses 1, 2, 3 respectively.
Table 6. Transposition rates of Osvaldo, Helena and Galileo in hybrid families and parental species.
| HYBRIDS | BC1 | BC2 | BC3 | |||||||||||||||||||||
| Ni | N | A | TR | Ni | N | A | TR | Ni | N | A | TR | |||||||||||||
| Osvaldo | ||||||||||||||||||||||||
| Family 1 | 9 | 7 | 37 | 1.7×10−2 | 7 | 18 | 37 | 5.2×10−3 | 53 | 40 | 41 | 1.6×10−2 | ||||||||||||
| Family 10 | 1 | 2 | 37 | 6.7×10−3 | 30 | 18 | 39 | 2.1×10−2 | 60 | 25 | 43 | 2.7×10−2 | ||||||||||||
| Family 13 | 1 | 2 | 37 | 6.7×10−3 | 2 | 15 | 37 | 1.8×10−3 | 22 | 36 | 37 | 8.3×10−3 | ||||||||||||
| Family 40 | 0 | 5 | 34 | 0 | 10 | 14 | 35 | 1.0×10−2 | 46 | 18 | 39 | 3.2×10−2 | ||||||||||||
| Helena | ||||||||||||||||||||||||
| Family 1 | 0 | 7 | 18 | 0 | 11 | 17 | 20 | 1.6×10−2 | 14 | 40 | 20 | 8.8×10−3 | ||||||||||||
| Family 10 | 0 | 2 | 18 | 0 | 5 | 19 | 20 | 6.5×10−3 | 4 | 33 | 24 | 2.5×10−3 | ||||||||||||
| Family 13 | 2 | 3 | 17 | 1.9×10−2 | 0 | 16 | 22 | 0 | 6 | 38 | 23 | 3.4×10−3 | ||||||||||||
| Family 40 | 0 | 5 | 21 | 0 | 2 | 13 | 23 | 3.3×10−3 | 3 | 16 | 24 | 3.9×10−3 | ||||||||||||
| Galileo | ||||||||||||||||||||||||
| Family 1 | 5 | 7 | 24 | 1.4×10−2 | 0 | 18 | 30 | 0 | 2 | 40 | 31 | 8.0×10−4 | ||||||||||||
| Family 10 | 0 | 2 | 35 | 0 | 0 | 17 | 39 | 0 | 0 | 30 | 40 | 0 | ||||||||||||
| Family 13 | 0 | 3 | 48 | 0 | 0 | 11 | 47 | 0 | 4 | 32 | 48 | 1.3×10−3 | ||||||||||||
| Family 40 | 7 | 5 | 52 | 1.4×10−2 | 5 | 14 | 55 | 3.3×10−3 | 0 | 24 | 56 | 0 | ||||||||||||
| PARENTAL SPECIES | ||||||||||||||||||||||||
| Osvaldo | Helena | Galileo | ||||||||||||||||||||||
| Ni | N | A | TR | Ni | N | A | TR | Ni | N | A | TR | |||||||||||||
| D. buzzatii | 3 | 28 | 74 | 7.2×10−4 | 6 | 28 | 54 | 2.0×10−3 | 0 | 26 | 84 | 0 | ||||||||||||
| D. koepferae | 0 | 24 | 73 | 0 | 0 | 19 | 47 | 0 | 24 | 87 | 0 | |||||||||||||
Ni: new insertions copy number, N: number of individuals, A: number of original insertions, TR: Transposition rate; BC1, BC2 and BC3 correspond to backcrosses 1, 2, 3 respectively.
Table 7. Comparison of the number of new insertions between hybrids and parental species by Mann-Whitney test.
| Parentals | BC1 | BC2 | BC3 | |||||||
| TEs | N | N | U | P value | N | U | P value | N | U | P value |
| Osvaldo | 53 | 16 | 206.5 | <10−3 ** | 65 | 1170.5 | <10−3 ** | 119 | 1330.5 | <10−3 ** |
| Helena | 47 | 17 | 372 | 0.439 | 65 | 1440 | 0.420 | 127 | 2772 | 0.278 |
| Galileo | 53 | 17 | 185 | <10−3 ** | 60 | 1431 | 0.031* | 126 | 3180 | 0.107 |
TEs: transposable elements, N: number of individuals, U: statistic value, *:P≤0.05.
**: P≤0.01; BC1, BC2 and BC3 correspond to backcrosses 1, 2, 3 respectively.
Table 8. Comparison of the number of new insertions between hybrid families by Kruskal-Wallis test.
| BC1 | BC2 | BC3 | |||||
| TEs | N | χ2 | P value | χ2 | P value | χ2 | P value |
| Osvaldo | 4 | 31.18 | <10−3** | 6.206 | 0.102 | 10.877 | 0.012* |
| Helena | 4 | 4.667 | 0.198 | 8.911 | 0.031* | 1.108 | 0.775 |
| Galileo | 4 | 10.768 | 0.013* | 17.623 | 0.001** | 6.875 | 0.076 |
TEs: Transposable elements; N: number of families, U: statistic value,
*:P≤0.05; P≤0.01; BC1, BC2 and BC3 correspond to backcrosses 1, 2, 3 respectively.
When transposition rates were calculated in hybrid males and females separately, a trend of increasing rates was seen in males, from families 13 and 1, compared to females (Table S3), but differences were only statistically significant for family 1 (P = 0.01*). Because hybrid females had been repeatedly backcrossed with D. buzzatii males, transpositions always occurred in the hybrid female germline, suggesting (but not proving) that the increase of transposition rates in males could be due to a higher male somatic transposition rate in these 2 families.
Helena Transposition Rates
In the case of the Helena element, we analyzed the 3′ end, obtaining insertion sizes ranging from 110 to 630 bp. These fragments included 107 bp of the 3′ Helena end plus a flanking DNA sequence. The number of insertions observed per hybrid family ranged between 17 and 25 (Table 5), some of which were new; e.g. 3 new insertions were found in BC2 from family 1, but only one insertion in the family B4 of D. buzzatii. The basal transposition rates of Helena, ranged from 0 to 8.2×10−3 for D. buzzatii, and 0 for D. koepferae. Helena showed an increase of transposition (10−2) in some families of the BC1 and BC2 generations, but this activity decreased to the basal transposition rate in the BC3 generation (Table 6). However, the number of new insertions in hybrids compared to parental species did not reach significance at statistical level (Table 7), neither were the differences between families, except in BC2 (Table 8). Comparisons of Helena transposition rates between males and females do not show a clear trend because the transposition increase was seen in males of a family and females of another.
Galileo Transposition Rates
The amplified insertions of Galileo contain a fragment of 182 bp of the 5′ end and a flanking DNA sequence ranging from 191 to 866 bp. Up to 7 new insertions (family 40, BC1) were found in the genome of hybrids, but no new insertions were detected in parental species (Table 6). While the basal transposition rate of Galileo for parental species is 0, this element shows high transposition activity (≈10−2) in 2 families of the BC1 generation showing significant differences from the parental species (Table 8), decreasing later in BC2 and BC3 (Table 6). When families were compared (Table 8) significantly differences in transposition rates were seen in BC1 and BC2, showing that the effect of hybridization on TE instability depends on the families analyzed.
Statistical analyses showed significant differences (P<10−3) of Osvaldo transposition rates between hybrids and parental species in the 3 backcrosses. In the case of Helena and in spite of the new insertions being detected in hybrids compared with parental species (only one new insertion in D. buzzatii, Tables S1), the differences were not significant (Table 7). For the Galileo element, significant differences were found in BC1 and BC2 (Table 7).
Expression of Osvaldo, Helena and Galileo
Transcription of Osvaldo, Helena and Galileo was examined in the testes, ovaries and carcasses of parental species D. buzzatii and D. koepferae by RT-PCR. Osvaldo and Helena were expressed in germinal and somatic tissues isolated from adult males and females. In the case of Galileo, no transcriptional activity was detected in germinal or somatic tissues of both species D. buzzatii and D. koepferae (Figure S1) suggesting that this element cannot move autonomously in the genome of our stocks, but probably needs assistance from a second TE for its mobilization.
Discussion
The joint analysis of AFLP patterns in 3 hybrid families, a strain of segmental hybrids and 4 families of parental species, shows the existence of genomic instability in the hybrid genomes, revealed by 72 instability markers plus a pseudogene. In contrast, only 4 and 6 instability markers were found in D. koepferae and D. buzzatii parental genomes, respectively. A high percentage (16, 29, 52 and 90% in BC1, BC2, BC3 and segmental hybrids, respectively) of them showed homology with TEs in hybrids probably originated by the TEs whose mobilization produce new insertions detected as new bands in the AFLP pattern. Another fraction of instability markers had homology with the D. mojavensis genome (the Drosophila sequenced genome phylogenetically closer to the species considered here) and probably originated by the suppression of a restriction site and/or by double-strand breaks that are usually associated to the loss of chromosomal segments or the rearrangement of genetic material [40].
It is noteworthy that 90% of the markers detected in segmental hybrids had homology with TE sequences. Because these hybrids have a portion of chromosome 4 (as detected by chromosomal asynapsis) of D. koepferae introgressed in the D. buzzatii genome, this region could be involved in TEs instability rendering this line unstable. This line was also maintained for 60 generations which would explain the accumulation of new TE copies. Hybrid families from BC3 generation and parental species showed significant differences in transposition instability (P = 0.045), but when families were considered separately, differences were only significant for family 40, despite all families showed an increase of transposition markers compared to parental species (only one new transposition marker detected). This result is probably due to the highest number of hybrids analyzed in this family and/or to an increasing segregation of instability markers only in certain hybrid families. Indeed, in previous studies Osvaldo transposition bursts have been preferentially found in some interspecific hybrids of D. buzzatii and D. koepferae [41]. The overall increase of genomic instability induced by transposition, as found in Drosophila interspecific hybrids, suggests that the stress produced by hybridization induces a significant activation of transposition in hybrid genomes. These observations are in agreement with studies on genomic instability of natural hybrids of Amaranthus, where the new AFLP hybrid markers detected in hybrids have homology with TEs associated to the mobilization of repetitive DNA [42]. Moreover, in the present study an increase of genomic instability in early generations of backcrossing was also observed. This is not uncommon because the level of introgression in the first backcross generation is the highest, decreasing in the next generations due to the increment of the proportion of the D. buzzatii genome through the repeating backcrossing with a D. buzzatii parental male. These results are in accord with those of Madlung et al. [43] who detected hybrid instability at the early generations of Arabidopsis interspecific crosses.
Osvaldo [41] is an active retrotransposon expressed in somatic and germinal tissues of D. buzzatii and D. koepferae stocks that we used. In situ hybridization, transposition rates and expression of Helena element, bolster the notion that this element is also an active element in the genomes of D. buzzatii and D. koepferae that has the capacity to move and create new insertions. An element that deserves special mention is Galileo, where no expression of transposase was found, suggesting that, in our stocks, most copies could be deleted showing passive activity mediated by a cooperative TE. New copies of Galileo have been noted in the hybrid genomes whose nucleotide sequences, amplified by transposon display, often contain other TE fragments in the same sequences, suggesting that their transposition may correspond to the activity of other autonomous TEs carrying inserted fragments of Galileo. Interestingly, something similar occurs when this element is screened in the genome of D. mojavensis, in which fragments of Galileo are frequently located next to other TEs.
Several attempts to induce transposition in Drosophila through hybridization have been attempted without success [26], [44] Nowadays, numerous examples of hybrid instability associated to TE activity are found in plants [23], [45] and, to a lesser extent, in insects [46], [47] and mammals [28], [29]. Our study constitutes the first genome-wide survey of hybrid instability showing that a high proportion of the instability markers detected in hybrids correspond to TE. Thus, an average of 32 and 90% of the markers detected in the three backcross generations and the segmental hybrids, respectively, correspond to a wide variety of TEs. The estimated transposition rates for Osvaldo and Helena in hybrids are higher than spontaneous estimated for some TEs in natural populations of Drosophila (10−4) [48], which can be directly associated to the genomic instability in the hybrid genomes. TEs have the capacity to create new copies in the genomes through many mechanisms, including excision, replication, insertion, and ectopic recombination. In D. melanogaster, for example, new chromosomal rearrangements and mutations can be produced by ectopic recombination between different copies of the hobo element [49], and in humans, ectopic recombination between Alu sequences seem to be important in producing deleterious mutations [50]. In our study, the new Galileo copies could also be attributed to ectopic recombination of Galileo long TIRs [51]; we consider that the new copies of the 3 TEs analyzed were probably produced by transposition because here a high number of copies observed were detected in heterochromatic regions, in which recombination rates are usually low. Elements outside heterochromatin are often active, hence more than 50% of mutations in Drosophila have been assigned to direct TEs insertion [52]. However, despite euchromatin harboring most of Drosophila genes, the effect of recombination cannot completely be ruled out because many genes are also found in heterochromatic regions [53].
In previous experiments, increases in transposition rate of Osvaldo were seen in interspecific hybrids (10−2) compared to parental species D. buzzatii (10−3) [54]. These results are in agreement with the transposition rates estimated for Osvaldo in the 3 backcross generations (10−2) and confirm its higher ability to mobilize in the hybrid genomes compared to its lower transposition rate in the parental species D. buzzatii (10−3) and D. koepferae (<10−3). However, in family 13 the transposition rate of Osvaldo is of the order of 10−3 in all backcross generations which supports the idea that bursts of transposition do not occur equally in all families of hybrids. The same was observed for Helena and Galileo, where differences in transposition rates were found between generations of backcrosses and hybrid families. This result is unsurprising considering that we introduce the genome of D. buzzatii in each backcross generation; despite the amount of the genome being introduced is the same, the region introgressed is different in each hybrid family. It is known that the mobility of some TEs is controlled by specific genomic loci; for example, gypsy, Idefix and Zam are controlled by the locus flamenco [55], [56] and P elements by a subtelomeric region of X chromosome [57]. These loci comprise fragmented and imperfect copies of retrotransposons that are the precursors of PIWI-interacting RNAs (piRNA) responsible of post-transcriptional TE control. Another result of note is the different activity of the 3 elements analyzed in hybrids; the most active element being Osvaldo, followed by Helena and then Galileo. These differences were also found across generations, indicating that a variation in TE stability exists depending on the element and the genetic background of the hybrid genome. In D. melanogaster, for example, there is a natural variation in TE stability depending on the genetic background [58]. In our case we hypothesized that TEs activation in hybrids could occur in a similar way to that in dysgenic crosses, due to the lack of specific maternal piRNAs Another hypothesis, not exclusive, could be the divergence of genes involved in the piRNA pathway. Recent findings in hybrids between D. melanogaster and D. simulans attributed the derepression of different TEs to divergence in piRNA genes rather than species-specific differences in piRNAs derived from TEs [47].
It is of note that, in the case of the Osvaldo retrotransposon, a trend towards an increase in transposition rate in hybrid males occurs. In other retrotransposons, e.g. copia element, transpositions are limited to male spermatocytes [59]. However, since hybrid males do not participate in hybrid crosses, we suggest that differences of transposition between sexes could be due to an increase of transpositions in the somatic line of males. Malone et al. [60] suggested a tissue-specific regulation of certain element classes correlated with tissue specific expression of piRNA clusters (genomic loci producing significant levels of piRNA). In Xenopus, a dramatic increase in the number of misexpressed genes found in hybrid females, compared to testes of hybrid males, suggests that divergence in female expression may be involved in sterility of hybrid males due to the inherent sensitivity of spermatogenesis [61]. We hypothesize that misregulation of genes implicated in the piwi pathway contribute to the instability of TEs in germinal lines. However, it is difficult to provide a valid explanation because we ignore the regulation mechanisms of Osvaldo and other elements.
Because different classes of TEs have been mobilized in hybrids, they could contribute to the genome reorganization either by transposition or/and ectopic recombination, producing deletions, duplications and inversions. Previous studies in D.buzzatii-kopeferae hybrids showed a high frequency of new chromosome rearrangements induced by introgressive hybridization [62]. Overall, our study contributed to a better understanding of the effects of hybridization in transposition release in hybrids, but the mechanisms triggering transposition are poorly understood and more knowledge about them would be of great importance to demonstrate that transposable elements are involved in speciation. Knowledge concerning the expression rates and TEs transcripts location could be the next step towards understanding the molecular mechanisms activating TEs in Drosophila hybrids.
Supporting Information
RT PCR of Osvaldo, Helena and Galileo in somatic and germinal tissues of D. buzzatii and D. koepferae .
(TIFF)
Osvaldo, Helena and Galileo matrices of transposon display experiment. Asterisks represent new insertions. The presence of an insertion is represented by 1 and the absence by 0.
(DOC)
Number of AFLP markers observed in each family and backcross generation.
(DOC)
Transposition rates for male and females BC3 hybrids.
(DOC)
AFLP instability markers with no homology to transposable elements.
(DOC)
Acknowledgments
We want to thank Christian Biémont for his valuable discussions and ideas at the beginning of this work, and to MJ Madison-Villar and an anonymous referee for the interesting comments contributing to the improvement of the manuscript.
Funding Statement
This work was supported by research grants CGL2006-13423-C02-01/02 from the Ministerio de Educación y Ciencia (Spain)(www.educacion.gob.es), grants CGL2009-12912-C03-01 and CGL2010-15395 from the Ministerio de Ciencia e Innovación (Spain)(www.educacion.gob.es), and grants 2005SGR 00995 and 2009SGR 636 from Generalitat de Catalunya (www.gencat.cat) to the Grup de Biologia Evolutiva. ANR-09-BLAN-0103 (www.agence-nationale-recherche.fr) Genemobile and Institut Universitaire de France and the CNRS GDRE Comparative Genomics. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Arnold M (1997) Evolution through genetic exchange. Oxford University Press.
- 2.Arnold M (2006) Natural hybridization and Evolution. Oxford University Press.
- 3.Pollock (1988) A morphometric analysis of a Pseudopanax hybrid swarm. University of Auckland.
- 4. Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, et al. (2003) Major ecological transitions in wild sunflowers facilitated by hybridization. Science (80-) 301: 1211–1216 Available: http://www.ncbi.nlm.nih.gov/pubmed/12907807. [DOI] [PubMed] [Google Scholar]
- 5. Nolte AW, Tautz D (2010) Understanding the onset of hybrid speciation. Trends Genet 26: 54–58 Available: http://linkinghub.elsevier.com/retrieve/pii/S0168952509002492. [DOI] [PubMed] [Google Scholar]
- 6.Fontdevila A (2004) Evolution: from Molecules to Ecosystems. In: Moya A, Font E, editors. Introgression and hybrid speciation via transposition. Oxford University Press. 182–194.
- 7. Michalak P (2010) An eruption of mobile elements in genomes of hybrid sunflowers. Heredity (Edinb) 104: 329–330 Available: http://europepmc.org/abstract/MED/20068587. [DOI] [PubMed] [Google Scholar]
- 8.Capy P, Langin T, Anxolabehere, Dominique Bazin C (1998) Dynamics and Evolution of Transposable Elements. Thomson Learning.
- 9. Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285 Available: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 10.García Guerreiro MP (2012) What makes transposable elements move in the Drosophila genome? Heredity (Edinb) 108: 461–468. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3330689&tool=pmcentrez&rendertype=abstract. Accessed 14 October 2013. [DOI] [PMC free article] [PubMed]
- 11. Ginzburg LEVR, Bincham PM, Yoo S (1984) On the theory of speciation induced by transposable elements. Genetics 107: 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ish-Horowicz D (1982) Transposable elements, hybrid incompatibility and speciation. Nature 299: 676–677 Available: 10.1038/299676a0. [DOI] [PubMed] [Google Scholar]
- 13. Rubin GM, Kidwell MG, Bingham PM (1982) The molecular basis of P-M hybrid dysgenesis: The nature of induced mutations. Cell 29: 987–994 Available: http://linkinghub.elsevier.com/retrieve/pii/0092867482904627. [DOI] [PubMed] [Google Scholar]
- 14. Bingham PM, Kidwell MG, Rubin GM (1982) The molecular basis of P-M hybrid dysgenesis: The role of the P element, a P-strain-specific transposon family. Cell 29: 995–1004 Available: http://linkinghub.elsevier.com/retrieve/pii/0092867482904639. [DOI] [PubMed] [Google Scholar]
- 15. Picard (1976) Non-mendelian female sterility in Drosophila melanogaster: hereditary transmission of I factor. Genetics 85: 107–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yannopoulos G, Stamatis N, Monastirioti M, Hatzopoulos P, Louis C (1987) hobo is responsible for the induction of hybrid dysgenesis by strains of Drosophila melanogaster bearing the male recombination factor 23.5MRF. Cell 49: 487–495 Available: http://linkinghub.elsevier.com/retrieve/pii/009286748790451X. [DOI] [PubMed] [Google Scholar]
- 17. Petrov DA, Schutzman JL, Hartl DL, Lozovskaya ER (1995) Diverse transposable elements are mobilized in hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci U S A 92: 8050–8054 Available: http://europepmc.org/abstract/MED/7644536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, et al. (2008) An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392 Available: http://europepmc.org/abstract/MED/19039138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akkouche A, Grentzinger T, Fablet M, Armenise C, Burlet N, et al. (2013) Maternally deposited germline piRNAs silence the tirant retrotransposon in somatic cells. EMBO Rep 14: 458–464. Available: http://www.ncbi.nlm.nih.gov/pubmed/23559065. Accessed 4 October 2013. [DOI] [PMC free article] [PubMed]
- 20. Rebollo R, Horard B, Hubert B, Vieira C (2010) Jumping genes and epigenetics: Towards new species. Gene 454: 1–7 Available: http://www.sciencedirect.com/science/article/pii/S0378111910000296. [DOI] [PubMed] [Google Scholar]
- 21. Josefsson C, Dilkes B, Comai L (2006) Parent-Dependent Loss of Gene Silencing during Interspecies Hybridization. Curr Biol 16: 1322–1328 Available: http://www.sciencedirect.com/science/article/pii/S0960982206016289. [DOI] [PubMed] [Google Scholar]
- 22. Liu B, Wendel JF (2000) Retrotransposon activation followed by rapid repression in introgressed rice plants. Genome 43: 874–880 Available: http://www.nrcresearchpress.com/doi/abs/10.1139/g00-058. [PubMed] [Google Scholar]
- 23. Shan X, Liu Z, Dong Z, Wang Y, Chen Y, et al. (2005) Mobilization of the Active MITE Transposons mPing and Pong in Rice by Introgression from Wild Rice (Zizania latifolia Griseb.). Mol Biol Evol 22: 976–990 Available: http://mbe.oxfordjournals.org/content/22/4/976.abstract. [DOI] [PubMed] [Google Scholar]
- 24.Ungerer MC, Strakosh SC, Zhen Y (2006) Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. 16. Available: 10.1016/j.cub.2006.09.020. [DOI] [PubMed]
- 25.Shivaprasad P V, Dunn RM, Santos BA, Bassett A, Baulcombe DC (2012) Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J 31: 257–266. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3261569&tool=pmcentrez&rendertype=abstract. Accessed 8 October 2013. [DOI] [PMC free article] [PubMed]
- 26. Coyne JA (1989) Mutation rates in hybrids between sibling species of Drosophila. Heredity (Edinb) 63: 155–162 Available: 10.1038/hdy.1989.87. [DOI] [PubMed] [Google Scholar]
- 27. Hey J (1988) Speciation via hybrid dysgenesis: negative evidence from the Drosophila affinis subgroup. Genetica 78: 97–103 Available: 10.1007/BF00058840. [DOI] [Google Scholar]
- 28. Metcalfe CJ, Bulazel K V, Ferreri GC, Schroeder-Reiter E, Wanner G, et al. (2007) Genomic Instability Within Centromeres of Interspecific Marsupial Hybrids. Genet 177: 2507–2517 Available: http://www.genetics.org/content/177/4/2507.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O’Neill RJW, O’Neill MJ, Graves JAM (1998) Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393: 68–72 Available: 10.1038/29985. [DOI] [PubMed] [Google Scholar]
- 30. Labrador M, Fontdevila A (1994) High transposition rates of Osvaldo, a new Drosophila buzzatii retrotransposon. Mol Gen Genet 245: 661–674. [DOI] [PubMed] [Google Scholar]
- 31. Labrador M, Farré M, Utzet F, Fontdevila a (1999) Interspecific hybridization increases transposition rates of Osvaldo. Mol Biol Evol 16: 931–937 Available: http://www.ncbi.nlm.nih.gov/pubmed/10406110. [DOI] [PubMed] [Google Scholar]
- 32. Pinol J, Francino O, Fontdevila A, Cabré O (1988) Rapid isolation of Drosophila high molecular weight DNA to obtain genomic libraries. Nucleic Acids Res 16: 2736 Available: http://nar.oxfordjournals.org/content/16/6/2736.short. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vela D, Guerreiro MP, Fontdevila A (2011) Adaptation of the AFLP technique as a new tool to detect genetic instability and transposition in interspecific hybrids. Biotechniques 50: 247–250 Available: http://europepmc.org/abstract/MED/21548908. [DOI] [PubMed] [Google Scholar]
- 34. Zampicinini G, Blinov A, Cervella P, Guryev V, Sella G (2004) Insertional polymorphism of a non-LTR mobile element (NLRCth1) in European populations of Chironomus riparius (Diptera, Chironomidae) as detected by transposon insertion display. Genome 47: 1154–1163 Available: http://europepmc.org/abstract/MED/15644974. [DOI] [PubMed] [Google Scholar]
- 35.Akkouche A, Rebollo R, Burlet N, Esnault C, Martinez S, et al. (2012) tirant, a Newly Discovered Active Endogenous Retrovirus in Drosophila simulans: 3675–3681. doi: 10.1128/JVI.07146-11. [DOI] [PMC free article] [PubMed]
- 36. García Guerreiro MP, Fontdevila A (2007) The evolutionary history of Drosophila buzzatii. XXXVI. Molecular structural analysis of Osvaldo retrotransposon insertions in colonizing populations unveils drift effects in founder events. Genetics 175: 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kruskal WH, Wallis WA (1952) Use of Ranks in One-Criterion Variance Analysis. J Am Stat Assoc 47: 583–621 Available: http://www.jstor.org/stable/2280779. [Google Scholar]
- 38. Jurka J, Kapitonov V V, Pavlicek A, Klonowski P, Kohany O, et al. (2005) Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110: 462–467 Available: http://europepmc.org/abstract/MED/16093699. [DOI] [PubMed] [Google Scholar]
- 39. Vieira C, Biémont C (1996) Geographical variation in insertion site number of retrotransposon 412 in Drosophila simulans. J Mol Evol 42: 443–451. [DOI] [PubMed] [Google Scholar]
- 40. Salomon S, Puchta H (1998) Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J 17: 6086–6095 Available: 10.1093/emboj/17.20.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pantazidis A, Labrador M, Fontdevila A (1999) The retrotransposon Osvaldo from Drosophila buzzatiidisplays all structural features of a funcrional retrovirus. Mol Biol Evol 16: 909–921. [DOI] [PubMed] [Google Scholar]
- 42. Steinau AN, Skinner DZ, Steinau M (2003) Mechanism of extreme genetic recombination in weedy Amaranthus hybrids. Weed Sci 51: 696–701 Available: http://wssajournals.org/doi/abs/10.1614/P2002-159. [Google Scholar]
- 43. Madlung A, Tyagi AP, Watson B, Jiang H, Kagochi T, et al. (2005) Genomic changes in synthetic Arabidopsis polyploids. Plant J 41: 221–230 Available: 10.1111/j.1365-313X.2004.02297.x. [DOI] [PubMed] [Google Scholar]
- 44. Coyne JA (1986) Meiotic segregation and male recombination in intespecific hybrids of Drosophila. Genetics 114: 485–494 Available: http://www.genetics.org/content/114/2/485.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kawakami T, Strakosh SC, Zhen Y, Ungerer MC (2010) Different scales of Ty1/copia-like retrotransposon proliferation in the genomes of three diploid hybrid sunflower species. Heredity (Edinb) 104: 341–350 Available: 10.1038/hdy.2009.182. [DOI] [PubMed] [Google Scholar]
- 46. Labrador M, Farré M, Utzet F, Fontdevila A (1999) Interspecific hybridization increases transposition rates of Osvaldo. Mol Biol Evol 16: 931–937 Available: http://mbe.oxfordjournals.org/content/16/7/931.abstract. [DOI] [PubMed] [Google Scholar]
- 47.Kelleher ES, Edelman NB, Barbash DA (2012) Drosophila Interspecific Hybrids Phenocopy piRNA-Pathway Mutants. PLoS Biol 10. Available: 10.1371/journal.pbio.1001428. [DOI] [PMC free article] [PubMed]
- 48. Suh DS, Choi EH, Yamazaki T, Harada K (1995) Studies on the transposition rates of mobile genetic elements in a natural population of Drosophila melanogaster. Mol Biol Evol 12: 748–758 Available: http://mbe.oxfordjournals.org/content/12/5/748.abstract. [DOI] [PubMed] [Google Scholar]
- 49. Sheen F, Lim JK, Simmons MJ (1993) Genetic instability in Drosophila melanogaster mediated by hobo transposable elements. Genet 133: 315–334 Available: http://www.genetics.org/content/133/2/315.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roy MA, Carroll LM, Kass HD, Nguyen VS, Salem A–H, et al. (1999) Recently integrated human Alu repeats: finding needles in the haystack. Genetica 107: 149–161 10.1023/A:1003941704138 [DOI] [PubMed] [Google Scholar]
- 51. Marzo M, Puig M, Ruiz A (2008) The Foldback-like element Galileo belongs to the P superfamily of DNA transposons and is widespread within the Drosophila genus. Proc Natl Acad Sci 105: 2957–2962 Available: http://www.pnas.org/content/105/8/2957.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Finnegan DJ (1992) Transposable elements. Curr Opin Genet Dev 2: 861–867 Available: http://www.sciencedirect.com/science/article/pii/S0959437X0580108X. [DOI] [PubMed] [Google Scholar]
- 53. Hoskins R a, Smith CD, Carlson JW, Carvalho a B, Halpern A, et al. (2002) Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol 3: RESEARCH0085 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=151187&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Labrador M, Naveira H, Fontdevila A (1990) Genetic mapping of the Adh locus in the repleta group of Drosophila by in situ hybridization. J Hered 81: 83–86. [DOI] [PubMed] [Google Scholar]
- 55. Desset S, Meignin C, Dastugue B, Vaury C (2003) COM, a Heterochromatic Locus Governing the Control of Independent Endogenous Retroviruses From Drosophila melanogaster. Genet 164: 501–509 Available: http://www.genetics.org/content/164/2/501.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pélisson A, Song SU, Prud’homme N, Smith PA, Bucheton A, et al. (1994) Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J 13: 4401–4411 Available: http://europepmc.org/abstract/MED/7925283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stuart JR, Haley KJ, Swedzinski D, Lockner S, Kocian PE, et al. (2002) Telomeric P elements associated with cytotype regulation of the P transposon family in Drosophila melanogaster. Genetics 162: 1641–1654 Available: http://europepmc.org/abstract/MED/12524339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nuzhdin S V, Mackay TF (1995) The genomic rate of transposable element movement in Drosophila melanogaster. Mol Biol Evol 12: 180–181 Available: http://europepmc.org/abstract/MED/7877494. [DOI] [PubMed] [Google Scholar]
- 59.Vu W, Nuzhdin S (2011) Genetic variation of copia suppression in Drosophila melanogaster. Heredity (Edinb) 106: 207–217. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3183883&tool=pmcentrez&rendertype=abstract. Accessed 14 October 2013. [DOI] [PMC free article] [PubMed]
- 60. Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, et al. (2009) Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 Available: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malone JH, Michalak P (2008) Gene expression analysis of the ovary of hybrid females of Xenopus laevis and X. muelleri. BMC Evol Biol 8: 82. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2330042&tool=pmcentrez&rendertype=abstract. Accessed 14 October 2013. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RT PCR of Osvaldo, Helena and Galileo in somatic and germinal tissues of D. buzzatii and D. koepferae .
(TIFF)
Osvaldo, Helena and Galileo matrices of transposon display experiment. Asterisks represent new insertions. The presence of an insertion is represented by 1 and the absence by 0.
(DOC)
Number of AFLP markers observed in each family and backcross generation.
(DOC)
Transposition rates for male and females BC3 hybrids.
(DOC)
AFLP instability markers with no homology to transposable elements.
(DOC)