Abstract
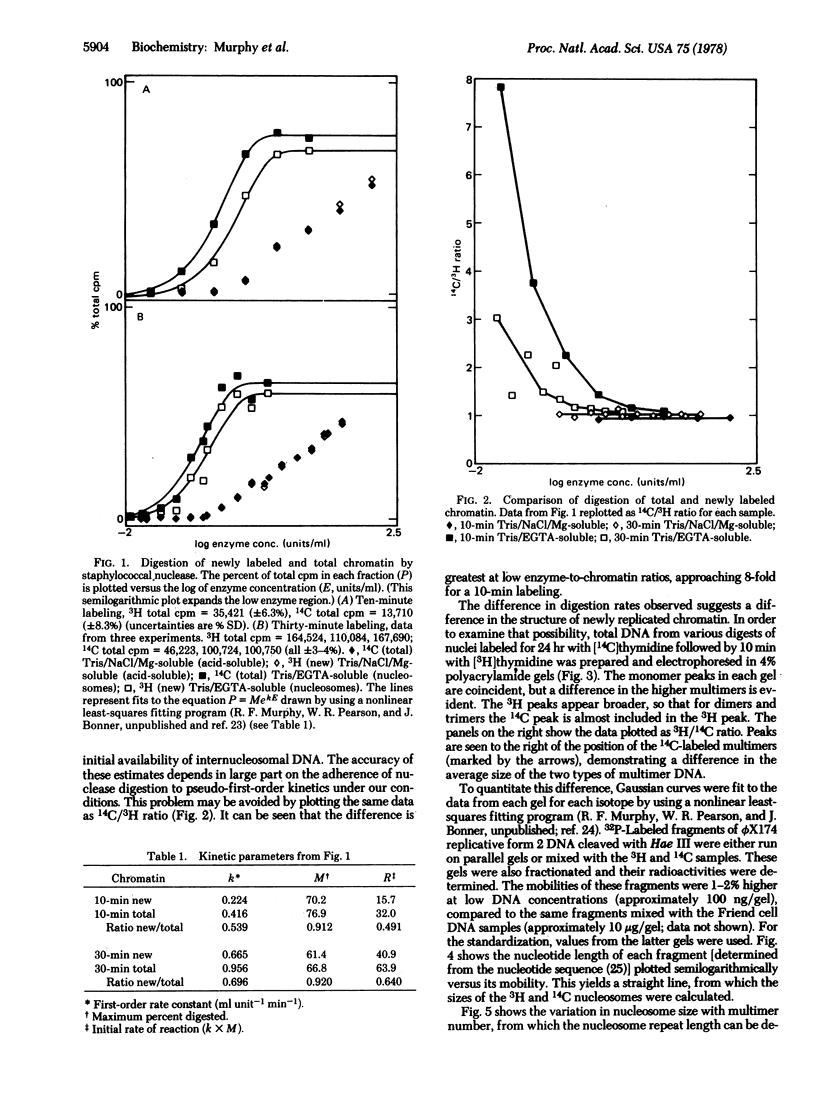
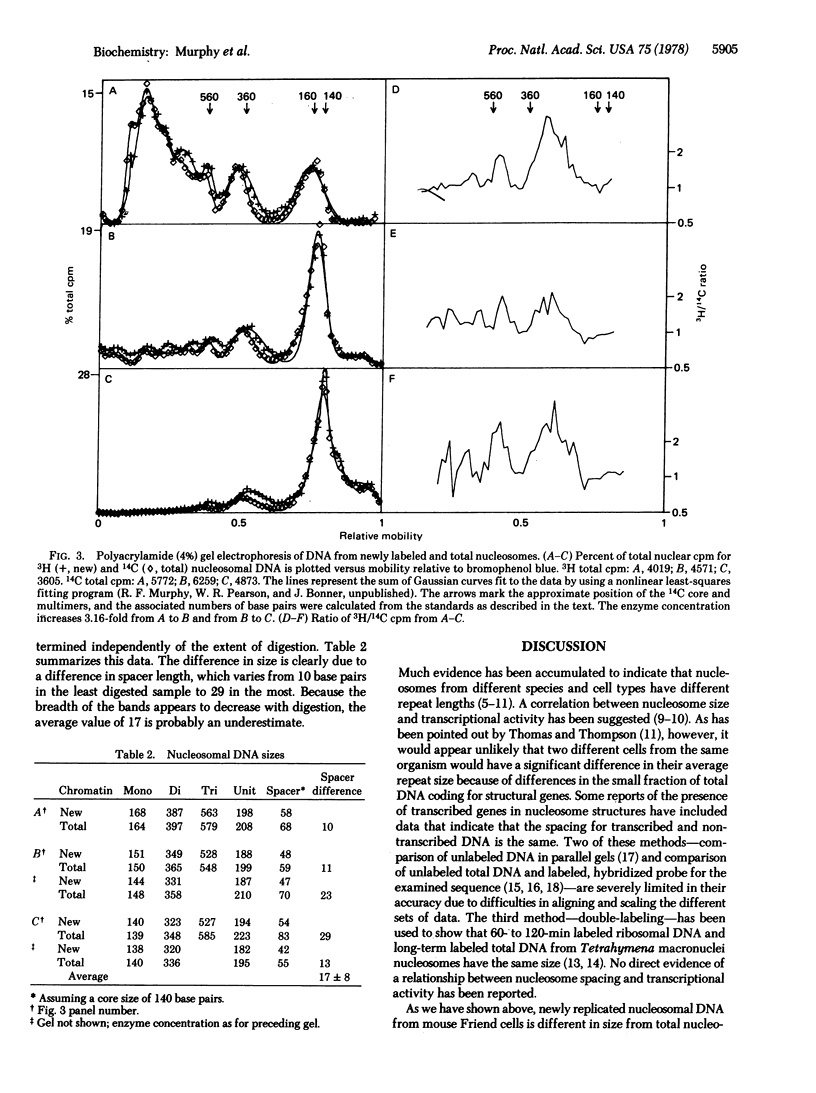
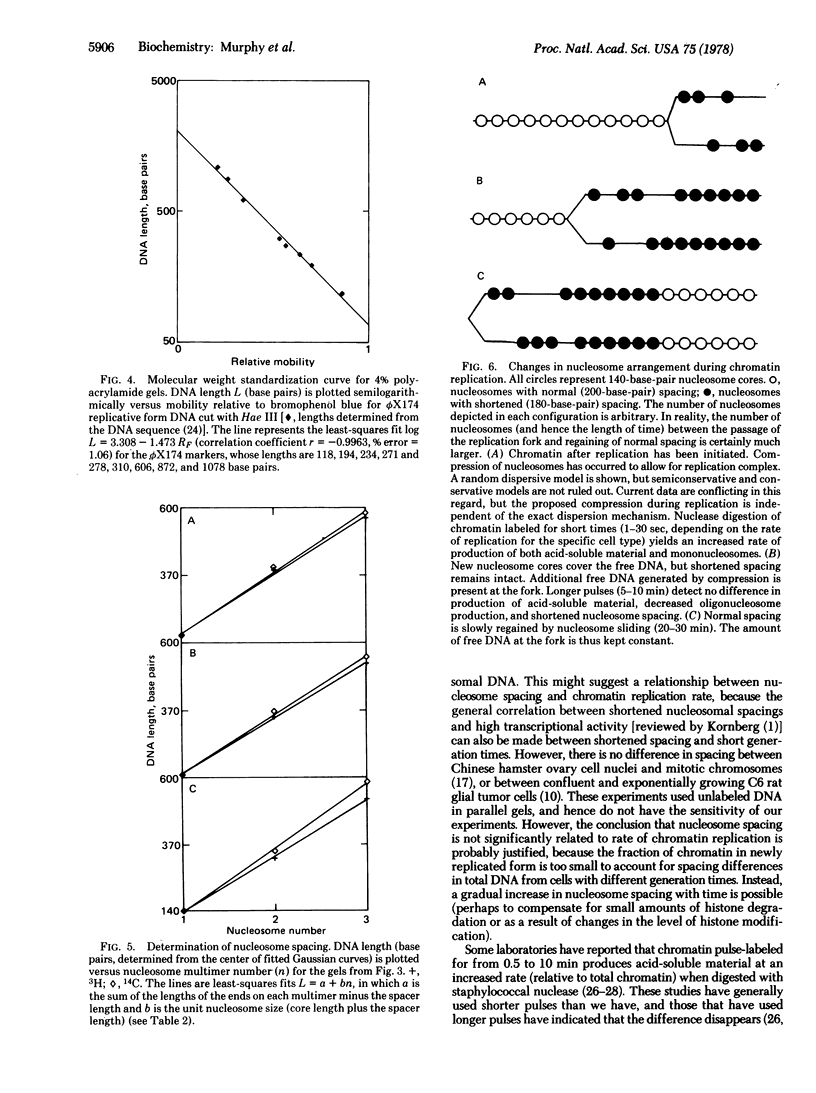
Chromatin from Friend leukemia cells labeled with [14C]thymidine for 24 hr followed by [3H]thymidine for 10 min is converted into nucleosomes by staphylococcal nuclease at only half the rate that total chromatin is converted. Polyacrylamide gel electrophoresis of nucleosomal DNA from cells labeled for 24 hr with [14C]thymidine followed by 10 min with [3H]thymidine demonstrates that the internucleosomal spacer of newly replicated chromatin is approximately 20 base pairs shorter than that of total chromatin. The implications of this difference for models of chromatin structure are discussed.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bakayev V. V., Bakayeva T. G., Varshavsky A. J. Nucleosomes and subnucleosomes: heterogeneity and composition. Cell. 1977 Jul;11(3):619–629. doi: 10.1016/0092-8674(77)90079-4. [DOI] [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. Polynucleotide kinase exchange reaction: quantitave assay for restriction endonuclease-generated 5'-phosphoroyl termini in DNA. J Biol Chem. 1977 May 25;252(10):3176–3184. [PubMed] [Google Scholar]
- Bokhon'ko A., Reeder R. H. The subunit structure of mouse satellite chromatin. Biochem Biophys Res Commun. 1976 May 3;70(1):146–152. doi: 10.1016/0006-291x(76)91120-7. [DOI] [PubMed] [Google Scholar]
- Bostock C. J., Christie S., Hatch F. T. Accessibility of DNA in condensed chromatin to nuclease digestion. Nature. 1976 Aug 5;262(5568):516–519. doi: 10.1038/262516a0. [DOI] [PubMed] [Google Scholar]
- Burgoyne L. A., Mobbs J. D., Marshall A. J. Chromatin structure: a property of the higher structures of chromatin and in the time course of its formation during chromatin replication. Nucleic Acids Res. 1976 Dec;3(12):3293–3304. doi: 10.1093/nar/3.12.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton J. L., Bellard M., Chambon P. Biochemical evidence of variability in the DNA repeat length in the chromatin of higher eukaryotes. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4382–4386. doi: 10.1073/pnas.73.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton J. L., Hancock R., Oudet P., Chambon P. Biochemical and electron-microscopic evidence that the subunit structure of Chinese-hamster-ovary interphase chromatin is conserved in mitotic chromosomes. Eur J Biochem. 1976 Nov 15;70(2):555–568. doi: 10.1111/j.1432-1033.1976.tb11047.x. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Melton D. A. The length of nucleosome-associated DNA is the same in both transcribed and nontranscribed regions of chromatin. Nature. 1978 May 25;273(5660):317–319. doi: 10.1038/273317a0. [DOI] [PubMed] [Google Scholar]
- Hildebrand C. E., Walters R. A. Rapid assembly of newly synthesized DNA into chromatin subunits prior to joining to small DNA replication intermediates. Biochem Biophys Res Commun. 1976 Nov 8;73(1):157–163. doi: 10.1016/0006-291x(76)90510-6. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Levy A., Jakob K. M. Nascent DNA in nucleosome like structures from chromatin. Cell. 1978 Jun;14(2):259–267. doi: 10.1016/0092-8674(78)90112-5. [DOI] [PubMed] [Google Scholar]
- Lohr D., Corden J., Tatchell K., Kovacic R. T., Van Holde K. E. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1977 Jan;74(1):79–83. doi: 10.1073/pnas.74.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Mathis D. J., Gorovsky M. A. Subunit structure of rDNA-containing chromatin. Biochemistry. 1976 Feb 24;15(4):750–755. doi: 10.1021/bi00649a005. [DOI] [PubMed] [Google Scholar]
- Morris N. R. A comparison of the structure of chicken erythrocyte and chicken liver chromatin. Cell. 1976 Dec;9(4 Pt 1):627–632. doi: 10.1016/0092-8674(76)90045-3. [DOI] [PubMed] [Google Scholar]
- Morris N. R. Nucleosome structure in Aspergillus nidulans. Cell. 1976 Jul;8(3):357–363. doi: 10.1016/0092-8674(76)90147-1. [DOI] [PubMed] [Google Scholar]
- Noll M. Differences and similarities in chromatin structure of Neurospora crassa and higher eucaryotes. Cell. 1976 Jul;8(3):349–355. doi: 10.1016/0092-8674(76)90146-x. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Davidson E. H., Britten R. J. A program for least squares analysis of reassociation and hybridization data. Nucleic Acids Res. 1977 Jun;4(6):1727–1737. doi: 10.1093/nar/4.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper P. W., Celis J., Kaltoft K., Leer J. C., Nielsen O. F., Westergaard O. Tetrahymena ribosomal RNA gene chromatin is digested by micrococcal nuclease at sites which have the same regular spacing on the DNA as corresponding sites in the bulk nuclear chromatin. Nucleic Acids Res. 1976 Feb;3(2):493–505. doi: 10.1093/nar/3.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Seale R. L. Assembly of DNA and protein during replication in HeLa cells. Nature. 1975 May 15;255(5505):247–249. doi: 10.1038/255247a0. [DOI] [PubMed] [Google Scholar]
- Seale R. L. Nucleosomes associated with newly replicated DNA have an altered conformation. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2717–2721. doi: 10.1073/pnas.75.6.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadafora C., Bellard M., Compton J. L., Chambon P. The DNA repeat lengths in chromatins from sea urchin sperm and gastrule cells are markedly different. FEBS Lett. 1976 Oct 15;69(1):281–285. doi: 10.1016/0014-5793(76)80704-1. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Furber V. Yeast chromatin structure. FEBS Lett. 1976 Jul 15;66(2):274–280. doi: 10.1016/0014-5793(76)80521-2. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Thompson R. J. Variation in chromatin structure in two cell types from the same tissue: a short DNA repeat length in cerebral cortex neurons. Cell. 1977 Apr;10(4):633–640. doi: 10.1016/0092-8674(77)90096-4. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Sargent T. D., Murphy R. F., Bonner J. Physical properties of chemically acetylated rat liver chromatin. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3244–3248. doi: 10.1073/pnas.74.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Removal of histone H1 exposes a fifty base pair DNA segment between nucleosomes. Biochemistry. 1976 Jul 27;15(15):3307–3314. doi: 10.1021/bi00660a022. [DOI] [PubMed] [Google Scholar]



