Abstract
NELF, a protein identified in migratory GnRH neurons, is predominantly nuclear and alternatively spliced. However, specific NELF splice variants expressed in immortalized GnRH neuronal cell lines from mouse and human are not known. RNA from migratory (GN11 and NLT) and postmigratory (GT1-7) cells in mouse, and (FNCB4-hTERT) cells in human was subjected to RT-PCR. RT-PCR products were cloned, electrophoresed on denaturing gradient gels and sequenced. In addition, quantitative RT-PCR was performed using variant-specific primers. Western blot and immunofluorescence using confocal microscopy were performed for selected variants. Nelf variant 2 (v2), which contains a nuclear localization signal (NLS), was the predominant variant in all mouse and human GnRH neurons. Variants without a NLS (v3 in mouse; v4 in human) were identified. In mouse, v2 protein expression was nuclear, while v3 was non-nuclear. In mouse GnRH neurons, six Nelf splice variant transcripts were identified, including three previously unreported variants. In human, four NELF variant transcripts were observed. In both mouse and human, nuclear and non-nuclear variant transcript and protein were identified, explaining variable NELF cellular localization.
Keywords: Nasal embryonic LHRH factor (NELF), GnRH neurons, NLT cells, GT1-7 cells, GN11 cells, hypogonadotropic hypogonadism, NSMF
Introduction
A critical factor in the development and maturation of the hypothalamic-pituitary-gonadal (HPG) axis is the proper migration of GnRH neurons. GnRH neurons arise from the olfactory placode region where they migrate to the hypothalamus and send projections to the median eminence (Schwanzel-Fukuda et al. 1996; Wierman et al. 2004). Many factors or molecules act as guidance cues for proper migration of GnRH neurons (Wray 2010). Impairment of GnRH function results in human disease. When GnRH expression, secretion, and/or action are perturbed, delayed puberty due to idiopathic hypogonadotropic hypogonadism occurs (IHH). Since sense of smell is normal in this situation, it is often termed normosmic hypogonadotropic hypogonadism (nHH). Kallmann Syndrome (KS) is due to the failure of migration of GnRH and olfactory neurons and results in a phenotype combining both IHH and anosmia (Bhagavath et al. 2006).
Mutations in key genes account for ~40% of IHH/KS in patients (Layman 2013). These mutations affect the development, migration, pulsatile release/secretion and signaling of GnRH neurons. Identified genes with mutations include KAL1, NR0B1, GNRHR, FGFR1, KISS1R, TACR3, TAC3, FGF8, CHD7, PROKR2, PROK2, GNRH1, NELF, WDR11, PCSK1, LEP, and LEPR (Layman 2013). Recently, mutations in HS6ST1 and SEMA3A have been identified in nHH/KS patients (Tornberg et al. 2011; Hanchate et al. 2012), as have mutations in additional genes (Miraoui et al. 2013; Pingault et al. 2013). The inheritance of nHH/KS may be X-linked recessive, autosomal dominant, autosomal recessive, sporadic or digenic/oligogenic in a small proportion of patients (Sykiotis et al. 2010; Quaynor et al. 2011).
Knockdown of the nHH/KS gene nasal embryonic LHRH factor (NELF), which is also known as NMDA receptor synaptonuclear signaling and neuronal migration factor (NSMF), has been shown to impair GnRH neuron migration in vitro (Palevitch et al. 2009; Xu et al. 2010), but the mechanism by which this occurs is not well understood. This gene was differentially isolated from migratory GnRH neurons and found to be expressed ubiquitously in the brain but most prominently in the cortex (Kramer et al. 2000). Similarly, the rat ortholog, known as the Jacob protein and 98% identical in protein sequence, is most highly expressed in the cortex and the hippocampus (Mikhaylova et al. 2013). NELF is predominantly a nuclear protein in rats, as the majority of this protein is found in the nucleus and around the nuclear membrane (Dieterich et al. 2008; Mikhaylova et al. 2013). Immunofluorescence studies have confirmed the same location in mice (Xu et al. 2010) , although NELF has also been found in extra-nuclear areas of mouse cells such as the cytoplasm and plasma membrane (Kramer and Wray 2000; Kramer et al. 2001; Palevitch et al. 2009). Studies from our laboratory in which immortalized GnRH neurons were transfected with NELF knockdown constructs showed markedly impaired GnRH neuronal migration (Xu et al. 2010). In addition, Nelf knockdown in zebrafish demonstrated that GnRH neurons are directed to locations outside the forebrain (Palevitch et al. 2009).
Human NELF gene structure and a rare missense variant of undetermined significance were first reported in 2004 (Miura et al. 2004). Functional NELF mutations have been identified in IHH/KS patients (Pitteloud et al. 2007; Xu et al. 2011). However, since very few monogenic human NELF mutations have been identified, the exact inheritance pattern is yet to be defined. At least one patient has been reported to have biallelic, compound heterozygous NELF mutations (Xu et al. 2011) while heterozygous mutations are often accompanied by a mutation in a second gene (Pitteloud et al. 2007; Quaynor et al. 2011) Molecular features of NELF, such as its binding and interacting partners, its release, and signaling are yet to be discovered. Since NELF possesses a nuclear localization signal (NLS) and its three-dimensional structure contains a putative zinc finger domain, it is likely that NELF is a transcriptional factor (Xu et al. 2010). There is also evidence that the rat ortholog is involved in NMDA signaling in the brain (Dieterich et al. 2008; Mikhaylova et al. 2013).
Currently, five NELF variants have been included in the National Center for Biotechnology Information (NCBI) database for both mouse and human, but which ones are actually expressed in GnRH neurons are not known. Because GnRH neurons are few in number and are scattered throughout the brain, immortalized GnRH neuronal cell lines have been generated from transgenic mice that may be migratory (GN11 and NLT)(Radovick et al. 1991) or postmigratory (GT1-7) (Mellon et al. 1990). In 2010, Xu et al. observed that Nelf mRNA expression was not different in migratory vs. postmigratory immortalized GnRH neuronal cell lines, but NELF protein was more highly expressed in migratory cells (Xu et al. 2010). In addition, mouse and rat NELF orthologs exhibited several bands by western blot analysis, suggesting the presence of multiple variants (Dieterich et al. 2008; Xu et al. 2010; Mikhaylova et al. 2013). We therefore hypothesized that the mRNA/protein discordance and protein expression patterns could be due to variable expression of different splice variants of NELF. We also hypothesized that cellular localization of NELF in the nucleus or cytoplasm could be due to expression of a translated variant either containing or lacking the NLS. The purpose of the present study was to determine which NELF splice variants were present in immortalized migratory (NLT and GN11) and post-migratory (GT1-7) mouse GnRH neurons and FNCB4-hTERT GnRH neuronal cell lines in humans, as well as determining the subcellular localization of the most frequent splice variants.
Materials and Methods
NELF Primer Design for Colony PCR
The putative human and mouse NELF variant sequences were obtained by searching the mouse genome databases available at UCSC Genome Browser. Using the provided reference sequences, five mouse Nelf variants (Table 1A) and five human NELF variants (Table 1B) were identified in silico. Primers were designed to selectively amplify variant-specific regions of mouse Nelf and human NELF using Oligo 6.0 or Primer 3 software (Figure S1). These primers flanked the variable regions of each variant such that each could be specifically differentiated except v2 vs. v4. Therefore to differentiate v2 vs. v4, a pair of primers was designed to flank the v4-specific sequence (Figure S2). However, this primer set will also amplify the other variants in addition to v4, but the frequency of v4 was able to be determined.
Human Nelf variant classification
| Variant # | Exon 5* | Exon 6* | Exon 8* | AA | RefSeq# |
|---|---|---|---|---|---|
|
| |||||
| 1 | + | + | + | 530 | NM_001130969 |
| 2 | − | + | + | 528 | NM_015537 |
| 3 | + | − | + | 507 | NM_001130970 |
| 4 | − | − | + | 505 | NM_001130971 |
| 5 | + | + | − | 500 | NM_001178064 |
Compared to Variant 1(NM_001130969)
Mouse Nelf variant from colony PCR in percent (the number of clones is shown in parentheses)
| Cell type | V1 | V2 | V3 | V4 | V5 | V6 | V7 | V8 | Total |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| 5% | 72.1% | 7.4% | 6.6% | 1.9% | 6.9% | ||||
| NLT* | (13) | (186) | (19) | 0 | 0 | (17) | (5) | (18) | 258 |
| 5.6% | 73.8% | 5.6% | 5.3% | 6.6% | 1.6% | ||||
| GN11* | (17) | (222) | (16) | 0 | 0 | (21) | (20) | (5) | 301 |
| 7.8% | 76.9% | 4.6% | 4.3% | 1.4% | 4.9% | ||||
| GT1-7** | (27) | (267) | (16) | 0 | 0 | (15) | (5) | (17) | 347 |
| Total | 57 | 675 | 51 | 0 | 0 | 53 | 30 | 40 | 906 |
Migratory cells are GN11 and NLT cells.
Post-migratory cells are Gt1-7 cells.
RT-PCR and Cloning
Immortalized GnRH neuronal cells NLT(Radovick et al. 1991), GN11(Radovick et al. 1991), and GT1-7 (Mellon et al. 1990) in mouse and FNCB4-hTERT (Vannelli et al. 1995) in human were cultured as described previously. GnRH neuronal cells were kindly provided by Sally Radovick (GN11 and NLT), Pamela Mellon (GT1-7), and Soo-Hyun Kim (FNCB4-hTERT). RNA from each of the four neuronal cell lines was extracted using TRI ®Reagent. Reverse-transcription Polymerase Chain Reaction (RT-PCR) was performed with 100ng of RNA from each of the four cell lines using SuperScript™ III One-Step RT-PCR with Platinum®Taq. Negative controls (no RNA or no RT) were used to rule out contamination. RT-PCR products were confirmed by agarose gel electrophoresis, and then cloned into Top 10F’ cells using the Invitrogen pCR 2.1 TOPO TA cloning vector, and incubated at 37° C. After 18 hours, 20-40 single colonies with the correct insert were randomly picked per LB-ampicillin plate using Xgal. The selected colonies were then screened for inserts by PCR and analyzed by agarose gel electrophoresis for the correct size bands.
Denaturing Gradient Gel Electrophoresis and Sanger Sequencing
Confirmed colony PCR products were subjected to denaturing gradient gel electrophoresis (DGGE), as described previously (Layman et al. 1997), to determine the variant present in each colony. Variants were then confirmed by DNA sequencing. Briefly, the PCR products were purified by ethanol precipitation and sequenced using the BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Purified sequencing reactions were run on an ABI 310 automated DNA sequencer and analyzed using BLAST (NCBI, Bethesda MD).
Plasmid Constructs for Nelf Variant 2 and 3
To clone full length mouse Nelf variants 2 and 3, HindIII and Csp45 I restriction recognition sites were added to both forward and reverse Nelf primers, respectively. The KOZAK sequence before the start codon was also added to facilitate translation (Kozak 1987). Total mRNA was extracted from NLT cells and converted to cDNA using SuperScript™ III First Strand cDNA synthesis kit (Invitrogen; Carlsbad, CA). This was followed by PCR using Platinum®Taq, and the resulting product was transformed into Top10’F competent cells. Colonies were screened and confirmed by DNA sequencing. Correct inserts were then identified after digestion with appropriate enzymes and ligated into pcDNA 3.1(+) myc-his, which were transiently transfected into NLT cells by lipofection.
Immunoblotting and immunofluorescence experiments were performed to confirm the size and the location of the cloned mouse NELF variants using c-Myc antibody [c-Myc.A7 (sc-56634)] from Santa Cruz Biotechnology Inc., Dallas, TX as described previously (Xu et al. 2010). For immunofluorescence, TO-PRO®-3 Iodide from Life Technologies (Grand Island, New York) was used for nuclear staining. NLT cells were viewed using laser-scan-confocal microscopy (Zeiss Axiovert LSM 510, Carl Zeiss, Jena, Germany). For v2 and v3 protein expression, random fields were selected from cells transfected with each variant and scored by two different observers. NLT cells were quantitated according to the number of cells for which there was greater nuclear than cytoplasmic expression (N>C) vs. cytoplasmic greater than nuclear (C>N) expression.
RT-qPCR
First strand cDNA was synthesized using 1000ng of RNA from each of the three mouse cell lines according to the directions from High Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Foster City, CA). RT-qPCR was performed three times in triplicate using 10ng of this cDNA using variant-specific primers to detect variants 1-5 (primer sequence is shown in Supplemental Table 1). The reaction was performed using GeneRead qPCR SYBR Green Mastermix (QIAGEN, Valencia, CA) and run on an Applied Biosystems 7300 Real-time PCR System (Life Technologies, Carlsbad, CA). The relative expression of the different Nelf variants in migratory and post migratory cell lines was normalized to v1 after normalization to β-actin (Actb).
Statistical Analysis
The prevalence of the most common splice variant in the three mouse cell lines, as well as the proportion of cells with N>C expression vs. C>N protein expression, was compared using chi square analysis.
Results
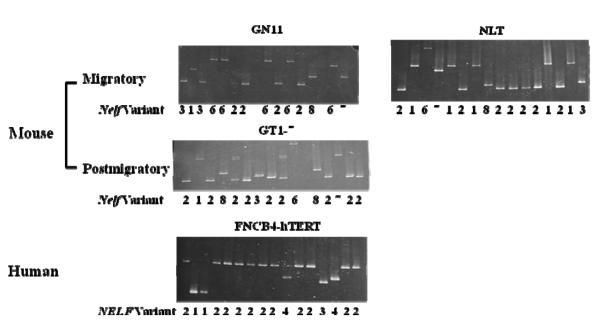
The RT-PCR product fragment size did not differ by agarose gel electrophoresis for any of the variants (data not shown). By colony PCR, a total of 906 clones using the initial primer set for mouse and 308 clones for human were analyzed. DNA from individual colonies picked from each plate after transformation did not show any size difference on agarose gel electrophoresis. However, NELF variants could be differentiated by DGGE of the PCR products from individual colonies because of differences in melting temperatures, which were then confirmed by DNA sequencing. NELF variants are shown for each of the three mouse (GN11, GT1-7 and NLT) and the single human (FNCB4-hTERT) GnRH cell lines (Figure 1).
Figure 1.
(A) Denaturing gradient gel results are shown for three mouse (GN11, NLT, and GT1-7) and human (FNCB4-hTERT) cells. Findings are shown for clones for each cell line; and the number below each lane indicates the variant identification (v1-3 and v6-8 for mouse; and v1-v4 for human).
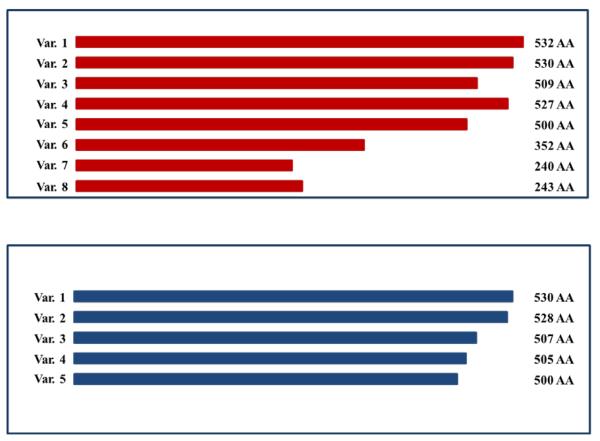
(B) Predicted protein size is shown for mouse variants. Although predicted for v4 and v5, no mRNA expression was observed for these variants. Only v2 and v3 have confirmed protein expression (the others are predicted).
(C) Predicted protein size is shown for human variants.
We confirmed three of the five variants (v1-3) in the UCSC database for mouse Nelf, but did not identify v4 or v5 in 906 clones. Although our initial primers could not differentiate v2 from v4 (because the sequence of this region was identical), no v4 was seen in any additional clones using primers that could differentiate v4 vs. all others: 0/90 clones in NLT; 0/83 in GN11; and 0/89 in GT1-7 cells. This finding makes it very unlikely that v4 is expressed in mouse GnRH neuronal cells. Three previously unreported variants included v6 and v7, both of which had an insertion of intron 4 (v6 with 295 nucleotides and v7 with 82 nucleotides); and v8, which had an insertion of 34 bases from intron 5 (Table 1). Although these three previously unreported variants contained additional nucleotides, they were all predicted to be translated by in silico analysis (Figure 1).
Mouse Nelf variant classification
| Variant# | Exon 5* | Exon 6* | Intron*
inserted |
# bp inserted |
Ansino Aeldx |
RefSeq# |
|---|---|---|---|---|---|---|
|
| ||||||
| 1 |
+ | + | − | − | 532 | N3M_00103986 |
| 2 |
− | + | − | − | 530 | NM_001039387 |
| 3 |
− | − | − | − | 509 | NM_020276 |
| 4 |
− | + | − | − | 527 | NM_001177654 |
| 5 |
− | + | − | − | 500 | NM_001177655 |
| 6 |
+ | + | 4 | 295 | 352 | |
| 7 |
+ | + | 4 | 92 | 240 | |
| 8 |
+ | + | 5 | 34 | 243 | |
Compared to Variant 1(NM_001039386): + and − indicate sequence if is present or absent
The variants in red are the novel variants identified and sequenced.
V4 has 9 additional nucleotides deleted (from exon 15) compared with V2.
V5 has exon 8 deleted.
Colony PCR findings from this study indicate that v2 is the most common (Table 2) variant in all three mouse cell lines regardless of migratory (GN11 and NLT) vs. postmigratory (GT1-7) status. There was no difference in the percentage of v2 in these three cell lines—72.1% in NLT, 73.8% in GN11, and 76.9% in GT1-7 cells. All the other variants in mouse cell lines were uncommon, ranging between 1.4%-7.8% (Table 2). Of interest, the v3 transcript was expressed in all three mouse cell lines. Since this splice variant lacks the sequence for the NLS, it could explain the finding of NELF protein localized outside of the nucleus.
Human Nelf variants from colony PCR in percent (the number of clones is shown in parentheses)
| FNCB4-hTERTcells | V1 | V2 | V3 | V4 | V5 | Total |
|---|---|---|---|---|---|---|
| 5.2%(16) | 66.6%(205) | 10%(31) | 18.2%(56) | 0 | 308 |
In addition to colony PCR, RT-qPCR was performed using splice variant-specific primers for v1-v5. Only v1-v3 were observed, and each was confirmed by DNA sequencing. Initially, v2 and v4 were seen in all three mouse cell lines, but upon DNA sequencing, only v2 was observed (no v4 transcripts were identified) indicating that v4 primers could amplify v2 (data not shown). Therefore, similar to colony PCR, v2 was the most common variant, and no v4 was seen. In addition, a small amount of v5 was observed by RT-qPCR, but upon DNA sequencing, v5 was not found (v2 was present). Therefore, colony PCR of cloned RT-PCR products and RT-qPCR yielded similar findings.
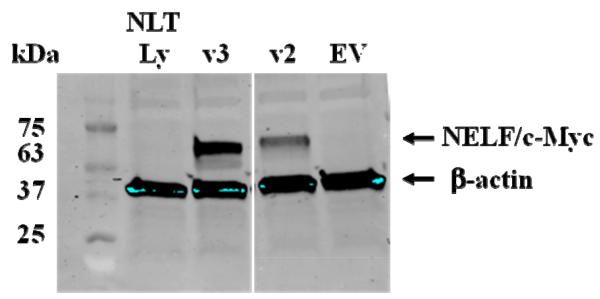
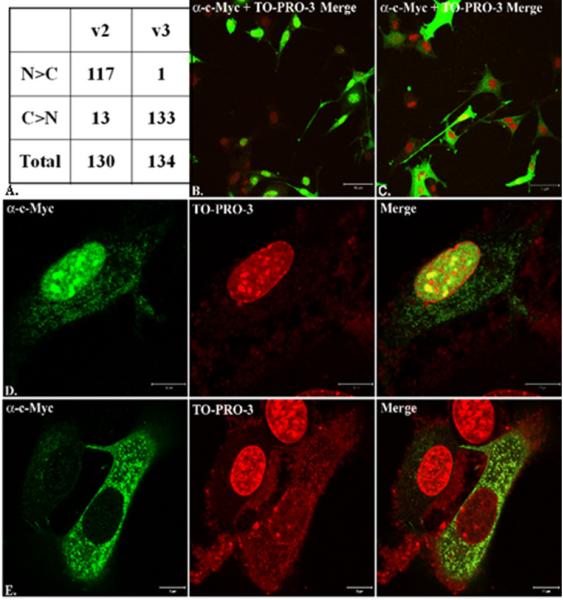
To determine protein expression and cellular localization, mouse v2 and v3 were cloned into pcDNA 3.1(+) myc-his and transfected into NLT cells. Proteins for both variants were expressed and analyzed by immunoblotting and immunofluorescence using the anti-Myc antibody. In NLT whole cell lysates, both v2 and v3 were expressed (Figure 2). Using confocal microscopy, 117 /130 (90%) of NLT cells transfected with v2 had principally nuclear localization, compared to 1/134 (0.7%) of v3 (Figure 3) (P<0.0001).
Figure 2.
Immunoblot analysis of mouse Nelf v2 and v3, which were cloned into mammalian expression vectors, transfected into NLT cells, and analyzed for exogenous NELF protein. The size for v2 is 63kDa, while v3 is 61kDa. Controls are shown as NLT Lys with no transfection (NLT Lysate) and EV (Empty Vector).
Figure 3.
Localization of mouse v2 (nucleus) and v3 (non-nuclear) by immunofluorescence using confocal microscopy. In (A), 130 NLT cells transfected with v2 and 134 cells transfected with v3 were quantified as being predominantly nuclear (N>C) or predominantly cytoplasmic (C>N). N = nuclear; C = cytoplasmic. Low 10X power views of v2 (B) and v3 (B) demonstrate cellular distribution. In addition, high power (40X) images of v2 (D) and v3 (E) are shown with c-Myc expression (indicating NELF), nuclear expression (TO-PRO-3), and merged images.
In human GnRH neuronal cells, we confirmed four of the five predicted human NELF variants from FNCB4-hTERT cells (v1-4). Similar to the mouse, the corresponding v5 was not observed in 308 clones from human GnRH neurons (Table 1). As in the mouse, v2 was by far the most common variant in human FNCB4-hTERT cells being found in 66.6% of clones, but in contrast to mouse, v4 was not only identified, it was the second most common variant at 18.2% in human GnRH cells. However, v4 in the human corresponds to v3 in the mouse since both variants lack sequences encoding the NLS.
Discussion
In this study, five Nelf splice variants (v1-v5) in mouse were predicted in silico, but we identified six variant transcripts (v1-3, v6-v8). Of the six identified in mouse, three variants were previously unreported. Variant 2 was the predominant one expressed in all mouse cell lines whether or not they were migratory (GN11 and NLT) or postmigratory (GT1-7) cells. Variant 2 only differs from v1 by the absence of six nucleotides within exon 5 in v2 such that it is 530 amino acids rather than 532. The expression of the other variants in mouse GnRH neurons was much lower and ranged from 1.4%-7.8%. Interestingly, neither predicted v4 nor v5 was found in any of the mouse GnRH neuronal cells.
Since v2 is the most common variant and is found in both migratory and postmigratory mouse GnRH neurons, it seems unlikely that this variant is only important for GnRH neuron migration. Variant 3 is expressed in mouse GnRH neurons, and this is the only one identified in mouse that lacks the NLS, encoded by the amino acid sequence RRKR. We hypothesized that migratory GnRH neurons would have a different Nelf variant expression profile compared to postmigratory neurons, but this was not observed. In previous studies of NELF knockdown, miRNA or morpholino targeted sequences were common to all splice variants so differential effects of splice variants could not be determined (Palevitch et al. 2009; Xu et al. 2010).
In addition to demonstrating mRNA expression in mouse GnRH neurons, we also showed that mouse v2 and v3 protein is expressed when cloned into a mammalian expression vector and transfected into migratory GnRH neurons. However since v3 lacked the nuclear localization signal, it was expressed at extranuclear sites in contrast to v2, which was principally expressed in the nucleus. This explains previous findings from other studies that NELF is predominantly expressed in the nucleus, but has also been aligned along the cell membrane and cytosol (Kramer and Wray 2001; Xu et al. 2010). Although we have localized v2 and v3 in immortalized GnRH neurons, the differential and temporal expression patterns and how these translate into function are not known in the living mouse. In rat, splice variants of the Jacob protein have also been characterized, with and without the NLS, as in the mouse (Kindler et al. 2009). Variants 1 and 2 in both the mouse and rat are 532 and 530 amino acids, respectively.
We were not able to identify v4 in any of the three immortalized mouse GnRH neuronal cell lines. However, three novel variant (v6-v8) transcripts were present in migratory and postmigratory GnRH cells in similar frequencies. Variants 6 and 7 differ by the insertion of intron 4 sequences, while v8 contains intron 5 sequences. Utilizing GENE Runner Software 3.05, v6-8 are predicted to be translated, but this will require further studies to determine if these proteins are expressed in GnRH neurons. Variants 6-8 are unlikely to be artifacts of cloning because they were identified all three mouse cell lines and in at least five clones.
In human, five NELF splice variants (v1-v5) were predicted in silico, but we identified four variants (v1-v4) in FNCB4-hTERT cells. FNCB4-hTERT cells were isolated and cloned from human fetal olfactory neuroepithelium and express both neuronal and olfactory markers (Vannelli et al. 1995). In addition, these cells were also shown to express GnRH and its receptor, providing evidence that they are migratory embryonic GnRH olfactory neuroblasts (Romanelli et al. 2004). Similar to mouse, v2 was also the dominant variant, occurring in 66.6% of colonies screened, while the percentage expression of the other variants ranged from 5%-19% (Table 2). Human v4 is similar to mouse v3 in that it lacks exons 5 and 6, which encode the NLS. Variant 5, which was not identified in human or mouse, lacks exon 8, suggesting that sequences encoded by this exon may have important functions in GnRH neurons.
In conclusion, we have identified six mouse Nelf splice variant transcripts, of which three were previously undescribed, and four human NELF splice variants in immortalized GnRH neuronal cell lines. In both mouse and human, nuclear-expressed v2 is the predominant splice variant expressed, while the others are all less common. However, non-nuclear v3 (mouse) and v4 (human) lack an NLS. In humans this non-nuclear variant is the second most common variant approximating 20% of clones. Importantly, both the nuclear v2 and the non-nuclear v3 proteins are also expressed following transfection in NLT cells suggesting they have biological importance in GnRH neurons. Endogenous expression of these splice variants could account for the previous mRNA/protein discordance and bands of different molecular weights observed by western blot analysis (Xu et al. 2010). This characterization of the major NELF splice variants should pave the way for future studies to characterize the exact function of each splice variant and NELF’s signaling mechanism(s).
Supplementary Material
Highlights.
The molecular basis is known for~35-40% of hypogonadotropic hypogonadism and 15% of hypergonadotropic hypogonadism in women
For endometriosis, polycystic ovary syndrome, and leiomyomata, no causative gene has been identified
Genetic testing can be used for clinical diagnosis in hypogonadism and in spontaneous ovarian hyperstimulation syndrome
Clinical genetic testing is not practical for endometriosis, polycystic ovary syndrome, and leiomyomata.
Acknowledgments
L.C.L. was supported from NIH grant HD33004 for this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure: The authors have nothing to disclose.
REFERENCES
- Bhagavath B, Podolsky RH, Ozata M, Bolu E, Bick DP, Kulharya A, et al. Clinical and molecular characterization of a large sample of patients with hypogonadotropic hypogonadism. Fertil Steril. 2006;85(3):706–713. doi: 10.1016/j.fertnstert.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Karpova A, Mikhaylova M, Zdobnova I, Konig I, Landwehr M, et al. Caldendrin-Jacob: a protein liaison that couples NMDA receptor signalling to the nucleus. PLoS Biol. 2008;6(2):e34. doi: 10.1371/journal.pbio.0060034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanchate NK, Giacobini P, Lhuillier P, Parkash J, Espy C, Fouveaut C, et al. SEMA3A, a gene involved in axonal pathfinding, is mutated in patients with Kallmann syndrome. PLoS Genet. 2012;8(8):e1002896. doi: 10.1371/journal.pgen.1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler S, Dieterich DC, Schutt J, Sahin J, Karpova A, Mikhaylova M, et al. Dendritic mRNA targeting of Jacob and N-methyl-d-aspartate-induced nuclear translocation after calpain-mediated proteolysis. J Biol Chem. 2009;284(37):25431–25440. doi: 10.1074/jbc.M109.022137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer PR, Wray S. Novel gene expressed in nasal region influences outgrowth of olfactory axons and migration of luteinizing hormone-releasing hormone (LHRH) neurons. Genes Dev. 2000;14(14):1824–1834. [PMC free article] [PubMed] [Google Scholar]
- Kramer PR, Wray S. Nasal embryonic LHRH factor (NELF) expression within the CNS and PNS of the rodent. Brain Res Gene Expr Patterns. 2001;1(1):23–26. doi: 10.1016/s1567-133x(01)00004-7. [DOI] [PubMed] [Google Scholar]
- Layman LC. The genetic basis of female reproductive disorders: Etiology and clinical testing. Mol Cell Endocrinol. 2013;370(1-2):138–148. doi: 10.1016/j.mce.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layman LC, Lee EJ, Peak DB, Namnoum AB, Vu KV, van Lingen BL, et al. Delayed puberty and hypogonadism caused by mutations in the follicle-stimulating hormone beta-subunit gene. N Engl J Med. 1997;337(9):607–611. doi: 10.1056/NEJM199708283370905. [DOI] [PubMed] [Google Scholar]
- Mellon PL, Windle JJ, Goldsmith PC, Padula CA, Roberts JL, Weiner RI. Immortalization of hypothalamic GnRH neurons by genetically targeted tumorigenesis. Neuron. 1990;5(1):1–10. doi: 10.1016/0896-6273(90)90028-e. [DOI] [PubMed] [Google Scholar]
- Mikhaylova M, Karpova A, Bar J, Bethge P, Yuanxiang P, Chen Y, et al. Cellular distribution of the NMDA-receptor activated synapto-nuclear messenger Jacob in the rat brain. Brain Struct Funct. 2013 doi: 10.1007/s00429-013-0539-1. [DOI] [PubMed] [Google Scholar]
- Miraoui H, Dwyer AA, Sykiotis GP, Plummer L, Chung W, Feng B, et al. Mutations in FGF17, IL17RD, DUSP6, SPRY4, and FLRT3 Are Identified in Individuals with Congenital Hypogonadotropic Hypogonadism. Am J Hum Genet. 2013;92(5):725–743. doi: 10.1016/j.ajhg.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Acierno JS, Jr., Seminara SB. Characterization of the human nasal embryonic LHRH factor gene, NELF, and a mutation screening among 65 patients with idiopathic hypogonadotropic hypogonadism (IHH) J Hum Genet. 2004;49(5):265–268. doi: 10.1007/s10038-004-0137-4. [DOI] [PubMed] [Google Scholar]
- Palevitch O, Abraham E, Borodovsky N, Levkowitz G, Zohar Y, Gothilf Y. Nasal embryonic LHRH factor plays a role in the developmental migration and projection of gonadotropin-releasing hormone 3 neurons in zebrafish. Dev Dyn. 2009;238(1):66–75. doi: 10.1002/dvdy.21823. [DOI] [PubMed] [Google Scholar]
- Pingault V, Bodereau V, Baral V, Marcos S, Watanabe Y, Chaoui A, et al. Loss-of-Function Mutations in SOX10 Cause Kallmann Syndrome with Deafness. Am J Hum Genet. 2013;92(5):707–724. doi: 10.1016/j.ajhg.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, Dwyer A, et al. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007;117(2):457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaynor SD, Kim HG, Cappello EM, Williams T, Chorich LP, Bick DP, et al. The prevalence of digenic mutations in patients with normosmic hypogonadotropic hypogonadism and Kallmann syndrome. Fertil Steril. 2011;96(6):1424–1430. doi: 10.1016/j.fertnstert.2011.09.046. e1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radovick S, Wray S, Lee E, Nicols DK, Nakayama Y, Weintraub BD, et al. Migratory arrest of gonadotropin-releasing hormone neurons in transgenic mice. Proc Natl Acad Sci U S A. 1991;88(8):3402–3406. doi: 10.1073/pnas.88.8.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanelli RG, Barni T, Maggi M, Luconi M, Failli P, Pezzatini A, et al. Expression and function of gonadotropin-releasing hormone (GnRH) receptor in human olfactory GnRH-secreting neurons: an autocrine GnRH loop underlies neuronal migration. J Biol Chem. 2004;279(1):117–126. doi: 10.1074/jbc.M307955200. [DOI] [PubMed] [Google Scholar]
- Schwanzel-Fukuda M, Crossin KL, Pfaff DW, Bouloux PM, Hardelin JP, Petit C. Migration of luteinizing hormone-releasing hormone (LHRH) neurons in early human embryos. J Comp Neurol. 1996;366(3):547–557. doi: 10.1002/(SICI)1096-9861(19960311)366:3<547::AID-CNE12>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Sykiotis GP, Plummer L, Hughes VA, Au M, Durrani S, Nayak-Young S, et al. Oligogenic basis of isolated gonadotropin-releasing hormone deficiency. Proc Natl Acad Sci U S A. 2010;107(34):15140–15144. doi: 10.1073/pnas.1009622107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornberg J, Sykiotis GP, Keefe K, Plummer L, Hoang X, Hall JE, et al. Heparan sulfate 6-O-sulfotransferase 1, a gene involved in extracellular sugar modifications, is mutated in patients with idiopathic hypogonadotrophic hypogonadism. Proc Natl Acad Sci U S A. 2011;108(28):11524–11529. doi: 10.1073/pnas.1102284108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannelli GB, Ensoli F, Zonefrati R, Kubota Y, Arcangeli A, Becchetti A, et al. Neuroblast long-term cell cultures from human fetal olfactory epithelium respond to odors. J Neurosci. 1995;15(6):4382–4394. doi: 10.1523/JNEUROSCI.15-06-04382.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierman ME, Pawlowski JE, Allen MP, Xu M, Linseman DA, Nielsen-Preiss S. Molecular mechanisms of gonadotropin-releasing hormone neuronal migration. Trends Endocrinol Metab. 2004;15(3):96–102. doi: 10.1016/j.tem.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Wray S. From nose to brain: development of gonadotrophin-releasing hormone-1 neurones. J Neuroendocrinol. 2010;22(7):743–753. doi: 10.1111/j.1365-2826.2010.02034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Bhagavath B, Kim HG, Halvorson L, Podolsky RS, Chorich LP, et al. NELF is a nuclear protein involved in hypothalamic GnRH neuronal migration. Mol Cell Endocrinol. 2010;319(1-2):47–55. doi: 10.1016/j.mce.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Kim HG, Bhagavath B, Cho SG, Lee JH, Ha K, et al. Nasal embryonic LHRH factor (NELF) mutations in patients with normosmic hypogonadotropic hypogonadism and Kallmann syndrome. Fertil Steril. 2011 doi: 10.1016/j.fertnstert.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.