Abstract
Heparan sulfate proteoglycans (HSPGs) are integral components of the lung. Changes in HSPGs have been documented in idiopathic pulmonary fibrosis (IPF). Many of the biological functions of HSPGs are mediated by heparan sulfate (HS) side chains, and little is understood about these side chains in the pathogenesis of IPF. The aims of this study were to compare HS structure between normal and IPF lungs and to examine how changes in HS regulate the fibrotic process. HS disaccharide analysis revealed that HS 6-O-sulfation was significantly increased in IPF lungs compared with normal lungs, concomitant with overexpression of HS 6-O-sulfotransferases 1 and 2 (HS6ST1/2) mRNA. Immunohistochemistry revealed that HS6ST2 was specifically expressed in bronchial epithelial cells, including those lining the honeycomb cysts in IPF lungs, whereas HS6ST1 had a broad expression pattern. Lung fibroblasts in the fibroblastic foci of IPF lungs expressed HS6ST1, and overexpression of HS6ST1 mRNA was observed in primary lung fibroblasts isolated from IPF lungs compared with those from normal lungs. In vitro, small interference RNA–mediated silencing of HS6ST1 in primary normal lung fibroblasts resulted in reduced Smad2 expression and activation and in reduced expression of collagen I and α-smooth muscle actin after TGF-β1 stimulation. Similar results were obtained in primary IPF lung fibroblasts. Furthermore, silencing of HS6ST1 in normal and IPF lung fibroblasts resulted in significant down-regulation of TβRIII (betaglycan). In summary, HS 6-O-sulfation is up-regulated in IPF with overexpression of HS6ST1 and HS6ST2, and overexpression of HS6ST1 in lung fibroblasts may regulate their fibrotic responses to TGF-β1.
Keywords: idiopathic pulmonary fibrosis, heparan sulfate, fibroblast, HS6ST, TGF-β1
Clinical Relevance
Heparan sulfate (HS) proteoglycans have been implicated in the pathogenesis of idiopathic pulmonary fibrosis (IPF). However, little is known about the structure and function of the HS side chains in this disease. We show in this study that HS 6-O-sulfation is significantly elevated in IPF lungs compared with normal lungs, with overexpression of HS 6-O-sulfotransferase 1 (HS6ST1) in the fibrotic foci in situ and in isolated primary IPF lung fibroblasts. Silencing of HS6ST1 in normal and IPF lung fibroblasts reduces TGF-β1–induced fibrotic response, which suggests that HS6ST1 could serve as a new therapeutic target in the treatment of IPF.
Idiopathic pulmonary fibrosis (IPF) is a progressive, debilitating, and ultimately fatal disease for which there is no effective therapy (1). Estimates of IPF prevalence and annual incidence in the United States range from 14 to 42.7 per 100,000 persons and 6.8 to 16.3 per 100,000 persons, respectively, with 3- and 5-year mortality rates at approximately 50 and 80% (2). The pathogenic mechanisms involved in the initiation and progression of IPF are poorly understood. The current paradigm suggests that recurrent epithelial cell injury leads to aberrant repair processes that result in dysregulation of the key cells in the fibrotic response, the myofibroblasts, allowing the fibrosis to proceed without constraint (3).
A hallmark of the histopathology of IPF is the presence of the fibroblastic foci, which are composed of fibroblasts with an activated myofibroblast phenotype. Myofibroblasts are a unique subpopulation of fibroblasts that express features of smooth-muscle differentiation, α-smooth muscle actin (SMA) (1, 2). The expression of α-SMA confers the myofibroblasts a contractile phenotype that contributes to the distortion of normal lung architecture and decreased lung compliance (4). Myofibroblasts are the effector cells that produce the extracellular matrix, including collagen, as shown in human and animal models of IPF (5, 6). The presence and the extent of the fibroblastic foci in patients with IPF have been shown to be one of the more reliable markers of a poor prognosis and early mortality (7). In addition, fibroblasts isolated from patients with IPF were shown to retain their fibrotic features in vitro even after many subcultivations (8–10).
TGF-β1 is the central regulator of fibroblast to myofibroblast differentiation in vitro and in vivo (11). TGF-β1 signals via the heterotetrameric complexes of the transmembrane type I and type II serine/threonine kinase receptors (TβRI and TβRII) (12). In the canonical TGF-β1 signaling pathway, activation of TβRI leads to phosphorylation of the receptor-specific Smads (Smad2 and Smad3) which then associate with the common mediator Smad4 and translocate to the nucleus, where they interact with other transcription factors to regulate gene expression. Activations of Smad2 and Smad3 have been shown to be required for optimal TGF-β1 responses in fibroblasts, including TGF-β1–induced expression of α-SMA and collagen I (13).
Heparan sulfate proteoglycans (HSPGs) are the major proteoglycans in alveolar basement membrane and on the cell surfaces (14, 15). In lung homogenates and in lavage fluid from patients with IPF, HSPG family members, such as syndecan-1 and syndecan-2, are up-regulated (16, 17). TGF-β1 induces syndecan-2 expression in primary human lung fibroblasts (17). Syndecan-4 expression is up-regulated in bleomycin-induced lung injury, and syndecan-4 null mice exhibit a dysregulated inflammatory response, increased myofibroblast recruitment, and interstitial fibrosis after bleomycin administration (18). In addition to alterations in the syndecan core proteins, heparan sulfate (HS) is increased in radiation-induced lung injury and in bleomycin-induced lung fibrosis in mice (19, 20). Changes in the HS sulfation pattern and its role in the development of lung fibrosis have not been carefully studied.
The HS side chains mediate many of the biological functions of the HSPGs (including the syndecans) through binding with numerous growth factors and cytokines, including fibroblast growth factors, vascular endothelial growth factor, and the profibrotic cytokine TGF-β1 (21, 22). HS polysaccharide chains contain repeating disaccharide units of uronic acid (UA, either D-glucuronic acid, GlcA, or L-iduronic acid, IdoA) linked to N-acetylglucosamine (GlcNAc). During HS biosynthesis in the Golgi, these disaccharides are further modified by epimerization of GlcA to IdoA and by sulfations at the N, 6-O, and 3-O positions of the GlcN and at the 2-O position of the UA residues (23). These modifications are tightly regulated, resulting in HS chains with highly distinct saccharide sequences and sulfation patterns (23). The aims of this study were to compare HS structure between normal and IPF lungs and to examine how changes in HS may regulate the fibrotic process with a focus on TGF-β1 signaling in lung fibroblasts.
Materials and Methods
Human Lung Samples
Normal and IPF lung samples were obtained from the Lung Tissue Research Consortium (LTRC; Concept Sheet 09–99–0012). The clinical data and specimens have been deidentified by the LTRC. The normal lung tissues were mostly noncancerous areas of the lungs from patients with lung cancer (2 men and 5 women; 56.3 ± 5.6 yr of age). The IPF lung tissues were from 15 patients with IPF (12 men and 3 women; 57.7 ± 1.1 yr of age) with reduced lung function (FVC < 80% predicted). Flash-frozen lung tissues were used for RNA extraction and HS disaccharide analysis, and formalin-fixed and paraffin-embedded lung tissue sections were used for immunohistochemistry.
HS Disaccharide Analysis
HS extraction and subsequent disaccharide analysis were performed at the Complex Carbohydrate Research Center at the University of Georgia using strong-anion exchange-HPLC with postcolumn derivatization and subsequent fluorescent detection (24).
Quantitative RT-PCR
Total RNA was isolated using Trizol (Invitrogen, Grand Island, NY). Reverse transcription and subsequent quantitative real-time PCR (qRT-PCR) analyses were performed as described (25). Primer sequences for collagen I, α-SMA, 36B4, and 18S were as described (22). Additional primers used were 5′-GGTCTCGTAGCAGGGTGAT-3′ (forward) and 5′-GACCGAGCTCACCAACTG-3′ (reverse) for HS6ST1 and 5′-GAGGATGGTGATGTAGTGGAA-3′ (forward) and 5′-CTCTTCTCCAGGTTCTCCAC-3′ (reverse) for HS6ST2.
Conventional PCR
Conventional PCR was performed to differentiate the splice variants of HS6ST2, using primers 5′-TGATCGTGTTCCTGCACATCCAGA-3′ (forward) and 5′-ATGCAGGGATGCTTTCCATGTTGC-3′ (reverse).
Immnunohistochemistry
Histostain-Plus Kits (Invitrogen) was used for detection of HS6ST1 and HS6ST2 essentially as described (25). Detailed procedures are provided in the online supplement.
Cell Culture
Normal and IPF lung fibroblasts were isolated as described (26). Written informed consent was obtained from all subjects in accordance with the University of Michigan Institutional Review Board, and cell lines were derived from patients in a blinded fashion without regard to clinical data except diagnosis. Cells were maintained in Dulbecco’s modified Eagle medium with 10% charcoal-stripped FBS, antibiotics, and glutamine. Commercially available normal human lung fibroblasts were obtained from Lonza (Walkersville, MD) and maintained in FGM-2 (Lonza). All studies were performed using cells between the fourth and ninth passages.
Small Interference RNA Transfection
Small interference RNA (siRNA) against human HS6ST1 was obtained from Ambion (Invitrogen). Reverse transfection was performed in 12-well plates (150,000 cells/ml/well) using Lipofectamine RNAiMAX reagent (Invitrogen) with a final concentration of 10 nM HS6ST1 siRNA or 10 nM negative control siRNA in FGM-2 without antibiotics. Transfection media were replaced with fibroblast basal medium + 0.2% BSA + antibiotics the next day. After another 24 hours (which was 48 h after transfection), cells were treated with TGF-β1 (0.5 ng/ml) (R&D Systems, Minneapolis, MN) in fibroblast basal medium + 0.2% BSA + antibiotics.
Western Blotting
The expression and activation of Smad2/3 and the expression of collagen I, α-SMA, and TβRI, -II, and -III were evaluated by Western blotting essentially as described (25). Detailed procedures are provided in the online supplement.
Statistical Analysis
Data were expressed as mean ± SEM. Statistical analyses were performed using unpaired Student’s t test for two groups and ANOVA followed by Bonferroni’s multiple comparison tests when more than two groups were compared. P < 0.05 was considered statistically significant.
Results
Up-Regulation of HS 6-O-Sulfation in IPF
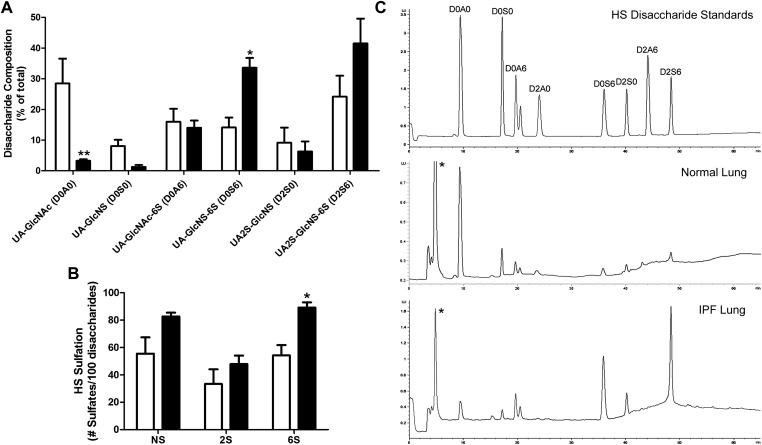
Three normal and three IPF lung samples were analyzed for HS disaccharide expression profiles. Sample selections were largely based on the size of the samples obtained from LTRC because relatively large amounts were needed for this analysis. The amounts of HS (μg/g wet tissue weight) extracted from the normal and IPF lungs were not significantly different (data not shown). The HS disaccharide compositions, however, were strikingly different between normal and IPF lungs (Figure 1). The IPF lungs contained markedly reduced levels of the unmodified UA-GlcNAc (3.27 ± 0.51% in IPF lungs vs. 28.48 ± 8.08% in normal lungs). This indicates that sulfation of HS in IPF lungs was markedly elevated. Indeed, HS from IPF lungs contained 219.7 ± 11.58 sulfates per 100 disaccharides, compared with 143.2 ± 28.39 sulfates per 100 disaccharide in the normal lungs (P < 0.05). Among the sulfated disaccharides, a significant increase was observed in the 6-O-sulfate containing UA-GlcNS-6S (33.59 ± 3.22% in IPF lungs vs. 14.14 ± 3.23% in the normal lungs). UA2S-GlcNS-6S was also increased in IPF lungs, although without reaching statistical significance. The increases in UA-GlcNS-6S and UA2S-GlcNS-6S led to a significant increase in the total 6-O-sulfate contents in IPF lungs compared with normal lungs (Figure 1B). In contrast, no significant differences were observed in the amount of N- or 2-O-sulfation. Representative chromatographs are shown in Figure 1C.
Figure 1.
Heparan sulfate (HS) disaccharide expression profiles of normal and idiopathic pulmonary fibrosis (IPF) lungs. (A) HS disaccharide composition (% of total) of normal (white bars) and IPF (black bars) lungs. (B) HS sulfation (number of N-sulfates [NS], 2-O-sulfates [2S], and 6-O-sulfates [6S] per 100 disaccharides) of normal (white bars) and IPF (black bars) lungs. *P < 0.05; **P < 0.01. (C) Representative chromatographs of HS disaccharide standards and HS disaccharides from normal and IPF lungs. *Unidentified peak, possibly HS monosaccharides. x Axis, elution time in minutes; y axis, fluorescent intensity, which corresponds to the quantity of each disaccharide.
Overexpression of HS6ST1 and HS6ST2 mRNA in IPF
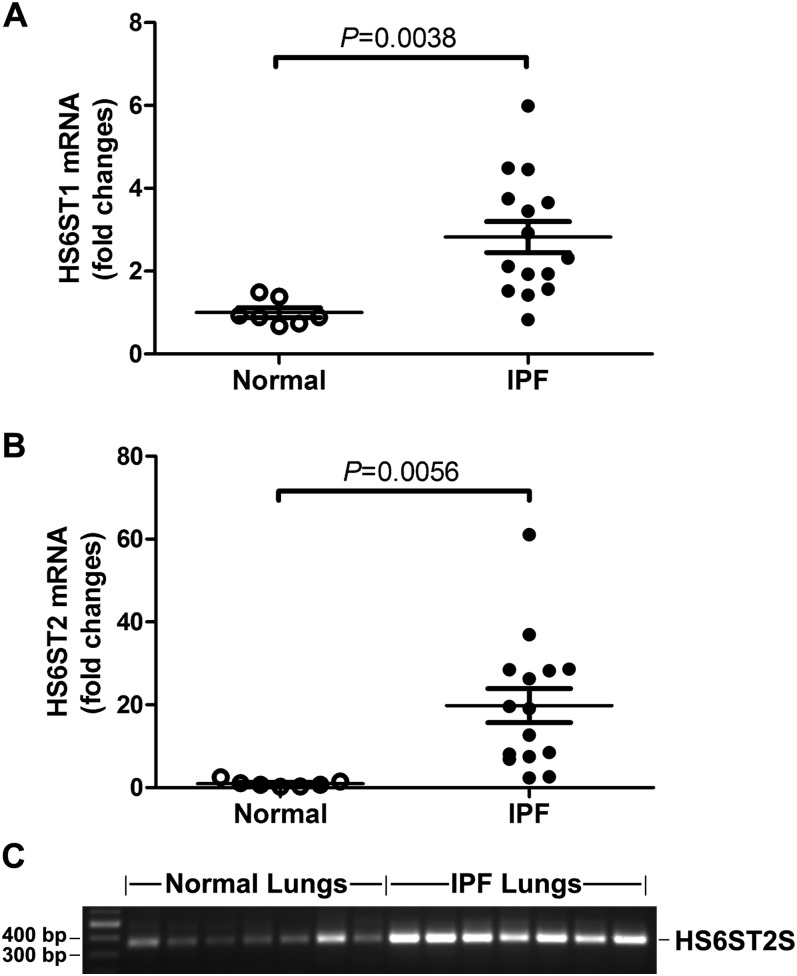
HS 6-O-sulfotransferases (HS6STs) catalyze the 6-O-sulfation of the GlcNAc/GlcNS residues in HS. In mammals, HS6STs exist in three isoforms (HS6ST1, -2, and -3) and in one alternatively spliced form (HS6ST2S) (27, 28). HS6ST2S is generated by alternative splicing in the coding regions of the HS6ST2 gene and lacks 40 amino acids encoded by exons 2 and 3. Despite this deletion, HS6ST2S retains 6-O-sulfotransferase activity not significantly different from that of HS6ST2 (28). Because of the up-regulation of HS 6-O-sulfation in the IPF lungs, we first performed qRT-PCR to examine the mRNA expression of the HS6STs in normal and IPF lungs. Our results showed that normal and IPF lungs expressed HS6ST1 and HS6ST2 mRNA and that, compared with normal lungs, HS6ST1 and HS6ST2 mRNA levels in the IPF lungs were increased 2.3- (P = 0.0038 compared with normal) and 18.6-fold (P = 0.0056 compared with normal), respectively (Figures 2A and 2B). These results were consistent with the increased HS 6-O-sulfation revealed by the HS structural analysis (Figure 1). HS6ST3 transcripts were not detected in normal or IPF lungs (data not shown). To examine which isoforms of HS6ST2 were expressed in normal and IPF lungs, we performed conventional RT-PCR with primers encompassing exons 2 and 3. Our results (Figure 2C) showed that normal and IPF lungs expressed HS6ST2S isoform (365 bp), not the HS6ST2 long form (485 bp).
Figure 2.
Overexpression of HS6ST1 and HS6ST2S mRNA in IPF lungs. (A) Quantitative real-time PCR analysis of mRNA expression of HS6ST1 and HS6ST2 in normal and IPF lungs. Fold changes were normalized to the expression of 18S RNA. (B) Conventional RT-PCR revealed that normal and IPF lungs express HS6ST2S splice variant (365 bp), not the HS6ST2 long form (485 bp).
Overexpression of HS6ST2S in the Bronchial Epithelial Cells in IPF
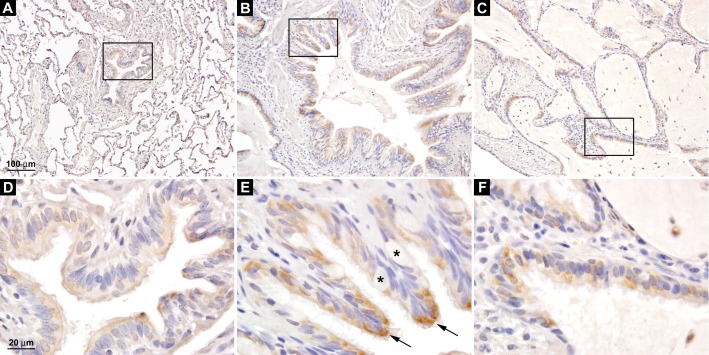
Because of the dramatic increase in HS6ST2S mRNA expression in the IPF lungs, we examined the cellular localization of HS6ST2S using standard immunohistochemistry. Our results showed that HS6ST2S protein was expressed in the bronchial epithelial cells. In normal lungs (n = 5), HS6ST2S was expressed at low levels in a subset of the ciliated bronchial epithelial cells (Figures 3A and 3D). In the IPF lungs (n = 7), in contrast, the majority of the ciliated bronchial epithelial cells (Figures 3B and 3E), including those lining the honeycomb cysts (Figures 3C and 3F), were found to express HS6ST2S, and the staining was more intense (compare Figures 3D and 3E). Compared with normal lungs, the bronchiolar component in the IPF lungs increased significantly in number and size (Figures 3A and 3B), consistent with previous reports (29). The epithelial cells lining the honeycomb cysts are of bronchiolar origin as they express high-molecular-weight cytokeratin, but not surfactant or CC10 antigens (29). Thus, the dramatic increase in HS6ST2S mRNA observed in the IPF lungs was likely the result of the increased expression of HS6ST2S in individual bronchial epithelial cells and the expansion of the bronchiolar structure in the IPF lungs. Alveolar epithelial cells (including type I and type II pneumocytes) in normal or IPF lungs were negative for HS6ST2S (see Figure E1A in the online supplement).
Figure 3.
Expression of HS6ST2S in ciliated bronchial epithelial cells. (A) HS6ST2S immunostaining in the normal lung. Boxed area is shown in D. (B and C) HS6ST2S immunostaining in IPF lungs. Boxed areas are shown in E and F. HS6ST2S was expressed in ciliated bronchial epithelial cells (arrow in E), whereas the mucus-producing goblet cells were negative for HS6ST2S (asterisks). A–C are at the same magnification, and D–F are at the same magnification. Images shown are representative of data from five normal and seven IPF lungs.
In addition to the bronchial epithelial cells, a small subset of alveolar macrophages and endothelial cells in normal and IPF lungs was found to express HS6ST2S protein (Figures E1B–E1D). HS6ST2S expression in the endothelial cells was often associated with inflammation (Figure E1C). These results indicate that HS6ST2S may play a role in inflammatory responses in endothelial cells and macrophages. No positive staining was detected with nonimmune rabbit IgG (Figure E1E).
The HS6ST2 antibody recognized a major band at approximately 80 kD of protein extracts from NIH-3T3 cells overexpressing human HS6ST2S, whereas the empty vector transfected cells expressed very low levels of HS6ST2S (Figure E1F), confirming the specificity of the HS6ST2 antibody.
Overexpression of HS6ST1 in IPF Lung Fibroblasts
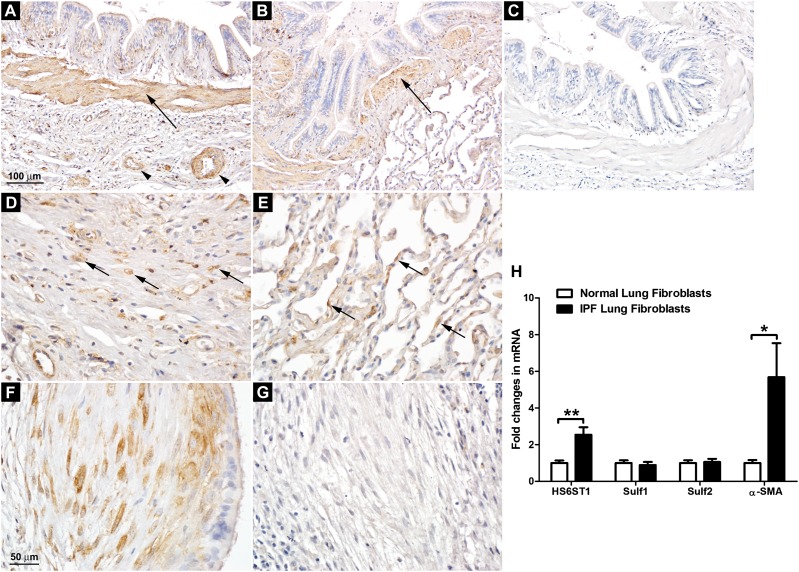
HS6ST1 was found to be expressed by multiple cell types, including smooth muscle cells, fibroblasts, and a subset of epithelial cells (not investigated further) in normal and IPF lungs, with the highest expression observed in smooth muscle cells (Figures 4A and 4B). Scattered HS6ST1 positivity was observed in the distorted interstitium of the IPF lungs (Figure 4D) and in the interalveolar septa of the normal lungs (Figure 4E), presumably from the resident fibroblasts. Because myofibroblasts are the effector cells in lung fibrosis, we focused on the fibroblastic foci in the IPF lungs. Our results showed that the fibroblasts/myofibroblasts in the fibroblastic foci expressed high levels of HS6ST1 (Figure 4F), but not HS6ST2S (Figure 4G). No positive staining was detected with nonimmune mouse IgG (Figure 4C).
Figure 4.
Overexpression of HS6ST1 in IPF lung fibroblasts. (A) HS6ST1 immunostaining in the IPF lung. (B) HS6ST1 immunostaining in the normal lung. (C) Control staining with nonimmune mouse IgG in the IPF lung. Smooth muscle cells surrounding the bronchi (arrows in A and B) and vascular structures (arrowheads in A) in normal and IPF lungs expressed high levels of HS6ST1. (D) HS6ST1 immunostaining in the interstitium of the IPF lung. (E) HS6ST1 immunostaining in the alveoli in the normal lung. Scattered HS6ST1 positivity was observed in the distorted interstitium of the IPF lungs (arrows in D) and in the interalveolar septa of the normal lungs (arrows in E). (F) HS6ST1 immunostaining in the fibroblastic focus in the IPF lung. (G) HS6ST2 immunostaining in the fibroblastic focus in the IPF lung. A–C are at the same magnification, and D–G are at the same magnification. Images shown are representative of data from five normal and seven IPF lungs. (H) Analysis of mRNA expression of HS6ST1, Sulf1, Sulf2, and α-smooth muscle actin (SMA) in primary normal (n = 4) and IPF (n = 5) lung fibroblasts by quantitative RT-PCR. Fold changes were normalized to the expression of the housekeeping gene 36B4. *P < 0.05; **P < 0.01.
We further examined the expression of HS6STs in primary lung fibroblasts isolated from normal and IPF lungs. Consistent with the immunohistochemistry data, IPF lung fibroblasts (n = 5) exhibited enhanced expression of HS6ST1 compared with lung fibroblasts isolated from normal lungs (n = 4) (Figure 4H). Neither normal nor IPF lung fibroblasts expressed detectable levels of HS6ST2 or HS6ST3 (data not shown). We also examined the mRNA expression of the HS 6-O-endosufatases, Sulf1 and Sulf2, in normal and IPF lung fibroblasts. Sulf1 and Sulf2 are the extracellular sulfatases that remove 6-O-sulfates from HS intrachain sites at the cell surface or in the extracellular matrix, thus fine-tuning HS–protein interactions (30). Our results showed that there were not significant differences in the expression of Sulf1 or Sulf2 between normal and IPF lung fibroblasts (Figure 4H). Consistent with their myofibroblast phenotype, IPF lung fibroblasts expressed significantly higher levels of α-SMA compared with normal lung fibroblasts (Figure 4H).
HS6ST1 Silencing Reduces TGF-β1 Signaling in Lung Fibroblasts
In a previous report, we showed that Sulf1 is a TGF-β1–responsive gene in normal human lung fibroblasts and that siRNA-mediated silencing of Sulf1 (which leads to increased HS 6-O-sulfation) results in enhanced TGF-β1 signaling (22). Because HS6ST1 catalyzes HS 6-O-sulfation, which is opposite to the 6-O-desulfation performed by Sulf1, we hypothesize that silencing of HS6ST1 would reduce TGF-β1 signaling in lung fibroblasts. Indeed, siRNA-mediated silencing of HS6ST1 led to reduced Smad2 activation (Figure 5A) and reduced expression of TGF-β1 target genes, including collagen I and α-SMA, at the mRNA and protein levels (Figures 5B and 5C). Total Smad2 levels were also reduced in HS6ST1-silenced lung fibroblasts (Figure 5A), which is in accordance with the increased total Smad2 levels in Sulf1 silenced lung fibroblasts reported in our previous study (22). In contrast, total and activated Smad3 were not significantly altered by silencing of HS6ST1.
Figure 5.
HS6ST1 silencing reduces TGF-β1 signaling in normal lung fibroblasts. (A) Analysis of phospho- and total Smad2/3 levels at 30 minutes after TGF-β1 stimulation (0.5 ng/ml) by Western blotting. Ratios of P-Smad2/glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (with TGF-β1 stimulation), T-Smad2/GAPDH (without TGF-β1 stimulation), and P-Smad3/T-Smad3 (with TGF-β1 stimulation) are shown. White bars, negative control (NC) siRNA–transfected cells; black bars, HS6ST1 siRNA–transfected cells. (B) Analysis of mRNA expression of HS6ST1, collagen I, and α-SMA by quantitative RT-PCR. Fold changes were normalized to the expression of the housekeeping gene 36B4. (C) Analysis of collagen I and α-SMA protein expression at 30 hours after TGF-β1 stimulation (0.5 ng/ml) by Western blotting. (D) Analysis of TβRI, -II, and -III expression at 48 hours after HS6ST1 siRNA transfection by Western blotting. *P < 0.05; **P < 0.01; ***P < 0.001. Data shown are representative of four independent experiments in duplicate.
Alterations in syndecan expression have been linked to changes in the expression levels of cell surface TGF-β1 receptors (31, 32). Therefore, we asked whether silencing of HS6ST1 would alter the expression of TGF-β1 receptors in lung fibroblasts. Our results showed that the expression of TβRIII, betaglycan, was significantly reduced in HS6ST1-silenced cells at the protein level, without changes in the expression of TβRI and TβRII (Figure 5D; Figure E2A). To further study the regulation of TβRIII expression by HS6ST1, we examined TβRIII mRNA in control and HS6ST1 siRNA–transfected lung fibroblasts, and our results showed that TβRIII mRNA was significantly down-regulated in HS6ST1 siRNA–transfected cells (Figure E2B), which was likely responsible for the down-regulation of TβRIII at the protein level (Figure 5D).
Using qRT-PCR, we confirmed the silencing of HS6ST1 in HS6ST1 siRNA–transfected lung fibroblasts (Figure 5B). Stimulation with TGF-β1 in control cells led to reduced HS6ST1 expression (< 20% of the levels in unstimulated cells). Thus, the reduced HS 6-O-sulfation in TGF-β1–treated lung fibroblasts reported previously (22) is likely the result of enhanced expression of Sulf1 and the reduced expression of HS6ST1.
We examined whether silencing of HS6ST1 could reduce TGF-β1 signaling in the IPF lung fibroblasts and obtained similar results (Figure 6). These data indicate that, although IPF lung fibroblasts exhibit the myofibroblast phenotype (Figure 4H), they continue to respond to TGF-β1 and that the TGF-β1 response in IPF lung fibroblasts could be dampened by silencing of HS6ST1, as in normal lung fibroblasts.
Figure 6.
HS6ST1 silencing reduces TGF-β1 signaling in IPF lung fibroblasts. (A) Analysis of phospho- and total Smad2/3 levels and the expression of TβRIII at 30 minutes after TGF-β1 stimulation (0.5 ng/ml) by Western blotting. Ratios of P-Smad2/GAPDH (with TGF-β1 stimulation), T-Smad2/GAPDH (without TGF-β1 stimulation), P-Smad3/T-Smad3 (with TGF-β1 stimulation), and TβRIII/GAPDH (without TGF-β1 stimulation) are shown in B. White bars, NC siRNA–transfected cells; black bars, HS6ST1 siRNA–transfected cells. (C) Analysis of collagen I and α-SMA protein expression at 30 hours after TGF-β1 stimulation (0.5 ng/ml) by Western blotting with quantifications shown in D. *P < 0.05; **P < 0.01; ***P < 0.001. HS6ST1 silencing was performed in four IPF lung fibroblast lines with similar results, and data from one IPF fibroblast line are shown.
Discussion
HS plays important roles in a variety of developmental, physiological, and pathological processes through interaction with a multitude of HS-binding growth factors and cytokines. HS–protein interactions depend on the amount and the positions of the O-sulfate groups, in particular, the 6-O-sulfates that form binding sites for proteins. HS 6-O-sulfation has been shown to be important in a number of developmental processes, including branching morphogenesis of the developing respiratory tract in Drosophila (33), branching of vascular structures in zebrafish embryos (34), and directing retinal axons at the optic chiasm (35) and lacrimal gland development in mice (36). In addition, increased HS 6-O-sulfation is associated with aging (37, 38), malignant transformation from human colon adenoma to carcinoma (39), and progression of chondrosarcomas (40). In this study, we show that HS 6-O-sulfation is dramatically increased in IPF lungs compared with normal lungs, concomitant with overexpression of HS6ST1 and HS6ST2S. In the effector cells of pulmonary fibrosis, the myofibroblasts, HS6ST1 is overexpressed and may regulate TGF-β1–induced fibrotic responses in these cells.
HS6ST1 is the major HS6ST isoform expressed in embryonic and adult lungs (27, 28). HS6ST1-deficient mice exhibit marked reduction of GlcNAc-6S and UA-GlcNS-6S residues in the HS isolated from the lung, and the defective lung morphology with enlarged alveolar spaces accompanied by abnormal elastin pattern suggestive of emphysematous changes (41, 42). These findings indicate that HS 6-O-sulfation regulated by HS6ST1 is important for normal lung development. Our current study suggests that HS 6-O-sulfation, likely regulated by HS6ST1 and HS6ST2S, is involved in pathological processes in the lung, in this case IPF.
The importance of HS 6-O-sulfation in regulating biological and pathological processes is underscored by the fact that two groups of enzymes regulate HS 6-O-sulfation status: the HS6STs, which add sulfates to the 6-O-position during HS biosynthesis, and the Sulfs, which remove the 6-O-sulfates in the extracellular milieu. In a previous study, we showed that Sulf1 is a TGF-β1 responsive gene in normal human lung fibroblasts and that siRNA-mediated silencing of Sulf1 (which leads to increased HS 6-O-sulfation) results in enhanced TGF-β1 signaling (22). In this study, we show that siRNA-mediated silencing of HS6ST1 (which leads to decreased HS 6-O-sulfation) results in reduced TGF-β1 signaling, opposite to the effect of Sulf1 silencing. In IPF lung fibroblasts, silencing of HS6ST1 results in similar inhibition of TGF-β1–induced fibrotic responses. These data suggest that HS6ST1 could be manipulated therapeutically to reduce the responsiveness of lung fibroblasts/myofibroblasts to TGF-β1 and possibly to prevent the progression of IPF.
HS 6-O-sulfation is regulated by TGF-β1 and is a regulator of the same pathway. On stimulation by TGF-β1, Sulf1 expression is induced (22) and HS6ST1 expression is suppressed (Figure 5B), leading to a reduction of HS 6-O-sulfation (22). Our previous and current studies suggest that this reduction of HS 6-O-sulfation serves as a negative feedback mechanism to restrain excessive TGF-β1 signaling. The regulation of TGF-β1 by HS 6-O-sulfation could occur through multiple pathways. First, HS 6-O-sulfation could alter TGF-β1 binding to the cell surface because TGF-β1 binds to HS and TGF-β1–HS interaction requires 6-O-sulfation (43). Although HS does not appear to be directly required for TGF-β1 binding and/or activation of its specific signal transducing receptors (44), binding of TGF-β1 to 6-O-sulfated HS at the cell membrane could help bring TGF-β1 to the close proximity of its signaling receptors and increase the local concentration of TGF-β1. Second, through unknown mechanisms, HS 6-O-sulfation regulates the expression of total Smad2. In lung fibroblasts with siRNA-mediated Sulf1 silencing, the expression of total Smad2 is increased (22), whereas in lung fibroblasts with silencing of HS6ST1, reduced expression of total Smad2 is observed, as shown in the current study. Last, HS6ST1 silencing leads to down-regulation of the TβRIII, betaglycan. Betaglycan is the most abundant TGF-β receptor at the cell surface, and membrane betaglycan has been shown to increase TGF-β1 binding to TβRII and to enhance cell responsiveness to TGF-β ligands (45). How silencing of HS6ST1 leads to down-regulation of betaglycan remains to be determined. Betaglycan is a transmembrane proteoglycan that carries HS and chondroitin sulfate chains; however, the glycosaminoglycan chains of betaglycan are not necessary for binding of TGF-β (45).
TGF-β activation is a complex process involving various protein interactions (46). A previous study reported reduced, rather than enhanced, TGF-β1 activity (as shown by less prominent P-Smad2 immunoreactivity) in the fibroblastic foci compared with the surrounding structures (47), which seems to be inconsistent with the high expression of HS6ST1 reported by the current study. The reduced P-Smad2 in the fibroblastic foci could be due to the high levels of the latent TGF-β–binding protein-1 found in these areas (47). The expression of latent TGF-β–binding protein-1 in lung fibroblasts has been shown to be inversely related to TGF-β1 activity (47).
The substrate specificity of HS6ST1 and HS6ST2 largely overlap, although individual isoforms exhibit a characteristic preference of the UA residue neighboring the GlcNS (27). As suggested by our study and by others (48), it is the tissue- or cell-type–specific expression that determines the unique biological functions of each isoform. In this study, we found that HS6ST2S is expressed in a subset of the bronchial epithelial cells in normal lungs and that its expression is greatly increased in the IPF lungs. In IPF, a progressive bronchiolar proliferation coincides with the loss of alveolar tissue and alveolar collapse (29). It is important to investigate whether the expression of HS6ST2S contributes to the bronchiolar metaplasia process of the distal lung in IPF. In addition, bronchiolar metaplasia is thought to be a preneoplastic lesion, and IPF is frequently associated with the development of lung cancer (49). On this note, HS6ST2S has been shown to be overexpressed in pancreatic cancer, and silencing of HS6ST2S leads to inhibition of cell invasion and migration and to reduced tumorigenesis (50). Whether the overexpression of HS6ST2S contributes to the increased incidence of lung cancer in patients with IPF remain to be elucidated.
Acknowledgments
Acknowledgments
This study used human lung specimens and data provided by the Lung Tissue Research Consortium, which is supported by the National Heart, Lung, and Blood Institute.
Footnotes
This work was supported by National Institutes of Health grants NHLBIR21 HL095865 (X.Y.), R03 HL096949 (X.Y.), NIH-NIGMS P20 GM103514 (former NCRR P20 RR018766), and NIH U01 HL111016 (E.S.W.). Heparan sulfate disaccharide analysis was supported in part by National Institutes of Health–funded Research Resource for Integrated Glycotechnology (P41GM103390) to the Complex Carbohydrate Research Center at University of Georgia.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2013-0204OC on August 20, 2013
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Kim R, Meyer KC. Therapies for interstitial lung disease: past, present and future. Ther Adv Respir Dis. 2008;2:319–338. doi: 10.1177/1753465808096948. [DOI] [PubMed] [Google Scholar]
- 2.Raghu G, Weycker D, Edelsberg J, Bradford WZ, Oster G. Incidence and prevalence of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2006;174:810–816. doi: 10.1164/rccm.200602-163OC. [DOI] [PubMed] [Google Scholar]
- 3.du Bois RM. Strategies for treating idiopathic pulmonary fibrosis. Nat Rev Drug Discov. 2010;9:129–140. doi: 10.1038/nrd2958. [DOI] [PubMed] [Google Scholar]
- 4.Zhang HY, Gharaee-Kermani M, Zhang K, Karmiol S, Phan SH. Lung fibroblast alpha-smooth muscle actin expression and contractile phenotype in bleomycin-induced pulmonary fibrosis. Am J Pathol. 1996;148:527–537. [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhn C, McDonald JA. The roles of the myofibroblast in idiopathic pulmonary fibrosis: ultrastructural and immunohistochemical features of sites of active extracellular matrix synthesis. Am J Pathol. 1991;138:1257–1265. [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang K, Rekhter MD, Gordon D, Phan SH. Myofibroblasts and their role in lung collagen gene expression during pulmonary fibrosis: a combined immunohistochemical and in situ hybridization study. Am J Pathol. 1994;145:114–125. [PMC free article] [PubMed] [Google Scholar]
- 7.King TE, Jr, Schwarz MI, Brown K, Tooze JA, Colby TV, Waldron JA, Jr, Flint A, Thurlbeck W, Cherniack RM. Idiopathic pulmonary fibrosis: relationship between histopathologic features and mortality. Am J Respir Crit Care Med. 2001;164:1025–1032. doi: 10.1164/ajrccm.164.6.2001056. [DOI] [PubMed] [Google Scholar]
- 8.Ramos C, Montaño M, García-Alvarez J, Ruiz V, Uhal BD, Selman M, Pardo A. Fibroblasts from idiopathic pulmonary fibrosis and normal lungs differ in growth rate, apoptosis, and tissue inhibitor of metalloproteinases expression. Am J Respir Cell Mol Biol. 2001;24:591–598. doi: 10.1165/ajrcmb.24.5.4333. [DOI] [PubMed] [Google Scholar]
- 9.Suganuma H, Sato A, Tamura R, Chida K. Enhanced migration of fibroblasts derived from lungs with fibrotic lesions. Thorax. 1995;50:984–989. doi: 10.1136/thx.50.9.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsson O, Diebold D, Fan D, Peterson M, Nho RS, Bitterman PB, Henke CA. Fibrotic myofibroblasts manifest genome-wide derangements of translational control. PLoS ONE. 2008;3:e3220. doi: 10.1371/journal.pone.0003220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phan SH. The myofibroblast in pulmonary fibrosis. Chest. 2002;122(Suppl):286S–289S. doi: 10.1378/chest.122.6_suppl.286s. [DOI] [PubMed] [Google Scholar]
- 12.Massagué J, Gomis RR. The logic of TGFbeta signaling. FEBS Lett. 2006;580:2811–2820. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 13.Ren M, Wang B, Zhang J, Liu P, Lv Y, Liu G, Jiang H, Liu F. Smad2 and Smad3 as mediators of the response of adventitial fibroblasts induced by transforming growth factor β1. Mol Med Rep. 2011;4:561–567. doi: 10.3892/mmr.2011.458. [DOI] [PubMed] [Google Scholar]
- 14.Smits NC, Shworak NW, Dekhuijzen PN, van Kuppevelt TH. Heparan sulfates in the lung: structure, diversity, and role in pulmonary emphysema. Anat Rec (Hoboken) 2010;293:955–967. doi: 10.1002/ar.20895. [DOI] [PubMed] [Google Scholar]
- 15.Sannes PL, Wang J. Basement membranes and pulmonary development. Exp Lung Res. 1997;23:101–108. doi: 10.3109/01902149709074023. [DOI] [PubMed] [Google Scholar]
- 16.Kliment CR, Englert JM, Gochuico BR, Yu G, Kaminski N, Rosas I, Oury TD. Oxidative stress alters syndecan-1 distribution in lungs with pulmonary fibrosis. J Biol Chem. 2009;284:3537–3545. doi: 10.1074/jbc.M807001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz XD, Mlakar LR, Yamaguchi Y, Su Y, Larregina AT, Pilewski JM, Feghali-Bostwick CA. Syndecan-2 is a novel target of insulin-like growth factor binding protein-3 and is over-expressed in fibrosis. PLoS ONE. 2012;7:e43049. doi: 10.1371/journal.pone.0043049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang D, Liang J, Campanella GS, Guo R, Yu S, Xie T, Liu N, Jung Y, Homer R, Meltzer EB, et al. Inhibition of pulmonary fibrosis in mice by CXCL10 requires glycosaminoglycan binding and syndecan-4. J Clin Invest. 2010;120:2049–2057. doi: 10.1172/JCI38644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenkrans WA, Jr, Penney DP. Cell-cell matrix interactions in induced lung injury: II. X-irradiation mediated changes in specific basal laminar glycosaminoglycans. Int J Radiat Oncol Biol Phys. 1985;11:1629–1637. doi: 10.1016/0360-3016(85)90215-9. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki T, Kojima S, Kubodera A. Uptake of 67Ga in the lung of mice during bleomycin treatment. Eur J Nucl Med. 1984;9:57–61. doi: 10.1007/BF00254437. [DOI] [PubMed] [Google Scholar]
- 21.Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 22.Yue X, Li X, Nguyen HT, Chin DR, Sullivan DE, Lasky JA. Transforming growth factor-beta1 induces heparan sulfate 6-O-endosulfatase 1 expression in vitro and in vivo. J Biol Chem. 2008;283:20397–20407. doi: 10.1074/jbc.M802850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 24.Toyoda H, Nagashima T, Hirata R, Toida T, Imanari T. Sensitive high-performance liquid chromatographic method with fluorometric detection for the determination of heparin and heparan sulfate in biological samples: application to human urinary heparan sulfate. J Chromatogr B Biomed Sci Appl. 1997;704:19–24. doi: 10.1016/s0378-4347(97)00478-7. [DOI] [PubMed] [Google Scholar]
- 25.Yue X, Lu J, Auduong L, Sides MD, Lasky JA. Overexpression of Sulf2 in idiopathic pulmonary fibrosis. Glycobiology. 2013;23:709–719. doi: 10.1093/glycob/cwt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White ES, Thannickal VJ, Carskadon SL, Dickie EG, Livant DL, Markwart S, Toews GB, Arenberg DA. Integrin alpha4beta1 regulates migration across basement membranes by lung fibroblasts: a role for phosphatase and tensin homologue deleted on chromosome 10. Am J Respir Crit Care Med. 2003;168:436–442. doi: 10.1164/rccm.200301-041OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Habuchi H, Tanaka M, Habuchi O, Yoshida K, Suzuki H, Ban K, Kimata K. The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for hexuronic acid adjacent to the targeted N-sulfoglucosamine. J Biol Chem. 2000;275:2859–2868. doi: 10.1074/jbc.275.4.2859. [DOI] [PubMed] [Google Scholar]
- 28.Habuchi H, Miyake G, Nogami K, Kuroiwa A, Matsuda Y, Kusche-Gullberg M, Habuchi O, Tanaka M, Kimata K. Biosynthesis of heparan sulphate with diverse structures and functions: two alternatively spliced forms of human heparan sulphate 6-O-sulphotransferase-2 having different expression patterns and properties. Biochem J. 2003;371:131–142. doi: 10.1042/BJ20021259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chilosi M, Poletti V, Murer B, Lestani M, Cancellieri A, Montagna L, Piccoli P, Cangi G, Semenzato G, Doglioni C. Abnormal re-epithelialization and lung remodeling in idiopathic pulmonary fibrosis: the role of deltaN-p63. Lab Invest. 2002;82:1335–1345. doi: 10.1097/01.lab.0000032380.82232.67. [DOI] [PubMed] [Google Scholar]
- 30.Lamanna WC, Kalus I, Padva M, Baldwin RJ, Merry CL, Dierks T. The heparanome: the enigma of encoding and decoding heparan sulfate sulfation. J Biotechnol. 2007;129:290–307. doi: 10.1016/j.jbiotec.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 31.Chen L, Klass C, Woods A. Syndecan-2 regulates transforming growth factor-beta signaling. J Biol Chem. 2004;279:15715–15718. doi: 10.1074/jbc.C300430200. [DOI] [PubMed] [Google Scholar]
- 32.Mytilinaiou M, Bano A, Nikitovic D, Berdiaki A, Voudouri K, Kalogeraki A, Karamanos NK, Tzanakakis GN. Syndecan-2 is a key regulator of transforming growth factor beta 2/Smad2-mediated adhesion in fibrosarcoma cells. IUBMB Life. 2013;65:134–143. doi: 10.1002/iub.1112. [DOI] [PubMed] [Google Scholar]
- 33.Kamimura K, Fujise M, Villa F, Izumi S, Habuchi H, Kimata K, Nakato H. Drosophila heparan sulfate 6-O-sulfotransferase (dHS6ST) gene: structure, expression, and function in the formation of the tracheal system. J Biol Chem. 2001;276:17014–17021. doi: 10.1074/jbc.M011354200. [DOI] [PubMed] [Google Scholar]
- 34.Chen E, Stringer SE, Rusch MA, Selleck SB, Ekker SC. A unique role for 6-O sulfation modification in zebrafish vascular development. Dev Biol. 2005;284:364–376. doi: 10.1016/j.ydbio.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 35.Pratt T, Conway CD, Tian NM, Price DJ, Mason JO. Heparan sulphation patterns generated by specific heparan sulfotransferase enzymes direct distinct aspects of retinal axon guidance at the optic chiasm. J Neurosci. 2006;26:6911–6923. doi: 10.1523/JNEUROSCI.0505-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qu X, Carbe C, Tao C, Powers A, Lawrence R, van Kuppevelt TH, Cardoso WV, Grobe K, Esko JD, Zhang X. Lacrimal gland development and Fgf10-Fgfr2b signaling are controlled by 2-O- and 6-O-sulfated heparan sulfate. J Biol Chem. 2011;286:14435–14444. doi: 10.1074/jbc.M111.225003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huynh MB, Morin C, Carpentier G, Garcia-Filipe S, Talhas-Perret S, Barbier-Chassefière V, van Kuppevelt TH, Martelly I, Albanese P, Papy-Garcia D. Age-related changes in rat myocardium involve altered capacities of glycosaminoglycans to potentiate growth factor functions and heparan sulfate-altered sulfation. J Biol Chem. 2012;287:11363–11373. doi: 10.1074/jbc.M111.335901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feyzi E, Saldeen T, Larsson E, Lindahl U, Salmivirta M. Age-dependent modulation of heparan sulfate structure and function. J Biol Chem. 1998;273:13395–13398. doi: 10.1074/jbc.273.22.13395. [DOI] [PubMed] [Google Scholar]
- 39.Jayson GC, Lyon M, Paraskeva C, Turnbull JE, Deakin JA, Gallagher JT. Heparan sulfate undergoes specific structural changes during the progression from human colon adenoma to carcinoma in vitro. J Biol Chem. 1998;273:51–57. doi: 10.1074/jbc.273.1.51. [DOI] [PubMed] [Google Scholar]
- 40.Waaijer CJ, de Andrea CE, Hamilton A, van Oosterwijk JG, Stringer SE, Bovée JV. Cartilage tumour progression is characterized by an increased expression of heparan sulphate 6O-sulphation-modifying enzymes. Virchows Arch. 2012;461:475–481. doi: 10.1007/s00428-012-1300-5. [DOI] [PubMed] [Google Scholar]
- 41.Habuchi H, Nagai N, Sugaya N, Atsumi F, Stevens RL, Kimata K. Mice deficient in heparan sulfate 6-O-sulfotransferase-1 exhibit defective heparan sulfate biosynthesis, abnormal placentation, and late embryonic lethality. J Biol Chem. 2007;282:15578–15588. doi: 10.1074/jbc.M607434200. [DOI] [PubMed] [Google Scholar]
- 42.Izvolsky KI, Lu J, Martin G, Albrecht KH, Cardoso WV. Systemic inactivation of Hs6st1 in mice is associated with late postnatal mortality without major defects in organogenesis. Genesis. 2008;46:8–18. doi: 10.1002/dvg.20355. [DOI] [PubMed] [Google Scholar]
- 43.Lyon M, Rushton G, Gallagher JT. The interaction of the transforming growth factor-betas with heparin/heparan sulfate is isoform-specific. J Biol Chem. 1997;272:18000–18006. doi: 10.1074/jbc.272.29.18000. [DOI] [PubMed] [Google Scholar]
- 44.Lyon M, Gallagher JT. Bio-specific sequences and domains in heparan sulphate and the regulation of cell growth and adhesion. Matrix Biol. 1998;17:485–493. doi: 10.1016/s0945-053x(98)90096-8. [DOI] [PubMed] [Google Scholar]
- 45.López-Casillas F, Wrana JL, Massagué J. Betaglycan presents ligand to the TGF beta signaling receptor. Cell. 1993;73:1435–1444. doi: 10.1016/0092-8674(93)90368-z. [DOI] [PubMed] [Google Scholar]
- 46.Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. J Cell Sci. 2003;116:217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- 47.Leppäranta O, Sens C, Salmenkivi K, Kinnula VL, Keski-Oja J, Myllärniemi M, Koli K. Regulation of TGF-β storage and activation in the human idiopathic pulmonary fibrosis lung. Cell Tissue Res. 2012;348:491–503. doi: 10.1007/s00441-012-1385-9. [DOI] [PubMed] [Google Scholar]
- 48.Habuchi H, Kimata K. Mice deficient in heparan sulfate 6-O-sulfotransferase-1. Prog Mol Biol Transl Sci. 2010;93:79–111. doi: 10.1016/S1877-1173(10)93005-6. [DOI] [PubMed] [Google Scholar]
- 49.Königshoff M. Lung cancer in pulmonary fibrosis: tales of epithelial cell plasticity. Respiration. 2011;81:353–358. doi: 10.1159/000326299. [DOI] [PubMed] [Google Scholar]
- 50.Song K, Li Q, Peng YB, Li J, Ding K, Chen LJ, Shao CH, Zhang LJ, Li P. Silencing of hHS6ST2 inhibits progression of pancreatic cancer through inhibition of Notch signalling. Biochem J. 2011;436:271–282. doi: 10.1042/BJ20110297. [DOI] [PubMed] [Google Scholar]