Abstract
Despite modern sequencing efforts, the difficulty in assembly of highly repetitive sequences has prevented resolution of human genome gaps, including some in the coding regions of genes with important biological functions. One such gene, MUC5AC, encodes a large, secreted mucin, which is one of the two major secreted mucins in human airways. The MUC5AC region contains a gap in the human genome reference (hg19) across the large, highly repetitive, and complex central exon. This exon is predicted to contain imperfect tandem repeat sequences and multiple conserved cysteine-rich (CysD) domains. To resolve the MUC5AC genomic gap, we used high-fidelity long PCR followed by single molecule real-time (SMRT) sequencing. This technology yielded long sequence reads and robust coverage that allowed for de novo sequence assembly spanning the entire repetitive region. Furthermore, we used SMRT sequencing of PCR amplicons covering the central exon to identify genetic variation in four individuals. The results demonstrated the presence of segmental duplications of CysD domains, insertions/deletions (indels) of tandem repeats, and single nucleotide variants. Additional studies demonstrated that one of the identified tandem repeat insertions is tagged by nonexonic single nucleotide polymorphisms. Taken together, these data illustrate the successful utility of SMRT sequencing long reads for de novo assembly of large repetitive sequences to fill the gaps in the human genome. Characterization of the MUC5AC gene and the sequence variation in the central exon will facilitate genetic and functional studies for this critical airway mucin.
Keywords: MUC5AC, single molecule real time sequencing, segmental duplication, PacBio, repetitive sequence
Mucins (MUC1–MUC22) are highly glycosylated proteins with important host-defense functions in lung and other organs. Secreted gel-forming mucins on epithelial surfaces contribute to the highly specialized biochemical matrices critical for epithelial protection and defense. In the airway, MUC5AC and MUC5B are major constituents of mucus, which is critical for mucociliary clearance, a key innate defense against respiratory insults (1). In the gut, MUC2, MUC5AC, and other mucins contribute to the barrier that maintains gastrointestinal homeostasis in the presence of the very acidic environment of the stomach and the huge bacterial burdens in the intestine (2–4). Like secreted (or gel-forming) mucins, transmembrane-anchored mucins contribute to airway host defense via the epithelial glycocalyx, where they can limit pathogen and particle access (5, 6).
The role of mucins in maintaining organismal homeostasis is highlighted by the diseases developing under conditions of defective mucin function. Loss of airway mucociliary clearance, which is dependent on secreted mucins, leads to severe respiratory diseases, such as chronic obstructive pulmonary disease and cystic fibrosis (CF). In mice, loss of Muc5AC increases susceptibility to intestinal nematodes and alters responses to ventilator-induced lung injury (7, 8). In the gut, expression, secretion, and glycosylation of MUC2/Muc2 contribute to inflammatory bowel diseases (9–12). New data point to the importance of mucin/pathogen interactions for gastric ulcers associated with Helicobacter pylori infection (13–15). Recently, mutations in MUC1 were shown to cause medullary cystic kidney disease type 1 (16).
Recent genetic studies further implicate mucin gene function in human disease. For example, a common polymorphism in the promoter of MUC5B is associated with idiopathic pulmonary fibrosis (17–19). In our own recent work, a specific 6.4-kb HinfI allele–length polymorphism was shown to associate with more severe lung disease in CF (20). MUC5AC and MUC5B are located in the subtelomeric, recombination-rich region on chromosome 11p15.5, which contains several mucin genes, from telomere to centromere, MUC6-MUC2-MUC5AC-MUC5B (21). The Genome Reference Consortium (GRCh37) and human genome reference (hg19) sequences have a gap between the 5′ and 3′ exons of MUC5AC (see Figure 1A for overview) that is at least partially comprised of an unusually large central exon encoding the tandem repeats (TR) (previously described in the literature as “variable number of tandem repeats”) and several highly similar cysteine-rich (CysD) regions (22, 23). The MUC5AC TR translates into an eight-amino-acid repeat rich in proline, threonine, and serine (PTS-TR). PTS-TR domains are characteristic of most mucin proteins and are often encoded by single exons representing the heavily O-glycosylated protein domains critical for mucin function (24). Southern blots have been used to describe the general nature of the genetic length variation in PTS-TR regions (25, 26), but specific sequence details of the polymorphic alleles are not available. Because of their highly repetitive and complex nature, these regions for many mucin genes are inadequately or inaccurately defined (27) and, in the case of MUC5AC, contribute to a sequence gap in GRCh37/hg19.
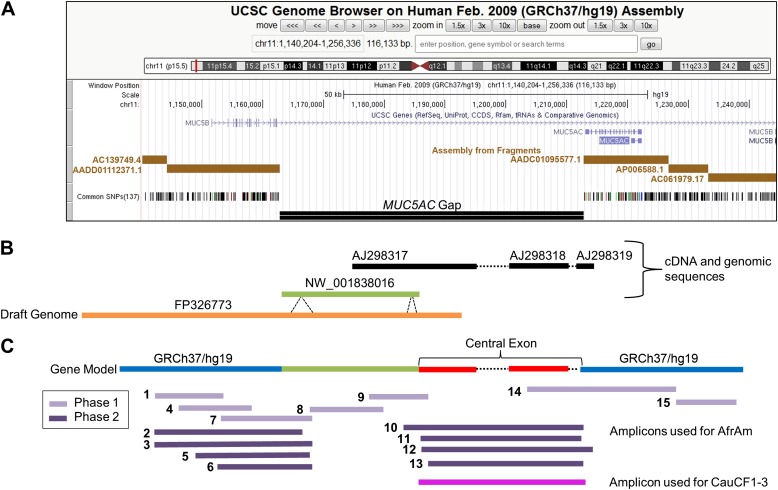
Figure 1.
MUC5AC genomic region. Current status and research design to cover the MUC5AC genomic gap. (A) Annotation tracks for the MUC5AC region excerpted from the University of California Santa Cruz genome browser (http://genome.ucsc.edu) (GRCh37/hg19) with notes added to emphasize the gap. The current gap in the MUC5AC gene is situated between a set of exons (blue vertical bars along arrowed line) that are 5′ (the 5′ exons are incorrectly annotated to MUC5B) and a set of 3′ exons (that are correctly annotated to MUC5AC). The entire region is in general disarray. (B) Available sequences used to inform the selection of PCR primers to characterize the gene, with a focus on filling the gap. The PCR primer selection was based on the use of the human reference alternative assembly genomic scaffold sequence NW_001838016 and other existing cDNA and genomic sequences. The High Throughput Genomics (http://www.ncbi.nlm.nih.gov/genbank/htgs) working draft sequence FP326773 became available during the course of this work, and it differs from NW_001838016 in length in the regions indicated. Together, these two sequences provide information for the 5′ end of the gap and contribute to the gene model. From previous efforts, there is strong evidence that the 3′ end portion of the gap consists of the MUC5AC large central exon. The available sequences in the large central exon region that were used to inform the PCR strategy consisted of one partial mRNA (AJ298317) and two partial genomic PCR sequences (AJ298318 and AJ298319) (22). These sequences were used in conjunction with NW_001838016 and FP326773 to generate a gene model, which was consistent with previous efforts (17). (C) Schematic representation of the overlapping PCR products used in contig development for de novo assembly of the MUC5AC gene from the African American (AfrAm) subject and the region of focus for white subjects with cystic fibrosis (CauCF1–3). The AfrAm individual was sequenced in two phases (Phase 1 and 2) as described in Materials and Methods (further details are provided in Table E2).
The deficiency in genomic references and poor annotation has led to a failure to fully represent mucins, such as MUC5AC, on exome chips, gene expression microarrays, and high-density single nucleotide polymorphism (SNP) panels. Thus, there is poor representation in genome-wide studies, and results from global expression studies are often inaccurate. In addition, large-scale genomic efforts (e.g., the Encyclopedia of DNA Elements [ENCODE; http://encodeproject.org], which seeks to understand global gene expression regulatory elements), fail to adequately query regions that lack high-quality reference sequences.
Third-generation, single-molecule sequencing technology, such as single molecule real-time (SMRT) sequencing by Pacific Biosciences (PacBio), provides relatively long sequence reads and offers an opportunity to overcome some of the difficulties (28, 29) that have prevented the availability of a reliable MUC5AC gene reference. We have successfully used this technology to (1) confirm the 5′ and 3′ structure of MUC5AC; (2) determine the missing sequence across the public GRCh37/hg19 gap in MUC5AC and demonstrate that it contains a single, large, complex central exon; and (3) characterize the structural variants, insertion/deletion (indel) variants, and SNPs among four subjects in this highly polymorphic exon. The results provide a solid basis for future efforts to define MUC5AC functional genetic variants for biological, mechanistic, and genetic association studies.
Materials and Methods
Selection of Subjects
Four subjects were analyzed, one nonCF African American (AfrAm) subject available through Duke University’s airway cell repository (30) and three white F508del homozygote CF subjects (labeled CauCF1, CauCF2, and CauCF3) from the University of North Carolina/Case Western Reserve University Genetic Modifier Study population (31). Relevant features of the subjects together with the DNA source and isolation strategy are listed in Table E1 in the online supplement. The three white subjects with CF were selected to represent common haplotypes on the basis of the MUC5AC HinfI fragment length and SNP genotypes previously reported (20). This study was conducted in accordance with institutional review board approvals.
Gene Model and Confirmation in the AfrAm Subject
DNA from the AfrAm subject was used to confirm a gene model built from existing genomic scaffold sequences (17, 22) (Figure 1). On the basis of this model, 11 pairs of primers were designed to produce seven overlapping products (Phase 1). In Phase 2, eight pairs of PCR primers targeting two regions not well covered in Phase 1 were used (Figure 1C; Table E2). The PCR primer pairs were designed using Primer-Blast from the National Center for Biotechnology Information (NCBI) (32) (Table E2). Additional details are provided in the online supplement.
Amplifying the Central Repetitive Region of MUC5AC in Subjects CauCF1, -2, and -3
There are two reasons for the focus on subjects CauCF1, -2, and -3: 1) to determine the sequence across the predicted central exon and 2) to define the polymorphic nature of allele sizes demonstrated for these three subjects on Southern blots for this region. A pair of unique primers was designed to amplify the central repetitive region in MUC5AC (Figure 1C). Additional details can be found in the online supplement.
Sequence Assembly
Details of the methods applied for sequencing runs, sequence assembly, and sequence processing are provided in the online supplement. Sequencing reads greater than 3 kb from the AfrAm individual were analyzed using the assembly tools in SMRT Portal (ALLORA; V1 Pacific Biosciences, Menlo Park, CA). Alignment of multiple contigs and merging of two contigs were accomplished by using Vector NTI (Life Technologies, Grand Island, NY). Alignment of genomic DNA and mRNAs was performed using Spidey from NCBI (http://www.ncbi.nlm.nih.gov/spidey/), and translation of DNA to protein was performed using a web-based translation tool (http://insilico.ehu.es/translate/) (33). Alternatively, for the CauCF subjects, SMRT sequencing reads were filtered by size, and de novo assembly was conducted using the current production release of MIRA v3.4 (http://www.chevreux.org/projects_mira.html) (34) with custom tuned parameters and minimum read length of 1 kb.
Confirmation of Duplication of 5′ Region Conserved Duplicon in CauCF3
A PCR method described in the online supplement was developed to confirm the duplication containing two extra internal CysD domains at the 5′ portion of the central repetitive region in CauCF3. In addition, direct genomic DNA sequencing from CauCF3 was conducted. Details of the sequencing and the subsequent analyses of this enriched DNA are given in the online supplement. Briefly, BsbI-digested genomic fragments from CauCF3 were size selected (∼ 9–15 kb) on agarose gels and purified via electroelution. Because BsbI is predicted to cut outside the central repetitive region, the size selection would leave the region intact while enriching the sample for the DNA of interest.
Results
Defining the MUC5AC Sequence and Genomic Gap in an African-American Subject
Recent builds of the human genome reference (GRCh37/hg19) have a gap where MUC5AC resides (Figure 1). On the basis of the data presented here, an accurate picture of this genomic region has been obtained, including all intron/exon sequences (Figures 2 and E1). Specifically, de novo assembly of the AfrAm subject sequence (Figure E1), consisting of two large contigs (contigs 1 and 2) (Figure 2A and Table E3), confirms the general structure of the gene model, which used various pieces of available sequence (Figure 1B). Sequencing of the AfrAm subject (Figure E1) confirmed the integrity of the High Throughput Genomics Center (http://www.ncbi.nlm.nih.gov/genbank/htgs) working draft FP326773 (Figure 1B).
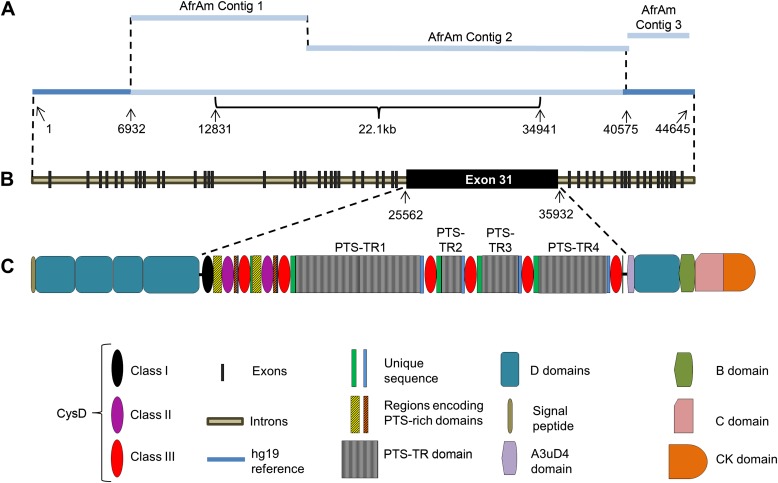
Figure 2.
MUC5AC gene defined in an AfrAm subject. (A) Schematic representation of de novo assembled sequence contigs produced from Pacific Biosciences sequencing of pooled PCR product sequences from the AfrAm subject. The relative sizes of the contigs are shown roughly to proportion, and the predicted gap size for the reference genome is 22.1 kb based on the location of flanking GRCh37/hg19 sequences. (B) Schematic representation of MUC5AC gene showing exon locations. The large central exon is predicted to be exon 31, containing nine CysD domains and the tandem repeat (PTS-TR) sequences. (C) Schematic MUC5AC mRNA protein translation showing the major protein domains and their relationship to the entire gene and the central exon. The central exon consists of a 5′ region characterized by one Class I CysD domain, duplicated pairs of Class II and Class III CysD domains, and adjacent homologous sequence, which is rich in prolines, threonines, and serines (PTS region). The 3′ half of the central exon has a different structure, which is characterized by Class III CysD domains and adjacent unique sequences separated by PTS-TR units (TR1–4) of 24-bp imperfect repeats. Other protein features shown were previously defined (22). C domain = von Willebrand factor type C domain; CK domain = C-terminal cysteine knot domain; D domain = von Willebrand factor type D domain. The definition of the CysD domain classes is provided in the text.
The MUC5AC gene contains 49 exons in the AfrAm individual (Figures 2B and E1). Location and sequences of the intron–exon boundaries, intron and exon sizes, and splice junction sequence are provided in Figure E1 and Table E4. After additional processing of the sequence (details are provided in the online supplement), the full-length predicted mRNA sequence was translated into 5,654 amino acids and had the structural organization/features as presented in Figure 2C. Most of the translation aligned well to p98088, the Uniprot MUC5AC reference protein sequence primarily translated from mRNA sequences (http://www.uniprot.org/uniprot/P98088); however, differences between the two sequences are noted in Figure 3 (blue and green horizontal bars). The sequences encoding these amino acids represented the sequences linking the previously available sequences (22, 35), essentially filling the gaps between AJ298317 and AJ298319 (Figures 1B and 1C, dashed horizontal lines). We confirmed the original hypothesis that the 3′ region of the genomic gap in GRCh37/hg19 consisted of one large central repetitive exon (exon 31) encompassing the TR (PTS-TR) and nine CysD domains. The CysD domains are characterized in this study into three classes on the basis of sequence alignment (Figure E2), a concept that has previously been proposed (22) but was expanded on for discussion purposes in this manuscript. The 5′ CysD domains combined with adjacent flanking sequences are referred to as “5′ region conserved duplicon” and the 3′ CysD domain of the central exon, combined with adjacent flanking sequences, is referred to as a “3′ region conserved CysD segment” throughout this paper (Figure 3).
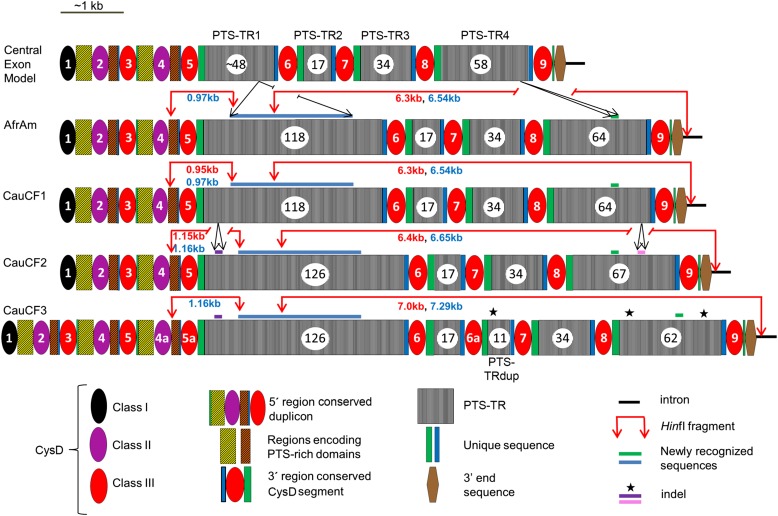
Figure 3.
Genetic variants and organization of the MUC5AC central exon. Sequence schematics representing the MUC5AC central exon from the AfrAm subject and three white subjects with cystic fibrosis produced by de novo assembly were compared, as a group and individually, with the central exon model (22), and the results are shown. All four subjects in this study have larger PTS-TR1 (extra 1.9 kb, blue bar) and PTS-TR4 (extra 216 bp, green bar) regions than the draft genome model sequence (Figure 1B). The increase in the PTS-TR lengths, when compared with the previously known model, more specifically shown in the central exon model of this figure, effectively link the previously available genomic fragments (Figure 1) into one unit and complete the central exon sequence. Indels are shown as purple bars, pink bars, or black stars. The three classes of CysD domains are shown (colored ovals). The duplication of the CysD domains in CauCF3, as compared with other subjects, is indicated by CysD4a, CysD5a, and CysD6a. HinfI sites are shown by red arrows, and the small and large HinfI fragment lengths identified by Southern blots are shown in red text, which are very similar to the in silico sizes (blue text).
De Novo Sequence Assembly from Three White Subjects with CF Produced Consensus Contigs Representing Two Alleles for the Large Central Exon from Each Subject
Alignment of the sequences obtained from the PCR amplicons in the central exon (exon 31) region of three white subjects (CauCF1, CauCF2, and CauCF3) confirmed their identity as MUC5AC sequences (data not shown). MIRAv3.4 assembler software successfully produced two consensus alleles for each CauCF individual when the filtering and processing criteria described in the methods were applied (Figures E3 and E4; Table E3). These consensus contigs were obtained even under circumstances when the dominant PCR product, before library preparation, was not full length (Figure E3A). Despite its highly repetitive nature, the central exon sequences obtained from de novo assembly from all three CauCF individuals were considered to be highly reliable for the following reasons: (1) the sequences were verified to contain a long open reading frame (ORF) encoding the PTS-TRs and CysD domains, consistent with previously published results and available protein sequences (22) (Figures 3 and E5); (2) the predicted HinfI fragments sizes produced from the de novo assembly closely matched previously obtained Southern blot data (Table E1), with slight differences likely reflecting the inability to precisely determine the size of fragments on Southern blots, especially in this large size range (20); and (3) the presence of canonical exon 3′-splice junction with GT donor sequence for the large central exon as predicted if the ORF would be continued via splicing to exon 32 (data not shown).
The DNA and protein sequence accession identification numbers, deposited into NCBI, are listed in Table 1. The lengths of the central exon in the four subjects were 10.25, 10.25, 10.5, and 12.25 kb for the AfrAm, CauCF1, CauCF2, and CauCF3 subjects, respectively. Thus, the extremely large central exon of MUC5AC joins the ranks of other mucins (i.e., MUC16 > 21; MUC4 12.7; MUC12 14.9; MUC17 12.2; and MUC5B 10.9 kb) and large proteins, such as titin (TTN > 17 kb), as one of the longest exons in the human genome (36). For illustrative purposes, we have defined at least 26 domains or subdomains (including unique sequences) for the AfrAm, CauCF1, and CauCF2 subjects and 34 for CauCF3 (Table E5), all of which likely contribute to genetic diversity, evolutionary history, and/or protein function. Although the PTS-TR region clearly consists of repeated units of 24 nucleotides (eight amino acids), the majority of PTS-TRs are not identical, with considerable sequence diversity. However, there are commonly seen identical repeat units among the different PTS-TR regions (TR1–TR4) (Tables E5 and E6).
Table 1:
DNA and Protein Accession Identification Numbers in the National Center for Biotechnology Information
| Contig Name | DNA ID | Protein ID |
|---|---|---|
| AfrAm | KC800812 | AGR44427 |
| CauCF1 c1* | KC821598 | AGI42861 |
| CauCF1 c2* | KC821599 | AGI42862 |
| CauCF2 c1* | KC821600 | AGI42863 |
| CauCF2 c2* | KC821601 | AGI42864 |
| CauCF3 c1* | KC821602 | AGI42865 |
| CauCF3 c2* | KC821603 | AGI42866 |
Definition of abbreviations: AfrAm, African American; CauCF, white subject with cystic fibrosis.
Alleles produced and sequenced for each CauCF subject are shown (c1 and c2).
Genetic Variants in the Large Central Exon
Successful generation of de novo contigs, coupled with alignments, allowed for the identification of the genetic variants in the central exon across the four individuals (Figure 3; details provided in Figure E5). Several types of genetic variants were identified, including SNPs, PTS-TR insertions/deletions (indels), and segmental duplications. The sequence structure of the AfrAm subject was highly similar to CauCF1. The AfrAm and CauCF1 subjects differed from the CauCF2 subject by the insertion of eight and three 24-bp repeat units in PTS-TR1 and PTS-TR4, respectively. Although the AfrAm and CauCF1 and -2 subjects contained nine CysD domains, similar to the central exon model, the CauCF3 subject contained two segmental duplications that introduced three additional CysD domains: an additional Class II domain (labeled CysD4a) and two additional Class III domains (labeled CysD5a and CysD6a) (Figures 3 and E6). The first segmental duplication giving rise to CysD4a and CysD5a likely arose from an extra duplication of one 5′ region–conserved duplicon containing one Class II and one Class III domain accompanied by flanking interspersed sequences. The second segmental duplication likely arose from a duplication occurring after PTS-TR2 that included CysD6a and the PTS-TRdup (Figure 3).
Confirmation of an Extra 5′ Region–Conserved Duplicon in CauCF3
Because the MUC5AC structural variations presented in CauCF3 (Figure 3) have not been reported previously, we sought to confirm their presence through additional testing (Figure 4). First, we were able to obtain PCR fragments of the expected size consistent with the duplication of the 5′ region–conserved duplicon of CauCF3 (Figure 4A). End sequencing of the PCR product isolated from the gel confirmed the fragment’s identity at the correct position in MUC5AC (data not shown). Direct SMRT sequencing of unamplified, enriched, BbsI-digested genomic DNA fragments from CauCF3 (see Materials and Methods and the online supplement) produced sequence reads spanning the central exon. Over a dozen individual sequences spanned the entire 5′ region of the central exon, and although each contained the expected sequencing errors from the PacBio platform, they all were shown to contain seven CysD domains as predicted from the de novo assembly of the SMRT Sequencing reads previously produced from the PCR amplicons (Figure 4B). Several other sequences aligned across the region containing CysD6a, confirming the presence of an additional duplication in that region. Moreover, when genomic sequence reads mapping to the entire MUC5AC gene model were used for de novo assembly, they produced a contig that reflected the structure of the 5′ region of the central exon that was determined from sequencing of the PCR amplicon (three Class II and three Class III CysD domains) (Figure 4C). Sequencing of genomic DNA, which was not subjected to amplification and therefore was not biased by potential PCR amplification related artifacts, conclusively demonstrates the existence of these duplication events in the individual being studied. The data presented in Figure 4 provide the necessary validation of the integrity of the de novo assembled contigs for CauCF3 and, importantly, they confirm the reliability of the overall strategy.
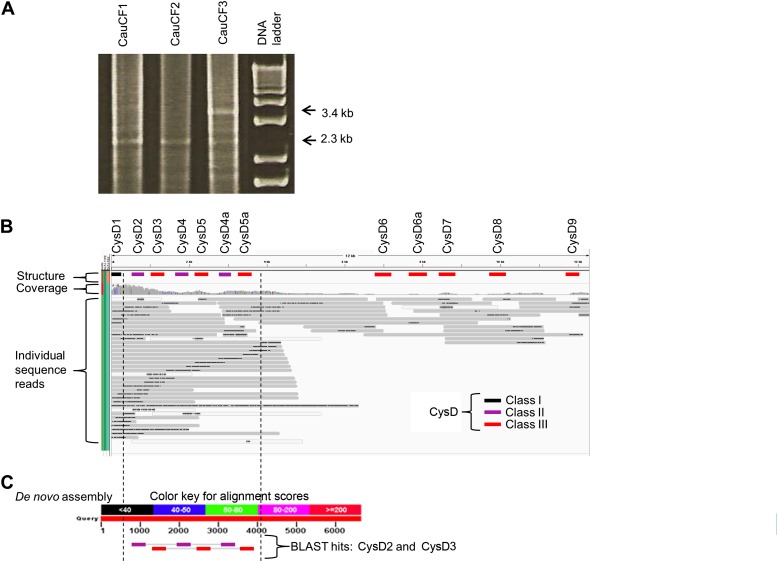
Figure 4.
Confirmation of structural variation in subject CauCF3 predicted from de novo assembly. (A) PCR primers located in CysD1 and PTS-TR1 were used to amplify genomic DNA. The expected increase in size (from 2.3 to 3.4 kb), consistent with the addition of a 5′ region conserved duplicon, was observed in CauCF3. (B) Long sequence reads from BbsI-enriched genomic DNA (see Materials and Methods) from CauCF3 were mapped to the de novo consensus contig 1 from CauCF3 using Burrows-Wheeler Aligner with custom parameters (49) and were visualized in the Integrated Genomics Viewer software (50, 51). The black, purple, and red bars (Class I, Class II, and Class III, respectively) indicate location of the 12 CysD domains on the de novo contig produced from the PCR amplification of the region from CauCF3. The gray horizontal bars show the individual BbsI-enriched genomic DNA sequence reads mapped to the contig. The black bars, within the gray sequence reads, indicate regions within the individual sequences that have a high indel content (consequence of SMRT Sequencing errors). Several individual sequence reads show the seven (CysD1–CysD5a) predicted CysD domains at the 5′ region. Several other individual sequence reads demonstrate the duplication of CysD6a in the 3′ region. (C) Although the coverage was not sufficient to produce a de novo assembly free of sequence errors across the entire region, six de novo assembled contigs were generated from the low-coverage genomic DNA reads that mapped to the previously determined MUC5AC gene model (not shown). BLAST of CysD domain sequences to one of these contigs demonstrates the arrangement of CysD domains expected from CysD4a-CysD5a duplication in CauCF3 contigs (purple: CysD2 aligning to Class II; red: CysD3 aligning to Class III). The dotted vertical lines mark the region in (B) and (C) for illustrative purposes.
Strong Linkage Disequilibrium between PTS-TR1 Genetic Variation and SNP rs28514396
Previous evaluation of the linkage disequilibrium (LD) structure of SNPs across the MUC5AC gene has demonstrated the existence of a few defined SNP “bins,” where bins are defined as SNPs that are in LD with each other (17). SNPs representative of these bins were previously shown to be in strong LD with PTS-TR allele–size modes (20). The insertion observed in PTS-TR1 of CauCF2 increases the size of a HinfI fragment from 0.97 to 1.16 kb (Figure 3, in silico blue values). Southern blots on a selected set of 276 white subjects with CF were conducted (data not shown) to test whether or not this PTS-TR1 HinfI-length polymorphism was in LD with nearby SNPs. The Southern blots showed the expected bimodal distribution of 0.95 or 1.15 kb (Southern blot red values are similar to the in silico predicted blue values in Figure 3) in the population. The analysis found that the 1.15-kb HinfI fragment including the insertion was in very strong LD with the minor allele of SNP rs28514396, which is approximately 7 kb downstream from the 3′ end of the central exon (Fisher’s test P value < 2.2 × 10−16) (Table 2). Thus, rs28514396 can be used to tag this PTS-TR1 genetic variant.
Table 2:
Association of Single Nucleotide Polymorphism rs28514396 Genotype with the 0.95/1.15kb HinfI Fragment*
| HinfI Genotype | rs28514396 Genotype |
||
|---|---|---|---|
| AA | AG | GG | |
| 0.95/0.95 kb | 93 (98)† | 2 (2) | 0 (0) |
| 0.95/1.15 kb | 2 (2) | 126 (98) | 0 (0) |
| 1.15/1.15 kb | 0 (0) | 3 (6) | 50 (94) |
Detected by Southern blot in subjects with cystic fibrosis from the University of North Carolina/Case Western Reserve University Genetic Modifiers Study (N = 276).
Values are n (%). Fisher’s test P value < 2.2 × 10−16.
Discussion
De Novo Assembly Provides a Reference Genome for Human MUC5AC and Identifies Genetic Variation in the Large, Repetitive Central Exon
Using high-fidelity, long PCR coupled with SMRT sequencing and state-of-the-art assembly algorithms, this work reports the complete sequence of the human MUC5AC genomic region from an AfrAm individual and the sequences of the large central repetitive region from three white subjects with CF. These sequences relied entirely on de novo assembly of high-coverage long sequence reads. De novo assembly had previously been impossible using sequences of shorter read lengths from Sanger and second-generation sequence technologies in this type of genomic gap. Significantly, the sequence (1) spanned the current gap in the GRCh37/hg19 genome reference (Figures 1 and 2), (2) solidified the intron–exon structure (Figure 2), (3) identified genetic variations associated with HinfI PTS-TR fragment length polymorphisms assessed by Southern blots (20) (Figure 3, data not shown for AfrAm subject), (4) identified additional significant structural variation generated by segmental duplications of CysD domains (Figure 3), (5) characterized the major functional domains coupled to genetic variation and genome stability (Figure E5; Table E5), (6) confirmed the integrity of the SMRT sequencing strategy by identification of segmental duplications with direct sequencing of genomic DNA (Figure 4), and (7) identified strong LD of a specific indel in the PTS-TR1 with an intronic SNP (Table 2).
Successful De Novo Assembly Using Long SMRT Sequencing Reads through the Highly Repetitive Central Exon
The SMRT technology provides long sequence reads from single DNA templates (37). The C2 polymerase and C2 sequencing chemistry used for this work generated average read lengths of 2.5 kb and approximately 5% of reads approach 15 kb. These features were likely critical for the success of the de novo assembly of the MUC5AC central exon in this report. On the other hand, the SMRT technology is reported to produce sequences with an error rate ranging from 10 to 15% (38), which is higher than error rates observed in second-generation sequencing approaches. Mean error rates in the data reported here were found to be around 10% (Figure E7). Because the errors are randomly distributed, they could be effectively dealt with bioinformatically by obtaining high coverage coupled with consensus error correction, ultimately leading to an estimated final error rate of less than 1/10,000 for the sequences reported here. We estimate that greater than 30× coverage for each base is desired when using SMRT sequencing for similar projects.
The methods used could achieve base level resolution in identifying SNPs between diploid alleles from each subject by nearly 50% representation of each allele among the contigs. In addition, independent sequencing of unamplified genomic DNA fragments supported the results obtained when PCR amplicons were sequenced. In conclusion, our results demonstrated successful application of the technology to produce de novo assembled contigs in difficult regions as long as sufficient long-read coverage is obtained to allow consensus correction. Optimizing the efficiency of amplification in the PCR, with special attention paid to DNA quality and environment, including gel purification of expected amplicons, should be considered in future studies.
Characteristics of the MUC5AC Gene and Central Exon Genetic Variants
The MUC5AC sequences produced in this report confirmed the original hypothesis that the MUC5AC gap contained a single, unusually large, exon encoding the previously described PTS-TR sequences and CysD domains (22, 23). The final consensus contigs for the central repetitive region translated into the predicted long ORFs in CauCF1, -2, and -3 (Figure E5) and in the AfrAm subject after additional sequence processing (details are provided in the online supplement). In addition, the assemblies produced identified diploid features of the alleles of CauCF1, -2, and -3 (Figure E4).
Although indels and SNPs in the central exon among the four individuals were identified, the most notable genetic variants detected were two separate segmental duplications (39) in CauCF3, the subject that carries the longest HinfI fragment (7.0 kb) (Figure 3, red arrows). A duplication across two CysD domains from Class II and Class III (including the flanking PTS-rich regions) occurred in the 5′ non–PTS-TR region of the exon in CauCF3 (Figure 3). This structural variant was confirmed by the expected increase in PCR-fragment size in the region and by SMRT sequencing of moderately enriched genomic DNA (Figure 4). Another duplication in the 3′ region, consisting of CysD6a and PTS-TRdup (Figure 3), was found in CauCF3, which primarily explains the increase in HinfI fragment size observed on Southern blots.
The occurrence of MUC5AC genetic variation in the central exon is likely driven by genome instability (40) due to high similarity in the PTS-TR sequences and among the CysD domains. MUC5AC resides in the subtelomeric region adjacent to the end of the short arm of chromosome 11 (41) and specifically is found in the middle of an approximately 300 kb recombination-rich region among other secreted mucins (from telomere to centromere, MUC6-MUC2-MUC5AC-MUC5B). Genes in subtelomeric regions are often more genetically dynamic compared with genes in other regions and are thought to be under strong evolutionary pressure influenced by host–environmental interactions (42). Although the data are limited to four individuals, the Class I, II, and III CysD domains seem to have different evolutionary outcomes. For example, Class III CysD domains and flanking unique sequences have expanded within the PTS-TR region, likely through PTS-TR–mediated duplications, and the 5′ region conserved duplicon (composed of Class II and Class III domains and the flanking regions coding for PTS-rich domains) has been replicated two (AfrAm, CauCF1, and CauCF2) or three (CauCF3) times. However, no similar duplication for the Class I domain was observed (Figure 3). Furthermore, the sequences immediately flanking the 5′ end of PTS-TR1 through the 3′ end of PTS-TR4 (CysD5a and CysD9 in CauCF3; CysD5 and CysD9 in all other subjects) are different from the sequences immediately flanking the CysD6 through CysD8 domains (as shown in the comparison for CauCF3 in Figure E6C; data not shown for other subjects), a feature that likely helps to prevent deletion of the entire PTS-TR region during structural change.
There are other interesting features in the MUC5AC PTS-TR1 through PTS-TR4 regions. First, all of the polymorphic features (segmental duplications, SNPs, and PTS-TR indels) in this region occur in the context of one large exon where the integrity of the ORF must be maintained. Second, the only exception to the 24-bp nucleotide repeat unit is the 14th repeat of PTS-TR3, which is only 21 bp (Table E5, red asterisk). Although these data clearly support the concept of the eight-amino-acid “repeat unit” in MUC5AC, these individual PTS-TRs are quite variable, with no one sequence accounting for more than 21% of the total within the PTS-TR regions (Table E6 and data not shown). Despite this variation, the proline-threonine-serine rich nature of the PTS-TRs is maintained (sustaining the potential for O-linked glycosylation), and negative selection against glycosylation-disruptive mutations most likely contributes to this phenomenon.
Intronic SNPs Can Tag PTS-TR Length Variation in the MUC5AC Central Exon
Subjects CauCF1, -2, and -3 were selected to represent common haplotypes in the white CF population, and further details of genetic variation among the subjects in the central repetitive region can now be provided. CauCF1 contains the most common PTS-TR allele (the 6.3-kb allele as defined in Reference 20), and it is found on chromosomes that contain primarily major alleles of available SNPs across the entire gene region. This supports the use of the structure outlined for CauCF1, which is the same as the AfrAm subject, as a reasonable “working reference sequence” to which other identified alleles can be compared. Previous work on the LD structure across the MUC5AC gene has demonstrated that the minor alleles of SNPs in the MUC5AC region can be divided into “bins” based on their LD (17). The insertion of eight repeat units in PTS-TR1 of CauCF2 and CauCF3 generates an increase in the HinfI fragment size, and we speculated that this specific insertion may be tagged with SNPs included on our previous GWAS panels (43). This speculation was supported by Southern blot data conducted on 276 subjects that specifically analyzed the smaller HinfI fragment size. The HinfI fragment size polymorphism was found to be in strong LD with nearby SNPs; specifically it was tagged by rs28514396 (Table 2). In previous work, SNPs rs35705491, rs3087562, and rs13380 were shown to be in strong LD with the HinfI PTS-TR allele size mode carried on CauCF3 (20), which we now know differs from smaller alleles in part by the duplication in the 3′ region between PTS-TR2 and CysD7 followed by the deletion of the first six PTS-TR units (Figure 3, black star in PTS-TRdup). Moreover, we noticed two deletions of one and four PTS-TR units in the PTS-TR4 region of this subject (Figure 3, black stars in PTS-TR4). These genetic variants, and other similar types of variations, likely account for the larger mode of the bimodal distribution in MUC5AC PTS-TR allele sizes previously demonstrated (20, 25). We do not have enough data to place the extra 5′ region conserved duplicon and the PTS-TR variants on specific haplotypes, but further efforts with additional subjects should provide this information. More diversity and informative features will surely be discovered as more individuals are sequenced and analyzed.
Functional Consequences of CysD Domain Copy Number Variation in the Central Exon
The CysD domains, which were shown by this work to vary in number between individuals, are highly conserved across many levels of eukaryotic taxonomies (42, 44). Although the 10 cysteines within these domains are thought to be involved in intramolecular disulfide bonds (45), the specific function of these domains has not been established. CysD domains are common to secreted mucins, including all secreted mucin genes found on human chromosome 11 (MUC2, MUC5AC, and MUC5B) (23, 42). Although they vary in number among these mucins, they are consistently found lying adjacent to or scattered within the PTS-TR regions, where they are reported to be less glycosylated compared with their highly glycosylated PTS-TR neighbors (46). Most of the available literature on the function of the CysD domains comes from work on the CysD domain of MUC2, where it has been shown that recombinant domains form noncovalent dimers, probably via hydrophobic interactions, suggesting involvement of the domains in cross-linking of mucus gels after protein secretion interactions (45, 47). CysD domains from MUC5AC have also been reported to form noncovalent, pH-independent dimers (47), although another paper reported predominantly monomers (48). In several lower organisms, the CysD domains in the mucin genes are encoded in separate exons, a mechanism that is thought to increase polymorphic variants in the mRNA via alternative splicing (23, 42). Given the available information on these domains, it is likely that variation in total number (as seen in this manuscript) will alter the density of mucus gel crosslinks, altering the mesh size, permeability, or binding properties of mucus and potentially influencing disease pathophysiology.
Conclusions and Relevance
We have used PacBio’s SMRT sequencing to resolve the sequence gap in human MUC5AC and to identify genetic variations, including segmental duplications, in the large central exon. Future studies to annotate these structural variations and other genetic variants in a larger population will set the stage for focused genetic association studies in a variety of disease cohorts where MUC5AC is predicted to have a key role. These diseases include common respiratory conditions (CF, COPD, asthma, and idiopathic pulmonary fibrosis), inflammatory bowel disease, susceptibility to Heliocobacter (the most common infection worldwide), parasitic infections (such as nematode infections) (7), and tumor development. The strategies used in this study may also be relevant for studies involving other genetic regions with highly repetitive sequences and for filling other genomic gaps that still exist in reference genomes.
Acknowledgments
Acknowledgments
The authors thank Quinn Langdon and Piotr A. Mieczkowski of the University of North Carolina High Throughput Sequencing Center for their assistance in SMRT Sequencing; Evan Eichler, Karyn Meltz Steinberg, Mehmet Kesimer, and Rui Cao for resources and technical assistance; Syanne Olson for editorial assistance and Beth Godwin for administrative assistance; Fred A. Wright, Fei Zou, and Ann Harris for insightful discussions; and the subjects who contributed specimens, the Genetic Modifier Study group in the North American CF Genetic Modifier Consortium, and all the study collaborators and participant CF sites.
Footnotes
This work was supported by United States Cystic Fibrosis Foundation grants R026-CR11 (W.K.O.), STONE08G0 (J.R.S.), and KNOWLES00A0 (M.R.K.); by National Institutes of Health grants RR00046 (M.R.K.), NIDDK P30 DK 065,988 (W.K.O.), NHLBI P01 HL 110,873 (W.K.O.), NHLBI P01 HL 68,890 (M.R.K.), and NHLBI R01 HL095396 (M.R.K.); by the University of North Carolina University Cancer Research Fund (C.J.); and by the Duke Center for Pediatric Lung Disease (J.A.V.).
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2013-0235OC on September 6, 2013
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Rose MC, Voynow JA. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol Rev. 2006;86:245–278. doi: 10.1152/physrev.00010.2005. [DOI] [PubMed] [Google Scholar]
- 2.Hansson GC. Role of mucus layers in gut infection and inflammation. Curr Opin Microbiol. 2012;15:57–62. doi: 10.1016/j.mib.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodríguez-Piñeiro AM, Bergström JH, Ermund A, Gustafsson JK, Schuette A, Johansson ME, Hansson GC. Gastrointestinal mucus proteome reveals Muc2 and Muc5ac accompanied by a set of core proteins: 2. Studies of mucus in mouse stomach, small intestine, and colon. Am J Physiol Gastrointest Liver Physiol. 2013;305:G348–G356. doi: 10.1152/ajpgi.00047.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008;1:183–197. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stonebraker JR, Wagner D, Lefensty RW, Burns K, Gendler SJ, Bergelson JM, Boucher RC, O’Neal WK, Pickles RJ. Glycocalyx restricts adenoviral vector access to apical receptors expressed on respiratory epithelium in vitro and in vivo: role for tethered mucins as barriers to lumenal infection. J Virol. 2004;78:13755–13768. doi: 10.1128/JVI.78.24.13755-13768.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Button B, Cai LH, Ehre C, Kesimer M, Hill DB, Sheehan JK, Boucher RC, Rubinstein M. A periciliary brush promotes the lung health by separating the mucus layer from airway epithelia. Science. 2012;337:937–941. doi: 10.1126/science.1223012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hasnain SZ, Evans CM, Roy M, Gallagher AL, Kindrachuk KN, Barron L, Dickey BF, Wilson MS, Wynn TA, Grencis RK, et al. Muc5ac: a critical component mediating the rejection of enteric nematodes. J Exp Med. 2011;208:893–900. doi: 10.1084/jem.20102057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koeppen M, McNamee EN, Brodsky KS, Aherne CM, Faigle M, Downey GP, Colgan SP, Evans CM, Schwartz DA, Eltzschig HK. Detrimental role of the airway mucin Muc5ac during ventilator-induced lung injury. Mucosal Immunol. 2013;6:762–775. doi: 10.1038/mi.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boltin D, Perets TT, Vilkin A, Niv Y. Mucin function in inflammatory bowel disease: an update. J Clin Gastroenterol. 2013;47:106–111. doi: 10.1097/MCG.0b013e3182688e73. [DOI] [PubMed] [Google Scholar]
- 10.Larsson JM, Karlsson H, Crespo JG, Johansson ME, Eklund L, Sjövall H, Hansson GC. Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm Bowel Dis. 2011;17:2299–2307. doi: 10.1002/ibd.21625. [DOI] [PubMed] [Google Scholar]
- 11.Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, Taupin D, Thornton DJ, Png CW, Crockford TL, Cornall RJ, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. 2008;5:e54. doi: 10.1371/journal.pmed.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, Büller HA, Dekker J, Van Seuningen I, Renes IB, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi M, Lee H, Nakayama J, Fukuda M. Roles of gastric mucin-type O-glycans in the pathogenesis of Helicobacter pylori infection. Glycobiology. 2009;19:453–461. doi: 10.1093/glycob/cwp004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niv Y, Boltin D. Secreted and membrane-bound mucins and idiopathic peptic ulcer disease. Digestion. 2012;86:258–263. doi: 10.1159/000341423. [DOI] [PubMed] [Google Scholar]
- 15.Lindén S, Mahdavi J, Hedenbro J, Borén T, Carlstedt I. Effects of pH on Helicobacter pylori binding to human gastric mucins: identification of binding to non-MUC5AC mucins. Biochem J. 2004;384:263–270. doi: 10.1042/BJ20040402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirby A, Gnirke A, Jaffe DB, Barešová V, Pochet N, Blumenstiel B, Ye C, Aird D, Stevens C, Robinson JT, et al. Mutations causing medullary cystic kidney disease type 1 lie in a large VNTR in MUC1 missed by massively parallel sequencing. Nat Genet. 2013;45:299–303. doi: 10.1038/ng.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, Loyd JE, Fingerlin TE, Zhang W, Gudmundsson G, Groshong SD, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. 2011;364:1503–1512. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stock CJ, Sato H, Fonseca C, Banya WA, Molyneaux PL, Adamali H, Russell AM, Denton CP, Abraham DJ, Hansell DM, et al. Mucin 5B promoter polymorphism is associated with idiopathic pulmonary fibrosis but not with development of lung fibrosis in systemic sclerosis or sarcoidosis. Thorax. 2013;68:436–441. doi: 10.1136/thoraxjnl-2012-201786. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Noth I, Garcia JG, Kaminski N. A variant in the promoter of MUC5B and idiopathic pulmonary fibrosis. N Engl J Med. 2011;364:1576–1577. doi: 10.1056/NEJMc1013504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo X, Pace RG, Stonebraker JR, Commander CW, Dang AT, Drumm ML, Harris A, Zou F, Swallow DM, Wright FA, et al. Mucin variable number tandem repeat polymorphisms and severity of cystic fibrosis lung disease: significant association with MUC5AC. PLoS ONE. 2011;6:e25452. doi: 10.1371/journal.pone.0025452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desseyn JL, Aubert JP, Porchet N, Laine A. Evolution of the large secreted gel-forming mucins. Mol Biol Evol. 2000;17:1175–1184. doi: 10.1093/oxfordjournals.molbev.a026400. [DOI] [PubMed] [Google Scholar]
- 22.Escande F, Aubert JP, Porchet N, Buisine MP. Human mucin gene MUC5AC: organization of its 5′-region and central repetitive region. Biochem J. 2001;358:763–772. doi: 10.1042/0264-6021:3580763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang T, Hansson GC, Samuelsson T. Gel-forming mucins appeared early in metazoan evolution. Proc Natl Acad Sci USA. 2007;104:16209–16214. doi: 10.1073/pnas.0705984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thornton DJ, Rousseau K, McGuckin MA. Structure and function of the polymeric mucins in airways mucus. Annu Rev Physiol. 2008;70:459–486. doi: 10.1146/annurev.physiol.70.113006.100702. [DOI] [PubMed] [Google Scholar]
- 25.Vinall LE, Hill AS, Pigny P, Pratt WS, Toribara N, Gum JR, Kim YS, Porchet N, Aubert JP, Swallow DM. Variable number tandem repeat polymorphism of the mucin genes located in the complex on 11p15.5. Hum Genet. 1998;102:357–366. doi: 10.1007/s004390050705. [DOI] [PubMed] [Google Scholar]
- 26.Fowler J, Vinall L, Swallow D. Polymorphism of the human muc genes. Front Biosci. 2001;6:D1207–D1215. doi: 10.2741/A674. [DOI] [PubMed] [Google Scholar]
- 27.Rousseau K, Swallow DM. Mucin methods: genes encoding mucins and their genetic variation with a focus on gel-forming mucins. Methods Mol Biol. 2012;842:1–26. doi: 10.1007/978-1-61779-513-8_1. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Davenport KW, Gu W, Daligault HE, Munk AC, Tashima H, Reitenga K, Green LD, Han CS. Improving genome assemblies by sequencing PCR products with PacBio. Biotechniques. 2012;53:61–62. doi: 10.2144/0000113891. [DOI] [PubMed] [Google Scholar]
- 29.English AC, Richards S, Han Y, Wang M, Vee V, Qu J, Qin X, Muzny DM, Reid JG, Worley KC, et al. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS ONE. 2012;7:e47768. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng S, Byrd AS, Fischer BM, Grover AR, Ghio AJ, Voynow JA. Regulation of MUC5AC expression by NAD(P)H:quinone oxidoreductase 1. Free Radic Biol Med. 2007;42:1398–1408. doi: 10.1016/j.freeradbiomed.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drumm ML, Konstan MW, Schluchter MD, Handler A, Pace R, Zou F, Zariwala M, Fargo D, Xu A, Dunn JM, et al. Gene Modifier Study Group. Genetic modifiers of lung disease in cystic fibrosis. N Engl J Med. 2005;353:1443–1453. doi: 10.1056/NEJMoa051469. [DOI] [PubMed] [Google Scholar]
- 32.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bikandi J, San Millán R, Rementeria A, Garaizar J. In silico analysis of complete bacterial genomes: PCR, AFLP-PCR and endonuclease restriction. Bioinformatics. 2004;20:798–799. doi: 10.1093/bioinformatics/btg491. [DOI] [PubMed] [Google Scholar]
- 34.Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14:1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meezaman D, Charles P, Daskal E, Polymeropoulos MH, Martin BM, Rose MC. Cloning and analysis of cDNA encoding a major airway glycoprotein, human tracheobronchial mucin (MUC5) J Biol Chem. 1994;269:12932–12939. [PubMed] [Google Scholar]
- 36.Bolisetty MT, Beemon KL. Splicing of internal large exons is defined by novel cis-acting sequence elements. Nucleic Acids Res. 2012;40:9244–9254. doi: 10.1093/nar/gks652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 38.Ewing B, Green P. Base-calling of automated sequencer traces using phred: II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 39.Marques-Bonet T, Girirajan S, Eichler EE. The origins and impact of primate segmental duplications. Trends Genet. 2009;25:443–454. doi: 10.1016/j.tig.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 41.Rousseau K, Byrne C, Griesinger G, Leung A, Chung A, Hill AS, Swallow DM. Allelic association and recombination hotspots in the mucin gene (MUC) complex on chromosome 11p15.5. Ann Hum Genet. 2007;71:561–569. doi: 10.1111/j.1469-1809.2007.00374.x. [DOI] [PubMed] [Google Scholar]
- 42.Desseyn JL. Mucin CYS domains are ancient and highly conserved modules that evolved in concert. Mol Phylogenet Evol. 2009;52:284–292. doi: 10.1016/j.ympev.2009.03.035. [DOI] [PubMed] [Google Scholar]
- 43.Wright FA, Strug LJ, Doshi VK, Commander CW, Blackman SM, Sun L, Berthiaume Y, Cutler D, Cojocaru A, Collaco JM, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet. 2011;43:539–546. doi: 10.1038/ng.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spada F, Steen H, Troedsson C, Kallesoe T, Spriet E, Mann M, Thompson EM. Molecular patterning of the oikoplastic epithelium of the larvacean tunicate Oikopleura dioica. J Biol Chem. 2001;276:20624–20632. doi: 10.1074/jbc.M100438200. [DOI] [PubMed] [Google Scholar]
- 45.Ambort D, van der Post S, Johansson ME, Mackenzie J, Thomsson E, Krengel U, Hansson GC. Function of the CysD domain of the gel-forming MUC2 mucin. Biochem J. 2011;436:61–70. doi: 10.1042/BJ20102066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thornton DJ, Howard M, Khan N, Sheehan JK. Identification of two glycoforms of the MUC5B mucin in human respiratory mucus: evidence for a cysteine-rich sequence repeated within the molecule. J Biol Chem. 1997;272:9561–9566. doi: 10.1074/jbc.272.14.9561. [DOI] [PubMed] [Google Scholar]
- 47.Bäckström M, Ambort D, Thomsson E, Johansson ME, Hansson GC. Increased understanding of the biochemistry and biosynthesis of MUC2 and other gel-forming mucins through the recombinant expression of their protein domains. Mol Biotechnol. 2013;54:250–256. doi: 10.1007/s12033-012-9562-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perez-Vilar J, Randell SH, Boucher RC. C-Mannosylation of MUC5AC and MUC5B Cys subdomains. Glycobiology. 2004;14:325–337. doi: 10.1093/glycob/cwh041. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]