Abstract
Formins are actin binding proteins conserved across species from plants to humans. The formin family is defined by their common FH2 domains. The 15 distinct human formins are involved in a broad range of cellular functions, including cell adhesion, cytokinesis, cell polarity, and cell morphogenesis. Their commonality is actin polymerization activity inherent to FH2 domains. While still requiring much study, biochemical activity of formins has been carefully described. In contrast, much less is known of their activities in complex living systems. With the diversity of the formin family and the actin structures that they affect, an extensive future of study beckons. In this study, we report the expression level of all 15 formins in 22 different human cell and tissue types using quantitative real-time PCR (qPCR). Identification of major themes in formin expression and documentation of expression profiles should facilitate the cellular study of formins.
Introduction
Formins are a class of actin-modifying proteins participating in cell adhesion, cytokinesis, cell polarity, and cell morphogenesis. Similar to other actin-assembly proteins such as Arp2/3, Spire or Cobl, formins have been shown to nucleate, cap, sever, bundle, and polymerize actin in vitro, but unlike other actin-modifying proteins, actin polymerizes linear actin filaments [Wallar and Alberts, 2003; Kovar, 2006; Goode and Eck, 2007; Chesarone et al., 2010]. Most formins have been genetically characterized in significant detail, allowing for functional analysis and classification based on domain structure. All formins contain a characteristic Formin Homology 2 (FH2) domain, which nucleates actin and remains associated with the fast-growing, barbed end of the actin filament throughout elongation Higgs, 2005; Kovar, 2006; Paul and Pollard, 2009]. The C-terminally located Formin Homology 1 (FH1) domain is proline rich and sequesters profilin-actin for facilitated use by the FH2 domain [Evangelista et al., 2003]. The FH2 domain forms a head-to-tail dimer, which is necessary for formins to carry out their actin-polymerizing function and moves with the growing filament, processively capping the barbed end and protecting it from capping proteins [Zigmond et al., 2003; Copeland et al., 2004; Campellone and Welch, 2010]. Formin function is regulated at several levels, including recruitment to specific cellular locations and the timing of the functional stages of autoinhibition, activation, and the actin nucleating and elongating activity [Goode and Eck, 2007; Aspenstrom, 2010; Chesarone, et al., 2010]. Formin function varies in efficiency between family members.
On a molecular level, the genomes of humans, mice, chickens, protists and yeast have been analyzed for the presence of FH2 domains and the resulting 15 formins identified in humans are the only existent human formins. These 15 formins are divided into seven groups based on domain similarities and phylogenetic relationships (Table 1) [Higgs and Peterson, 2005; Schönichen and Geyer, 2010].
Table 1. Human formin family nomenclature and sequence references.
The seven formin families with all their members are listed, as well as the alternate nomenclature for each formin. The 15 mammalian formins analyzed in this study are grouped into families based on FH2 phylogeny with gene names and accession numbers provided. The amino acid and genomic accession sequences used for alignment and oligonucleotide design are listed.
| Formin Family | Formin Name | Alternate Name(s) + References | Protein Accession # | Gene Name | Gene Accession # |
|---|---|---|---|---|---|
| Diaphanous formins (Dia) | Dia1 | Protein diaphanous homolog 1 = DIAP1, DIAPH1 Diaphanous-related formin 1 = DRF1 [Castrillon and Wasserman, 1994]. |
NP_005210 | diaphanous homolog 1 (DIAPH1) | NM_005219 |
| Dia2 | Protein diaphanous homolog 2 = DIAP2, DIAPH2 Diaphanous-related formin 2 = DRF2 [Chae et al., 1998] |
NP_006720 | diaphanous homolog 3 (DIAPH3) | NM_001042517 | |
| Dia3 | Protein diaphanous homolog 3 = DIAP3, DIAPH3 Diaphanous-related formin 3 = DRF3 [Peng et al., 2003] |
NP_001035982 | diaphanous homolog 2 (DIAPH2) | NM_007309 | |
|
| |||||
| formin-related proteins identified in leukocytes (FRL) | FRL1 | formin-like protein 1 = FMNL1 formin homology domain containing protein 4 = FHOD4 [Katoh and Katoh, 2003a] |
NP_005883 | formin-like 1 (FMNL1) | NM_005892 |
| FRL2 | formin-like protein 2 = FMNL 2 formin-like protein 3 = FRL3 formin homology domain containing protein 2 = FHOD2 [Block, et al., 2012; Harris, et al., 2010, Katoh and Katoh, 2003a] |
NP_443137 | formin-like 2 (FMNL2) | NM_052905 | |
| FRL3 | formin-like protein 3 = FMNL 3 formin-like protein 2 = FRL2 formin homology domain containing protein 3 = FHOD3 [Block, et al., 2012; Harris, et al., 2010, Katoh and Katoh, 2003a] |
Q8IVF7 | formin-like 3 (FMNL3) | NM_175736 | |
|
| |||||
| disheveled-associated activators of morphogenesis (DAAM) | DAAM1 | disheveled-associated activators of morphogenesis 1 = DAAM1 [Habas et al., 2001] |
NP_055807 | dishevelled associated activator of morphogenesis 1 (DAAM1) | NM_014992 |
| DAAM2 | disheveled-associated activators of morphogenesis 2 = DAAM2 [Katoh and Katoh, 2003b] |
NP_001188356 | dishevelled associated activator of morphogenesis 1 (DAAM1) | NM_015345 | |
|
| |||||
| delphilin | Delphilin | Glutamate receptor, ionotropic, delta 2-interacting protein 1 = GRID2IP [Miyagi et al., 2002 Katoh and Katoh, 2003c |
NP_001138590 | glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein (GRID2IP) | NM_001145118 |
|
| |||||
| inverted formins (INF) | INF1 | FH2 domain-containing protein 1 = FHDC1 [Katoh and Katoh, 2004a] |
Q9C0D6 | FH2 domain containing 1 (FHDC1) | NM_033393 |
| INF2 | WASp homology 2 (WH2) domain-containing formin 1 = WHIF1 [Chhabra and Higgs, 2006] |
NP_071934 | inverted formin, FH2 and WH2 domain containing (INF2) | NM_022489 | |
|
| |||||
| formin homology domain containing proteins (FHOD) | FHOD1 | formin homolog over-expressed in spleen 1 = FHOS1 [Westendorf et al., 1999] |
NP_037373 | formin homology 2 domain containing 1 (FHOD1) | NM_013241 |
| FHOD3 | formin homolog over-expressed in spleen 2 = FHOS2 [Westendorf et al., 1999] |
NP_079411 | formin homology 2 domain containing 3 (FHOD3) | NM_025135 | |
|
| |||||
| original formins (FMN) | FMN1 | FMN1 = Formin 1 [Katoh and Katoh, 2004b] |
NP_001096654 | formin 1 (FMN1) | NM_001103184 |
| FMN2 | FMN2 = Formin 2 [Katoh and Katoh, 2004c] |
NP_064450 | formin 2 (FMN2) | NM_020066 | |
Documentation of the cellular functions and distribution of formins lags far behind the advances of formin biochemistry and genetic identification. To facilitate the cellular study of formins, we have analyzed the expression levels of all 15 human formins in 22 different human cell and tissue types, using qPCR with Western blot confirmations where available.
Results
We performed specific quantitative reverse transcription PCR (RT-PCR) on total RNA equivalently isolated from human cells and tissues as described in Methods. Total RNA was harvested to ensure that all available message was sampled, since little is known of formin transcript activators or locale. Normalization of signal to total cellular RNA and a reference gene as applied in this study, are deemed the most reliable method of quantification [Bustin, SA 2002, Nolan et al., 2006]. Oligo(dT)20, identified as an appropriate and acceptable priming choice in “The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments,” was used for all RT reactions [Bustin et al., 2009]. Annealing to the C-terminally located Poly-A tail of intact mRNA makes Oligo(dT)20 a good priming choice, since the target region of the primers used for PCR is located toward the C-terminus of each formin [Ståhlberg et al., 2004].
The sites of specific oligonucleotide primer annealing were carefully chosen based on alignment of all formin protein sequences (Fig. 1A). Between the FH1 and FH2 domains of all human formins is a region of highly divergent sequence of generally consistent length (avg. length = 100.57 amino acids with a standard deviation of 13.39; % identity = 19.11%). Amplification across this region exhibited a high specificity for individual formins. Further, with a single exception, this region also represents a splice site for mRNA synthesis, bridging non-coding introns averaging 9000 bases. Incorporating the conserved homology of the FH1 and FH2 domain termini in this analysis permitted oligonucleotide design consistency important for comparison between formins. Average product size was 307 base pairs with a standard deviation of only 41 base pairs, varying only 13% in size, yet allowing single band selectivity (Fig. 1B). The qualitative PCR products (Fig. 1B) indicated no evidence of genomic DNA, and all products were as anticipated from RNA, as there were no products that included introns, even with 35 cycles of PCR, further demonstrating the lack of genomic DNA in RNA samples. End point melt-curve analysis confirmed the presence of single amplicons in each reaction.
Figure 1.
A: Formin alignments – Formin protein sequences from Table 1 were analyzed using COBALT. Amino acids highlighted in light grey are translations of forward primers and amino acids highlighted in black are translations of reverse primers used for PCR and qPCR as described in Methods. The vertical line indicates the start of the FH2 domain. The C-terminal end of the FH1 domain is not depicted in this alignment, and is located upstream of the amino acids indicating the forward primers used for qPCR (Fig. 1S). The underlined amino acids indicate the location of intron-exon borders between forward and reverse primers. For information regarding intron and extron length and position, see Methods.
B: PCR products – PCR performed on DNA from RT-PCR to demonstrate that the primer pairs for each formin, as listed in Methods, produce unique and specific products of predicted sizes. Product sizes are above each band in number of base pairs. PCR products of formin primers were analyzed using electrophoresis on a 2% agarose gel and visualized using ethidium bromide under ultraviolet light. The gel was photographed using Polaroid GelCam (Minnetonka, MN) and Fujifilm FP-3000B (Tokyo, Japan) instant black and white film. The resulting image was scanned using an HP ScanJet5300C (Palo Alto, CA) and processed using the Microsoft Scanning Wizard and Microsoft Powerpoint (Redmond, WA). A single example from U2OS cells of multiple trials in several cell types is shown as a representative example.
The detected levels of formins are presented as the percentage of expression of the 18S ribosomal subunit (Table 2). Based on previous experiments, expression levels above 30% of the 18S subunit translate to protein levels detectable via SDS PAGE and Western blotting. [Mersich et al., 2010] This expression level has been confirmed as detectable in Dia1, Dia2, FRL1, FHOD1 and FHOD3 [Mersich et al., 2010] (data not shown). Therefore, expression levels above 30% likely represent functional levels of the protein. Primer pairs used for qPCR produced the predicted product and were confirmed to yield qualitative results by PCR and agarose gel electrophoresis (Fig. 1B). Each of the cell and tissue types analyzed typically showed three to six highly expressed formins (Table 1). With the exception of neutrophils (discussed below), no cell type exhibited expression of all formin family members. Expression of some formins, such as FHOD3 and INF1, showed extreme variation between cell types, suggesting very specific functions. In contrast, some formins, termed homeostatic here, were expressed roughly equally in all cells. For qPCR, all reactions were run in triplicates, using DNA retro-transcribed from equal amounts of RNA, and results averaged, a standard approach detailed in “Properties of the reverse transcription reaction in mRNA quantification” [Ståhlberg et al., 2004].
Table 2. Expression level of human formins in cell and tissue types.
qRT- PCR for 15 human formins was performed on the cell and tissue types listed as described in Methods. The data are presented as the average of samples prepared in triplicates and reported as the percent of the signal achieved for levels of 18S RNA, p<0.05.
| HeLa | Macrophage | Cerebellum | U2OS | HCN | Skeletal muscle | THP | Diff. THP | U937 | K562 | Cardiac muscle | HaCaT | MDA | HK-2 | HUVEC | HEK | Meg-01 | Neutrophil | Platelet | Monocyte | HT1080 | Jurkat | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DAAM1 | 31 | 16 | 48 | 31 | 30 | 16 | 34 | 26 | 7 | 9 | 9 | 49 | 32 | 52 | 42 | 32 | 30 | 70 | 39 | 58 | 4 | 37 |
| DAAM2 | 23 | 9 | 43 | 22 | 38 | 4 | 36 | 8 | 0 | 6 | 9 | 66 | 15 | 62 | 16 | 14 | 5 | 71 | 23 | 32 | 0 | 39 |
| Delphilin | 18 | 1 | 23 | 5 | 17 | 19 | 19 | 11 | 0 | 0 | 0 | 27 | 8 | 35 | 12 | 5 | 0 | 81 | 19 | 44 | 2 | 57 |
| Dia1 | 50 | 34 | 42 | 38 | 38 | 14 | 46 | 37 | 21 | 27 | 15 | 50 | 47 | 53 | 44 | 44 | 56 | 91 | 51 | 60 | 15 | 46 |
| Dia2 | 40 | 9 | 26 | 32 | 36 | 18 | 36 | 16 | 14 | 16 | 8 | 44 | 38 | 47 | 31 | 31 | 31 | 62 | 24 | 25 | 3 | 30 |
| Dia3 | 27 | 20 | 35 | 28 | 31 | 33 | 40 | 29 | 18 | 4 | 5 | 38 | 33 | 48 | 37 | 30 | 33 | 81 | 33 | 45 | 0 | 37 |
| FHOD1 | 51 | 38 | 52 | 45 | 50 | 20 | 61 | 49 | 31 | 33 | 28 | 58 | 50 | 68 | 49 | 49 | 49 | 111 | 54 | 63 | 22 | 72 |
| FHOD3 | 13 | 0 | 39 | 14 | 27 | 32 | 16 | 3 | 0 | 0 | 23 | 48 | 37 | 49 | 35 | 23 | 6 | 68 | 17 | 29 | 13 | 41 |
| FMN1 | 40 | 22 | 37 | 27 | 29 | 21 | 54 | 37 | 34 | 31 | 28 | 54 | 43 | 59 | 40 | 53 | 26 | 94 | 48 | 46 | 2 | 71 |
| FMN2 | 30 | 5 | 50 | 13 | 33 | 9 | 34 | 19 | 63 | 42 | 3 | 62 | 18 | 47 | 18 | 18 | 17 | 81 | 32 | 24 | 0 | 46 |
| FRL1 | 38 | 26 | 41 | 24 | 26 | 20 | 33 | 33 | 11 | 7 | 6 | 30 | 32 | 45 | 19 | 25 | 29 | 87 | 38 | 60 | 5 | 52 |
| FRL2 | 40 | 31 | 53 | 19 | 33 | 18 | 31 | 41 | 6 | 0 | 14 | 37 | 38 | 46 | 45 | 36 | 35 | 73 | 25 | 34 | 11 | 40 |
| FRL3 | 44 | 35 | 54 | 32 | 35 | 24 | 41 | 46 | 18 | 16 | 11 | 37 | 46 | 47 | 44 | 37 | 39 | 77 | 45 | 54 | 12 | 41 |
| INF1 | 52 | 26 | 47 | 34 | 42 | 24 | 51 | 37 | 17 | 23 | 15 | 46 | 38 | 59 | 35 | 34 | 35 | 101 | 52 | 39 | 0 | 49 |
| INF2/WHIF1 | 40 | 28 | 34 | 31 | 44 | 10 | 41 | 37 | 9 | 9 | 4 | 35 | 37 | 49 | 43 | 23 | 6 | 68 | 17 | 29 | 13 | 41 |
Homeostatic Formins
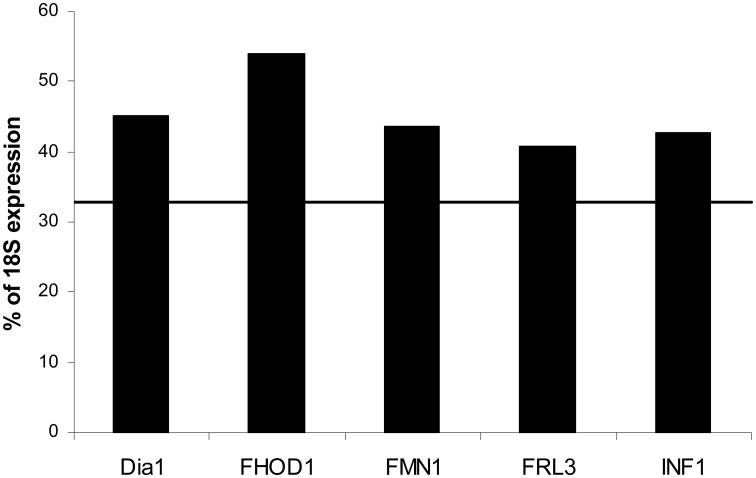
The level of formin expression varies greatly between different formins as well as between different cell and tissue types. Several formins exhibited a high level of expression in most cells and tissues analyzed (Fig. 2). The average expression of FHOD1, Dia1, FMN1, INF1 and FRL3 in the cells and tissues analyzed is above 40% of the expression of the 18S ribosomal subunit. The ubiquitous and consistent expression of these formins suggests potential roles in the production or maintenance of microfilament structures common to all cells, but does not rule out additional, specific activities. Dia1 is the most-studied of these formins to date on a structural as well as cellular level. Activated by the GTPase RhoA, and catalyzed by ROCK, Dia1 is an actin nucleator that has been identified to play a role in cell adhesion, migration, and stimulation of T-cells by dendritic cells [Li and Higgs, 2003; Tanizaki et al., 2010].
Figure 2. Homeostatic formins expressed at detectable levels in all cell and tissue types.
The level of each formin's expression in all cell and tissue types listed in Table 2 was analyzed. Five formins, Dia1, FHOD1, FMN1, FRL3, and INF1 were found to exhibit a consistently elevated level of expression in all cell and tissue types, suggesting functional roles common to all cells. Line indicates the average expression of all 15 formins in all cells. Data are reported as the average percentage of expression of 18S ribosomal subunit, p<0.05.
Least and most expressed formins
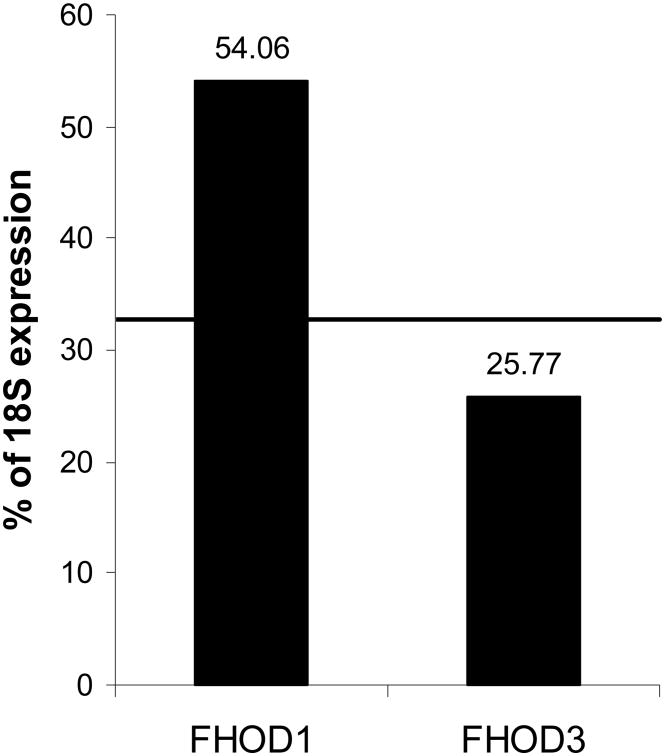
Averaged across all cell and tissue types analyzed, the most highly expressed human formin is FHOD1 and the least expressed formin we were able to detect reliably is FHOD3, with an expression of 54.06% and 25.77% of the 18S ribosomal subunit, respectively (Fig. 3). FHOD1 is expressed in all cell and tissue types analyzed, thus it is not surprising that it is the most highly expressed formin. Remarkably, the formin most structurally homologous to FHOD1, FHOD3 (50% identity, 67% similarity), is the least expressed formin, raising the question of why these two formins have such opposite expression profiles. This contrasting expression can be seen in the inverted expression patterns of FHOD1 and FHOD3 in skeletal and cardiac muscle. While FHOD1 is more highly expressed in cardiac muscle than skeletal muscle, 27.78% compared to 20.50% of the 18S ribosomal subunit, FHOD3 shows the opposite expression profiles, with 23.04% in cardiac and 31.87% in skeletal muscle (Fig. 1). These expression profiles have been confirmed by Western blotting (data not shown).
Figure 3. Least and most commonly expressed formins are closely related.
The levels of formin expression in all cell and tissue types listed in Table 2 were analyzed. Two formins, FHOD1 and FHOD3, were notable for consistently high and low expression, respectively. Line indicates the average expression of all 15 formins in all cells. Data are reported as the average percentage of expression of 18S ribosomal subunit, p<0.05. FHOD1 and FHOD3 data points have a p-value of <0.05.
Formins in hematopoietic versus non-hematopoietic cells and tissues
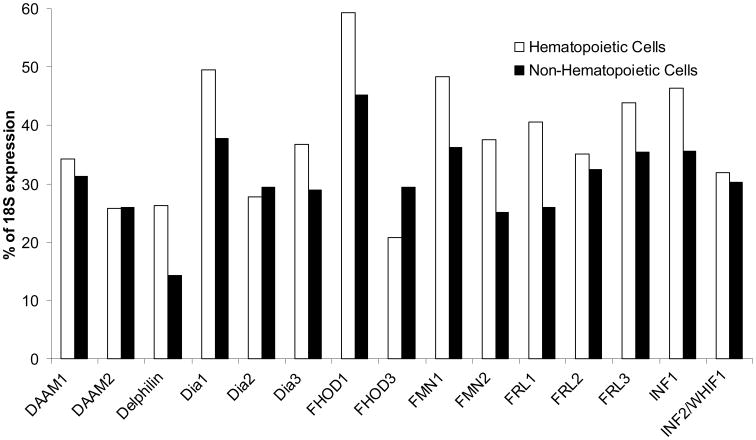
Formin levels in hematopoietic and non-hematopoietic cell types exhibit the expected elevated levels of expression of homeostatic formins, such as Dia1, FHOD1 and FMN1 (Fig. 4). Surprisingly, with the exception of FHOD3, formins have a higher average level of expression in hematopoietic cells than non-hematopoietic cells, despite the former typically exhibiting the greater microfilament organization. FHOD3 is closely related to FHOD1, yet their expression in hematopoietic cells versus non-hematopoietic cells is inverted when compared to the 14 other formins. This raises the question of why FHOD3 is the only formin more highly expressed in non-hematopoietic cells than in hematopoietic cells. FHOD3 has been found to promote myofibril maintenance in cardiac muscle, but no other specific cellular localization or function has been identified to provide insight into this finding [Taniguchi et al., 2009].
Figure 4. Formin family expression is elevated in hematopoietic cells.
qPCR was performed as described in Methods. Data are reported as the average percentage of expression of 18S ribosomal subunit, p<0.05, with experiments performed in triplicate and data within one standard deviation used for analysis. Non-hematopoietic cells and tissues include HeLa, cerebellum, U2OS, HCN, skeletal and cardiac muscle, HaCaT, MDA, HK-2, HUVEC, HEK, HT1080. All other cells and tissues are hematopoietic.
Mesenchymal, epithelial, and neural origins
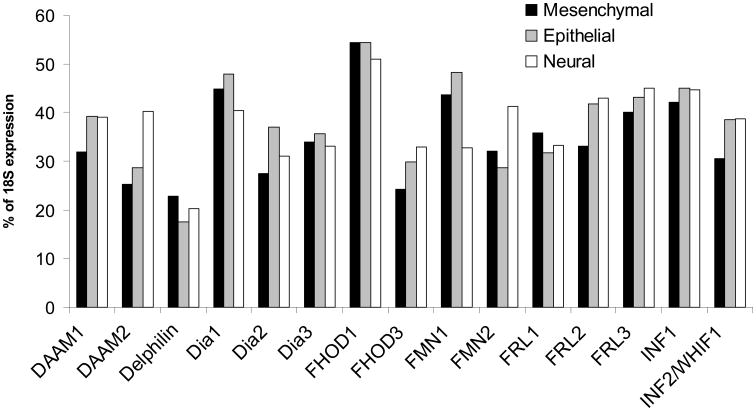
The average formin expression among mesenchymal, epithelial, and neural cell and tissue types follows the expected homeostatic pattern, with Dia1, FHOD1 and FMN1 having higher average levels of formin expression (Fig. 5). An unexpected finding was that the expression of each individual formin varies little between the three distinct cell and tissue embryonic origins. DAAM2, however, has a significantly higher level of expression in cells and tissues of neural origin than of mesenchymal or epithelial origin.
Figure 5. Expression of most formins is similar between cells and tissues of mesenchymal, epithelial and neural origins.
qPCR was performed as described in Methods. Data are reported as the average percentage of expression of 18S ribosomal subunit, p<0.05. Mesenchymal cells and tissues include MDA, macrophage, monocyte, THP, K562, U937, HT1080, neutrophil, platelet, Jurkat, megakaryocyte, U2OS, and cardiac and skeletal muscle. Epithelial cells include HeLa, HUVEC, HEK, and HK-2. Neural cells and tissues include HCN and cerebellum.
An analysis of DAAM1 and DAAM2 expression in the central nervous system of mouse and chick embryos showed that the two proteins have different expression profiles, with DAAM2 being expressed more highly in later developmental stages in the brain, while DAAM1 is expressed earlier overall. Kida et al., found little expression of DAAM2 in the spinal cord, while Lee and Deneen show that DAAM2 plays a role in dorsal patterning in the spinal cord through Wnt signaling [Kida et al., 2004; Lee and Deneen, 2012]. While no conclusion can be made about the localization of DAAM2 in the central nervous system based on the expression analysis presented here, the studies in chick and mice support the comparatively high level of expression of DAAM2 in neural tissues [Kida et al., 2004; Lee and Deneen, 2012].
Formin levels in cells and tissues
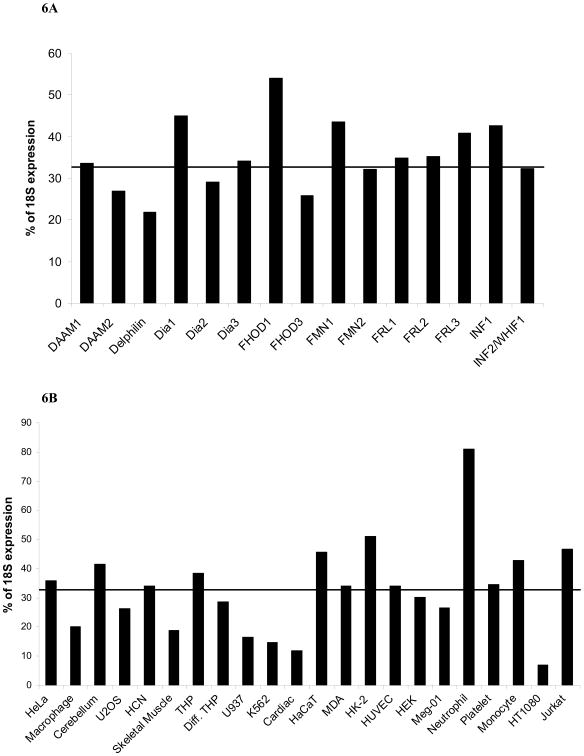
The level of formin expression for all 15 formins combined varies from barely detectable in HT1080 cells to 80% of 18S expression in neutrophils (Fig. 6A). The significantly higher level of formin expression in neutrophils was not predicted, but may be explained by considering their functions. Neutrophils are multipotent, nondividing cells that are recruited to sites of inflammation immediately after injury, and their function spans a variety of tasks including migration, phagocytosis, and degranulation. In order to carry out these tasks in a timely manner, it may be beneficial for neutrophils to have a large amount of RNA ready for translation. In turn, this reinforces a model where differentiation-based structures are constructed with the input of specific formins [Chhabra and Higgs, 2007]. Another explanation would be a lower level of 18S RNA in neutrophils than in the other cells and tissues that were analyzed, although this was not evident in qPCR of the 18S RNA. Furthermore, similar elevated expression was not seen in fully differentiated macrophages that execute comparable physiologic functions (Table 2). Regardless of the reason for the high level of expression of formins in neutrophils, all formins exhibit high message levels in neutrophils. When the expression level of individual formins is averaged for its total expression in all human cell and tissue types, we found a remarkably similar expression level for all formins with notable exceptions FHOD1 and FHOD3 discussed above and Delphilin (Fig. 6B).
Figure 6. A – Total expression of all formins varies in human cell and tissue types.
qPCR was performed as described in Methods. The level of expression of all formins in each individual cell and tissue type was averaged and is displayed as a percentage of expression of the 18S ribosomal subunit. The line indicates the average expression of all 15 formins in all cells. Data are reported as the average percentage of expression of 18S ribosomal subunit, p<0.05.
B – Variation of individual formin expression in human cell and tissue types. qPCR was performed as described in Methods. The total level of expression for each individual formin in all cell and tissue types was averaged and is displayed as a percentage of expression of the 18S ribosomal subunit. The line indicates the average expression of all 15 formins in all cells. Data are reported as the average percentage of expression of 18S ribosomal subunit, p<0.05.
Delphilin
Delphilin was first identified as a glutamate receptor δ2 (GluRδ2) interacting protein in parallel fiber-Purkinje cell postsynapses in the cerebellar cortex [Miyagi et al., 2002 Watanabe-Kaneko et al., 2007]. It was termed GRID2IP (glutamate receptor, ionotropic, delta 2-interacting protein 1) and delphilin from “Delta2-philic-protein”. Two differently palmitoylated isoforms of delphilin have been characterized and may be involved in controlling GluRδ2 signaling in Purkinje cells [Matsuda et al., 2006]. Delphilin could not be detected via PCR in any tissue and cell types analyzed. The primers to confirm the presence of delphilin were designed to encompass 289 base pairs located N-terminally of the poly-proline stretch and extending into the FH2 domain. This pair of primers did not yield a DNA product detectable via agarose gel electrophoresis using DNA obtained from U2OS cells, HeLa cells and HCN cells. Since delphilin has been reported in Purkinje cells located in the cerebellar cortex, DNA obtained from total human cerebellar RNA as described in Methods was also used to detect delphilin, but PCR did not yield a product [Miyagi et al., 2002]. Two additional sets of primers to detect delphilin in these cells and tissues also did not yield a product, one starting slightly C-terminally and ending slightly N-terminally of the original primers with a 426 base pair product, and one located at the C-terminal end of delphilin with a 401 base pair product (Fig. 1S).
Discussion
In this report we highlight several results to show the potential that exists in the field of formin research. The biochemical study of formins with regard to their actin polymerization activity and structure is quite advanced [Goode and Eck, 2007; Paul and Pollard, 2008, 2009]. However, the cellular localization and function of most formins remain elusive. As actin modifiers, formins are present in every cell type we analyzed, and quite predictably in every other cell and tissue, providing a plethora of in vitro model systems.
The formin field nomenclature and classification exhibited an expected extent of disorder, complicating the pursuit of this new field. The diligent efforts of several laboratories have brought to fruition a clear nomenclature that is universally accepted among formin researchers, making the sharing of data and ideas easier (Table 1). We see similarities between the newer field of formins and the more established field of integrins. The formin and integrin families have a comparable number of molecules that are present in a multitude of cells, all of which contain several members of the protein families, with some proteins being very specific, while others are partly redundant. Considering the widespread presence of formins and the importance of the actin cytoskeleton to numerous essential physiological processes, we predict that formins harbor decades of investigative research, yielding thousands of functionally descriptive publications, comparable to integrin research efforts.
It is important to note that some formins, such as FRL1, have already been ascribed multiple alternative splice forms [Colon-Franco et al., 2011]. While these isoforms were readily detectable through PCR using isoform specific primers, the variant nature of the spliced sequences prohibited confidence in the quantitative reliability of the oligonucleotides utilized (data not shown). Detailed analysis of individual formins should include evaluation of the specific role of alternatively spliced isoforms.
As the speed of formin research is increasing exponentially due to the growing realization of the significance and widespread implications of the formin field, we are excited to witness the future of this promising field unfold. The results presented in this paper and our experience in working with formins suggest that some formins serve more general functions, while others have highly specialized functions. A complete understanding of formins will involve not only the expression profiles discussed here, but also insight into the activation and localization of formins in cells. We hope that the information provided here can be a starting point for future studies on formins. To encourage the study of the cellular aspects of the formin field, we will continue to analyze human cell and tissue types using qPCR as they become available. Please contact the corresponding author if you are interested in providing us with cells or tissues.
Materials and Methods
Protein alignment
The 15 members of the human formin family of actin binding proteins (Table 1, Fig. 1A) were aligned using Constraint-based Multiple Alignment Tool (COBALT). For individual comparisons Basic Local Alignment Search Tool for proteins (BLASTp) was used with gapping of 10%.
Cells and RNA
The following cells were purchased from ATCC (Manassas, VA): HeLa (immortalized human cervical cancer cell line), Meg-01 (human megakaryocytes from chronic myelogenous leukemia), U2OS (human osteosarcoma cell line), HCN (human cortical neurons), THP-1 (acute human monocytic leukemia cell line), K562 (immortalized human myelogenous leukemia cell line), U937 (monocytic human cell line of myeloid lineage from histiocytic lymphoma), and HUVEC (human umbilical vein endothelial cells). Total human RNA of cerebellum, skeletal muscle, and cardiac tissue was purchased from Clontech Laboratories (Mountain View, CA). The following cells were gifts: HK-2 (immortalized proximal tubule epithelial cell line from human kidney) from Thomas R. Welch, M.D.; MDA-MB-231 (human breast cancer cell line from epithelial adenocarcinoma) and HT1080 (human fibrosarcoma cell line) from Christopher E. Turner, Ph.D.; Jurkat (immortalized line of human T-lymphocytes) from Andras Perl, M.D., Ph.D., all at SUNY Upstate Medical University in Syracuse, NY. HEK (primary human keratinocyte cell line from neonatal foreskin) and HaCaT (human immortalized keratinocyte cell line) were gifts from Andrew P. Kowalczyk, Ph.D. in the department of Cell Biology at Emory University in Atlanta, GA. Monocytes that were differentiated into macrophages and platelets and neutrophils were isolated from healthy human blood as previously described [Mersich et al., 2010; Blystone et al., 1997; Jones et al., 1998].
RNA purification
RNA was extracted and purified from 5×106 to 20×106 cells using RNeasy®Mini Kit from QIAGEN (Valencia, CA), in keeping with clinical laboratory standards for qPCR as laid out in “Quantification of mRNA using real-time RT-PCR” and “The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments.” [Bustin et al., 2009; Nolan et al., 2006] Cells grown in suspension were pelleted at 1000 rpm for 5 minutes using a Hermle Z400K centrifuge (Hermle Labortechnik, Wehingen, GERMANY) at room temperature (RT). For adherently grown cells, growth media was removed and cells were incubated at 37°C in 0.25% Trypsin-EDTA (1X), Phenol Red (GIBCO® Life Technologies, Grand Island, NY) for 5 minutes and pelleted at 1000 rpm for 5 minutes at RT. Cell pellets were disrupted by adding 600μl QIAGEN buffer RLT, containing a high concentration of guanidine isothiocycanate to facilitate RNA binding to a silica membrane in subsequent steps, and the resulting lysate was homogenized by passing it through a blunt 20-gauge needle 5 times. Subsequently 600μl 70% ethanol from Pharmaco-AAPER (Pharmaco, Brookfield, CT; AAPER, Shelbyville, KY) was added to the lysate and mixed by pipetting. The sample was then transferred to the RNeasy spin column, which was placed in a collection tube and centrifuged for 15 seconds at 14,000 rpm using an Eppendorf 5415C centrifuge (Hamburg, GERMANY). To remove non-specifically bound carbohydrates, proteins, fatty acids and RNA molecules smaller than 200 bases from the silica membrane, 700μl QIAGEN buffer RW1, containing guanidine salt and ethanol, was added to the column and centrifuged at 14,000 rpm for 15 seconds. Next, 500μl buffer RPE is added to the column and centrifuged at 14,000 rpm for 15 seconds, followed by another 500μl of buffer RPE and centrifugation at 14,000 rpm for 2 minutes. After placing the column in a new collection tube, it was centrifuged for 1 minute at 14,000 rpm to ensure the elimination of ethanol. 30μl RNase-free water were then added to the column, which was placed in a new collection tube and centrifuged at 14,000 rpm for 1 minute, and repeated with the eluate from this step.
RNA quantification
RNA was quanitified using Agilent RNA 6000 Nano Kit in the Agilent 2100 Bioanalyzer (Santa Clara, CA). RNA Nano 6000 reagents were warmed to RT for 30 minutes in the dark. Prior to running the experimental chip, the electrodes of the bioanalyzer were decontaminated by adding 350μl RNAseZap and 350μl RNAse-free DI water into the RNAseZap and RNAse-free DI water electrode cleaner chip, respectively. The RNAseZap electrode cleaner chip was placed into the bioanalyzer, and incubated with the lid closed for 1 minute, and the chip was removed and replaced with the RNAse-free DI water electrode cleaner chip, which was incubated for 10 seconds with the lid closed. After removing the chip the bioanalyzer was left open to dry for 10 seconds. To prepare the Gel Matrix, 550μl of gel was filtered through the filter column and centrifuged at 4000 rpm for 10 minutes, and once filtered, 65μl of gel were put into one tube. Next the Dye Concentrate was vortexed for 10 seconds and 1μl was added to the 65μl of filtered gel matrix. After vortexing thoroughly, the mixture was centrufiged at 13,000 g for 10 minutes. A new chip was placed on the chip priming station and 9μl of the gel-dye mixture were added to the G well, the plunger was pulled up to the 1ml mark, the station closed, and the plunger depressed and incubated for 30 seconds. A properly sealed syringe gasket was indicated by the plunger's retraction to 0.3ml and after 5 seconds the plunger was pulled back to 1ml and the priming station was opened. 9μl of gel-dye mixture were added to each G well and 5μl of nano marker buffer was added to each of the 12 sample wells and the ladder well. 1μl of 150 μg/ml RNA 6000 ladder was added to the ladder well and 1μl of sample into each sample well, and the chip was vortexed at 240 rpm for 1 minute before it was inserted into the bioanalyzer. The assay for total eukaryote RNA was selected in the 2100 Expert Software for each analysis, which calculated RNA quality and concentration, expressed as an RNA Integrity Number (RIN) using an algorithm based on a Bayesian analysis of nine distinct features of the electropherogram of 1208 samples of RNA of varying degrees of degradation using the Agilent 2100 bioanalyzer system. The RNA extracted from some cell types was found to have a low RIN, which was due to the low concentration of RNA in the sample, and not due to degraded RNA as can be seen from gels showing integrity of major 28s and 18S RNA (Fig. 2S). However, since the algorithm used to compute the RIN incorporates 9 features of the electropherogram, the low concentration yielded a low RIN. [Schroeder et al., 2006]
Reverse Transcription PCR (RT-PCR)
For reverse transcription PCR, 1ug of RNA, determined as described above, was used to prepare DNA using the SuperScript®III First Strand Synthesis System for RT-PCR from Invitrogen (St. Louis, MO) [Ståhlberg, et al., 2004]. For each reaction, 1μg of RNA, 2.5μl dNTP mix and 2.5μl oligo(dT)20 were added to a final volume of 25μl diethylpyrocarbonate-treated water and incubated first at 70°C for 5 minutes, and then on ice for 5 minutes. Following these incubations, 22.5μl 10× RT buffer, 20μl MgCl2, 10μl DTT and 5μl RNase OUT were added and the mixture was incubated at 42°C for 2 minutes. After adding 2.5μl of SuperScript®III, the mixture was incubated at 42°C for 50 minutes, 70°C for 15 minutes to terminate the reaction and on ice for 5 minutes. Next, 2.5μl of RNase H were added to degrade any remnant RNA and incubated at 37°C for 20 mintues.
Qualitative PCR
For each qualitative PCR reaction, 1μl each of specific forward and reverse primers at 20pM were mixed with 2ul of cDNA. PCR was run using MJ Research PTC-100 Programmable Thermal Controller from Bio-Rad (Hercules, CA). After 1 minute at 94°C, 94μl of the following was added: 10μl TAQ DNA Polishing Buffer B, 6μl MgCl2, 2μl dNTP, 0.8μl of 5,000 units/ml Taq DNA Polymerase, all from Fisher Scientific (Hampton, NH), 5μl dimethyl sulfoxide (DMSO) from Sigma-Aldrich (St. Louis, MO), and 71.2μl water, followed by a drop of mineral oil from Fisher Scientific. After a total of 2 minutes at 94°C followed 35 cycles of 0:30 at 55°C, 1:00 at 72°C, and 1:00 at 92°C, and one 2:00 cycle at 94°C.
Quantitative real-time PCR (qPCR)
Quantitative real-time PCR was performed using Eco Real-Time PCR System from Illumina, Inc. (San Diego, CA) and iQ™SYBR®Green Supermix from BIO-RAD (Hercules, CA). For each qPCR cell analysis 500μl of iQ™SYBR®Green Supermix was combined with 50μl of cDNA and 360μl of distilled water and mixed carefully. For each formin analyzed, 1μl of forward specific primer, 1μl of reverse specific primer, each at 1.25pM, the design of which is detailed in Methods, and 18μl of the cDNA mixture were combined. 1μL of forward and reverse 18S primer was used at a concentration of 10pM. The qPCR reaction was programmed as follows: 2:30 at 95°C, 40 cycles of 0:15 at 95°C, 0:20 at 55°C, 0:30 at 72°C, concluded with 0:15 each at 55°C, 60°C and 95°C. The data was analyzed and graphed using Microsoft Excel (Redmond, WA).
Primer design
Custom primer pairs were designed and synthesized for each formin to span the FH1 and FH2 domain linkage of each formin. The products of each primer pair range from 179 to 355 base pairs, averaging 307 bp. Oligonucleotide annealing locations are indicated in Figure 1A. Each set of formin primers, except for FMN1 spans at least one intron, with an average intron size of 9745 bp, and ranging from 83 bp to 107733 bp. The product sizes corresponded to the combined size of the exons, showing that no genomic DNA was present in the PCR reactions. Intron-exon boundaries in the primer product regions are indicated by an underlined amino acid. Oligo(dT)20 was used for RT-PCR, resulting in retro-transcription starting from the C-terminal end only. The primers used for qPCR and qualitative PCR are equidistant from the C-terminus, with the exception of the inverted formin INF1 and FMN1. A component of the oligo design stemmed from our search for comparable regions within target templates across all formins, following RNA and DNA alignments, as performed using COBALT.
Primers used for qPCR
DAAM1
Daam2
| NP_001188356 | 1068 aa | linear | PRI 09-SEP-2011 |
| NM_015345 | 6252 bp mRNA | linear | PRI 28-NOV-2011 |
| 5′- GGA GCG TGT CCC TGG CAC CGT ATG G -3′ | |||
| Bp 2048-2072 | |||
| AA 616-623 | |||
| 5′- CCA GCA TGT CCT TAG CAA GGT CCT CC -3′ | |||
| Bp 2318-2343 | |||
| AA 706-713 | |||
| product size 296 bp, spans exons 16-18, bridges introns sized 1086, 2577 | |||
Delphillin
| NP_001138590 | 1211 aa | linear | PRI 15-AUG-2011 |
| NM_001145118 | 4632 bp mRNA | linear | PRI 15-AUG-2011 |
| 5′- GGC ACC ATC TGG GGT CAG CTC GGG G -3′ | |||
| Bp 2527 - 2551 | |||
| AA 843 - 850 | |||
| 5′- GCC CGC ACA TCT CGC GCA GCT GCT GC -3′ | |||
| Bp 2790 – 2815 | |||
| AA 932 – 940 | |||
| product size 289 bp, spans exons 13-16, bridges introns sized 2077, 949, 1105 | |||
Delphillin
| NP_001138590 | 1211 aa | linear | PRI 15-AUG-2011 |
| NM_001145118 | 4632 bp mRNA | linear | PRI 15-AUG-2011 |
| 5′- GCG TCA AGC GCT TGC GGT GGG AAC AGG -3′ | |||
| Bp 2486-2512 | |||
| AA 831-838 | |||
| 5′- GGA CCA GTT CGT CCT GCA GGT GC -3′ | |||
| Bp 2889-2911 | |||
| AA 964-970 | |||
| product size 426 bp | |||
Delphillin
| NP_001138590 | 1211 aa | linear | PRI 15-AUG-2011 |
| NM_001145118 | 4632 bp mRNA | linear | PRI 15-AUG-2011 |
| 5′- GGA CAT GAG GTC AGA GGC TAT TGG(sequence = AACTGCTCCCACGA) -3′ | |||
| Bp 981-990 | |||
| AA 328-331 | |||
| 5′- CCA GCA CAG GAA CAT CGA CAC CC -3′ | |||
| Bp 1260-1282 | |||
| AA 421-427 | |||
| product size 301 bp | |||
Dia 1
Dia 2
| NP_006720 | 1101 aa | linear | PRI 04-MAR-2012 |
| NM_001042517 | 4812 bp mRNA | linear | PRI 21-NOV-2011 |
| 5′ – GATCAGACCTCATGAAATGACTG – 3′ | |||
| Bp 2178 - 2200 | |||
| AA 645 - 652 | |||
| 5′ – CGGTTGGCAGAGTCTATGATTCAG – 3′ | |||
| Bp 2461 - 2484 | |||
| AA 742 - 749 | |||
| product size 307 bp, spans exons 17-20, bridges introns sized 45073, 8525, 4318 | |||
Dia 3
| NP_001035982 | 1193 aa | linear | PRI 21-NOV-2011 |
| NM_007309 | 3782 bp mRNA | linear | PRI 04-MAR-2012 |
| 5′ – GGTCAAAGATTGAACCCACAG – 3′ | |||
| Bp 2324 - 2344 | |||
| AA 652 - 657 | |||
| 5′ – GCTGAGTGAGGCTTTAATTCAGAACC – 3′ | |||
| Bp 2622 - 2647 | |||
| AA 749 - 756 | |||
| product size 324 bp, spans exons 16-20, bridges introns sized 6944, 107733, 2124, 24432 | |||
FHOD1
FHOD3
FMN1
| NP_001096654 | 1196 aa | linear | PRI 30-JAN-2012 |
| NM_001103184 | 12355 bp mRNA | linear | PRI 30-JAN-2012 |
| 5′ – GCTGAAGAAGGGGGCTACCGC – 3′ | |||
| Bp 846 – 866 | |||
| AA 992 - 999 | |||
| 5′ – GGAGAGTGGGAGTGGCCTTCG – 3′ | |||
| Bp 1004 – 1024 | |||
| AA 1083 - 1090 | |||
| product size 179 bp | |||
FMN2
FRL1
FRL2
FRL3
| Q8IVF7 FRL3_HUMAN | 1028 aa | linear | PRI 22-FEB-2012 |
| NM_175736 | 11192 bp mRNA | linear | PRI 28-JAN-2012 |
| 5′- GCA CTG AAA CCC AAC CAG ATC AGT GGC -3′ | |||
| Bp 1966 - 1992 | |||
| AA 579 - 587 | |||
| 5′- GCG CAT CAG GCA CTC CAC GAA GTC C -3′ | |||
| Bp 2259 - 2292 | |||
| AA 676 - 686 | |||
| product size 327 bp, spans exons 16-18, bridges introns sized 289, 750 | |||
INF1
WHIF1
| NP_071934 | 1249 aa | linear | PRI 26-FEB-2012 |
| NM_022489 | 4725 bp mRNA | linear | PRI 26-FEB-2012 |
| 5′- GCT GCC ATC CAA CGT GGC ACG TGA GC -3′ | |||
| Bp 1856 - 1881 | |||
| AA 572 - 577 | |||
| 5′- CGT GCT TCT CGG GAA GGA GCT TAA GG -3′ | |||
| Bp 2168 - 2193 | |||
| AA 676 - 683 | |||
| product size 338 bp, spans exons 8-11, bridges introns sized 433, 83, 548 | |||
Data analysis
q RT-PCR analyses were run in triplicate and the results were averaged. Data within one standard deviation were used for the expression analysis. The percent of 18S expression of each formin was calculated as the inverted natural logarithm of the ratio of average formin expression to average 18S expression and multiplied by 100 (1 – LN (average formin expression / average 18S expression) × 100). ANOVA and T-tests were performed on raw data, p < 0.05 for specific comparisons. ANOVA was performed on all data points listed in Table 2 with a p-value < 0.05.
Supplementary Material
Acknowledgments
We would like to thank Thomas R. Welch, M.D. and Lisa W. Blystone for use and training of the Eco Real-Time PCR System, as well as Andrew P. Kowalczyk, Ph.D., Andras Perl, M.D., Ph.D., Christopher E. Turner, Ph.D. and Thomas R. Welch, M.D. for generously sharing cells with us. This work was supported by NIH DK79884 to S.D.B.
References
- Aspenstrom P. Formin-binding proteins: modulators of formin-dependent actin polymerization. Biochim Biophys Acta. 2010;1803:2174–182. doi: 10.1016/j.bbamcr.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Block J, Breitsprecher D, Kuhn S. Curr Biol. Vol. 22. Elsevier Ltd; 2012. FMNL2 drives actin-based protrusion and migration downstream of Cdc42; pp. 111005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blystone SD, Williams MP, Slater SE, Brown EJ. Requirement of integrin beta3 tyrosine 747 for beta3 tyrosine phosphorylation and regulation of alphavbeta3 avidity. J Biol Chem. 1997;272:4528757–28761. doi: 10.1074/jbc.272.45.28757. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:4611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29:123–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:4237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillon DH, Wasserman SA. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120:123367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- Chae SC, Inazu Y, Amagai A, Maeda Y. Biochem Biophys Res Commun. Vol. 252. Academic Press; 1998. Underexpression of a novel gene, dia2, impairs the transition of Dictyostelium cells from growth to differentiation; pp. 1278–283. [DOI] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:162–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:101110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J Biol Chem. 2006;281:3626754–26767. doi: 10.1074/jbc.M604666200. [DOI] [PubMed] [Google Scholar]
- Colon-Franco JM, Gomez TS, Billadeau DD. Dynamic remodeling of the actin cytoskeleton by FMNL1gamma is required for structural maintenance of the Golgi complex. J Cell Sci. 2011;124:Pt 183118–3126. doi: 10.1242/jcs.083725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland JW, Copeland SJ, Treisman R. Homo-oligomerization is essential for F-actin assembly by the formin family FH2 domain. J Biol Chem. 2004;279:4850250–50256. doi: 10.1074/jbc.M404429200. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Zigmond S, Boone C. Formins: signaling effectors for assembly and polarization of actin filaments. J Cell Sci. 2003;116:Pt 132603–2611. doi: 10.1242/jcs.00611. [DOI] [PubMed] [Google Scholar]
- Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Habas R, Kato Y, He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 2001;107:7843–854. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- Harris ES, Gauvin TJ, Heimsath EG, Higgs HN. Cytoskeleton (Hoboken) Vol. 67. Wiley-Liss, Inc; 2010. Assembly of filopodia by the formin FRL2 (FMNL3) pp. 12755–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN, Peterson KJ. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell. 2005;16:11–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SL, Wang J, Turck CW, Brown EJ. A role for the actin-bundling protein L-plastin in the regulation of leukocyte integrin function. Proc Natl Acad Sci U S A. 1998;95:169331–9336. doi: 10.1073/pnas.95.16.9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M, Katoh M. Identification and characterization of human FHDC1, mouse Fhdc1 and zebrafish fhdc1 genes in silico. Int J Mol Med. 2004a;13:6929–934. [PubMed] [Google Scholar]
- Katoh M, Katoh M. Identification and characterization of the human FMN1 gene in silico. Int J Mol Med. 2004b;14:1121–126. [PubMed] [Google Scholar]
- Katoh M, Katoh M. Characterization of FMN2 gene at human chromosome 1q43. Int J Mol Med. 2004c;14:3469–474. [PubMed] [Google Scholar]
- Katoh M, Katoh M. Identification and characterization of human FMNL1, FMNL2 and FMNL3 genes in silico. Int J Oncol. 2003a;22:51161–1168. [PubMed] [Google Scholar]
- Katoh M, Katoh M. Identification and characterization of human DAAM2 gene in silico. Int J Oncol. 2003b;22:4915–920. [PubMed] [Google Scholar]
- Katoh M, Katoh M. Identification and characterization of human GRID2IP gene and rat Grid2ip gene in silico. Int J Mol Med. 2003c;12:61015–1019. [PubMed] [Google Scholar]
- Kida Y, Shiraishi T, Ogura T. Identification of chick and mouse Daam1 and Daam2 genes and their expression patterns in the central nervous system. Brain Res Dev Brain Res. 2004;153:1143–150. doi: 10.1016/j.devbrainres.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Kovar DR. Molecular details of formin-mediated actin assembly. Curr Opin Cell Biol. 2006;18:111–17. doi: 10.1016/j.ceb.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Lee HK, Deneen B. Dev Cell. Vol. 22. Elsevier Inc; 2012. Daam2 is required for dorsal patterning via modulation of canonical Wnt signaling in the developing spinal cord; pp. 1183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Matsuda S, Gladding CM, Yuzaki M. Characterization of the delta2 glutamate receptor-binding protein delphilin: Splicing variants with differential palmitoylation and an additional PDZ domain. J Biol Chem. 2006;281:3525577–25587. doi: 10.1074/jbc.M602044200. [DOI] [PubMed] [Google Scholar]
- Mersich AT, Miller MR, Chkourko H, Blystone SD. Cytoskeleton (Hoboken) Vol. 67. Wiley-Liss, Inc; 2010. The formin FRL1 (FMNL1) is an essential component of macrophage podosomes; pp. 9573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi Y, Yamashita T, Fukaya M. Delphilin: a novel PDZ and formin homology domain-containing protein that synaptically colocalizes and interacts with glutamate receptor delta 2 subunit. J Neurosci. 2002;22:3803–814. doi: 10.1523/JNEUROSCI.22-03-00803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:31559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Energetic requirements for processive elongation of actin filaments by FH1FH2-formins. J Biol Chem. 2009;284:1812533–12540. doi: 10.1074/jbc.M808587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18:19–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Wallar BJ, Flanders A, Swiatek PJ, Alberts AS. Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr Biol. 2003;13:7534–545. doi: 10.1016/s0960-9822(03)00170-2. [DOI] [PubMed] [Google Scholar]
- Schonichen A, Geyer M. Fifteen formins for an actin filament: a molecular view on the regulation of human formins. Biochim Biophys Acta. 2010;1803:2152–163. doi: 10.1016/j.bbamcr.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhlberg A, Hakansson J, Xian X, Semb H, Kubista M. Properties of the reverse transcription reaction in mRNA quantification. Clin Chem. 2004;50:3509–515. doi: 10.1373/clinchem.2003.026161. [DOI] [PubMed] [Google Scholar]
- Wallar BJ, Alberts AS. The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 2003;13:8435–446. doi: 10.1016/s0962-8924(03)00153-3. [DOI] [PubMed] [Google Scholar]
- Watanabe-Kaneko K, Sonoda T, Miyagi Y, Yamashita T, Okuda K, Kawamoto S. The synaptic scaffolding protein Delphilin interacts with monocarboxylate transporter 2. Neuroreport. 2007;18:5489–493. doi: 10.1097/WNR.0b013e3280586821. [DOI] [PubMed] [Google Scholar]
- Westendorf JJ, Mernaugh R, Hiebert SW. Identification and characterization of a protein containing formin homology (FH1/FH2) domains. Gene. 1999;232:2173–182. doi: 10.1016/s0378-1119(99)00127-4. [DOI] [PubMed] [Google Scholar]
- Zigmond SH, Evangelista M, Boone C, Yang C, Dar AC, Sicheri F, Forkey J, Pring M. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol. 2003;13:201820–1823. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.