Abstract
The polarity of the conducting airway epithelium is responsible for its directional secretion. This is an essential characteristic of lung integrity and function that dictates interactions between the external environment (apical) and subepithelial structures (basolateral). Defining the directional secretomes in the in vitro human bronchial epithelial (HBE) differentiated model could bring valuable insights into lung biology and pulmonary diseases. Normal primary HBE cells (n = 3) were differentiated into respiratory tract epithelium. Apical and basolateral secretions (24 h) were processed for proteome profiling and pathway analysis. A total of 243 proteins were identified in secretions from all HBE cultures combined. Of these, 51% were classified as secreted proteins, including true secreted proteins (36%) and exosomal proteins (15%). Close examination revealed consistent secretion of 69 apical proteins and 13 basolateral proteins and differential secretion of 25 proteins across all donors. Expression of Annexin A4 in apical secretions and Desmoglein-2 in basolateral secretions was validated using Western blot or ELISA in triplicate independent experiments. To the best of our knowledge, this is the first study defining apical and basolateral secretomes in the in vitro differentiated HBE model. The data demonstrate that epithelial polarity directs protein secretion with different patterns of biological processes to the apical and basolateral surfaces that are consistent with normal bronchial epithelium homeostatic functions. Applying this in vitro directional secretome model to lung diseases may elucidate their molecular pathophysiology and help define potential therapeutic targets.
Keywords: bronchial epithelium, secreted proteins, secretome, proteome
Clinical Relevance
Developing the directional secretomes in the in vitro human bronchial epithelial differentiated model could bring valuable insights into lung biology and pulmonary diseases. Applying this in vitro directional secretome model to lung diseases may elucidate their molecular pathophysiology and help define potential therapeutic targets.
Epithelial cells line the mammalian respiratory, gastrointestinal, and reproductive tracts and form highly ordered polarized epithelia that have apical and basolateral compartments with unique functions (1, 2). The apical surfaces provide a primary protective barrier against pathogens and environmental insults. In general, apical receptors function as sensors to foreign pathogens and allergens, which then activate signaling pathways with various biochemical and biological consequences in the apical and/or basolateral compartments (3). Signaling activated by interactions at apical surfaces results in rapid secretion of chemokines and cytokines to provide early warning signals to the immune system, followed by secretion of innate immune defense responders. Signaling activated at apical surfaces can also trigger basolateral secretion, which can then propagate signals to cells in the underlying stroma (4, 5). In addition, polarized sorting of receptors (e.g., P2Y and ion channels) to the apical and/or basolateral surfaces regulates electrolyte transport and secretion across various epithelial tissues as specific receptors are functionally coupled to different signaling pathways (6). Thus, epithelial polarity is important for maintaining homeostasis within tissues, and polarized protein secretion may provide information on these mechanisms.
Polarity is especially important in the conducting airway epithelium, which is now recognized as a key element that plays an important role in responding to inflammation and remodeling in lung diseases (7). The apical airway epithelium responds to airborne bacteria, viruses, and environmental toxins and has been reported to affect local and systemic changes in the basal lamina (5). Secreted proteins are likely to affect interactions between the apical surfaces and airway environment, whereas basolateral secretions affect subepithelial structures, including fibroblasts, smooth muscles, and blood vessels. Remodeling of the airway epithelium occurs in many lung diseases (asthma [8], cystic fibrosis [CF] [9], and chronic obstructive airway diseases [10]) and may cause dysregulated signaling at the apical and/or basolateral compartments with subsequent clinical symptoms that may be reflected in airway secretions.
Recently, several laboratories have begun to analyze the secretomes of the airway system, including bronchoalveolar lavage fluid (BALF) and sputum, using proteomic techniques. This approach has the power to provide a global overview of differentially altered proteins between normal and diseased lungs. The protein patterns in BALF from patients with CF compared with those of healthy individuals implicated specific biological processes, including defense response and immunity, cellular proliferation and adhesion, wound repair, stress response, apoptosis, proteolysis, and complement activity in the pathophysiology of CF lung disease (11, 12). Other researchers have found changes in the sputum proteome of patients with CF that are related to clinical exacerbations (13) and response to treatment (14). Furthermore, proteomic studies of BALF of patients with asthma after allergen challenge show increased secretion of proteins associated with inflammation, eosinophilia, airway remodeling, tissue damage, and mucus production compared with normal subjects (15). However, studies of in vivo samples are typically complicated because patients may have unreported episodes of inflammation and/or changes due to clinical treatment. These factors make it difficult to delineate whether observed differences are intrinsic to the epithelium itself or secondary to the systemic response of the epithelium to external stimuli for any given disease.
An in vitro model of polarized airway epithelium can be generated from primary HBE cells that morphologically recapitulates the conducting airway epithelium of the respiratory tract (16–19). This model is devoid of inflammatory/immune response cells, thus eliminating confounding variables and providing a simple way to study epithelial specific pathways. This model maintains disease phenotypes observed in vivo when HBE cells are cultured from patients with CF (20) or individuals with asthma (21). Indeed, recent proteomic analysis of in vitro HBE secretions and lung secretions (induced by hypertonic saline exposure of healthy individuals) demonstrates that in vitro apical protein secretions are similar in identity and quantity to those found in in vivo samples (22). Although these data suggest that applying proteomic techniques to in vitro models will be a useful approach to study disease mechanisms and provide a unique opportunity for global understanding of protein interactions in lung diseases, the secretions from each cohort were pooled, so the opportunity to interrogate the most consistent proteins secreted by each cohort was lost.
Studies of secretions from differentiated HBE cells have been limited to analysis of apical secretions with no analysis of basolateral secretions. Recently, we demonstrated that in vitro differentiated HBE cells secreted cytokines basally after wounding and that specific cytokines levels were increased in asthmatic epithelia (23). Because the interplay between apical and basolateral compartments appears to be vital to polarity and normal epithelial function, we sought to characterize the individual directional secretomes from the epithelium of three normal donors. To the best of our knowledge, no study has characterized apical and basolateral secretomes in a human in vitro airway epithelium model. The data demonstrate that secretomes of differentiated HBE cells are directional in nature and that fundamentally different processes predominate in the apical and basal compartments.
Materials and Methods
A description of cell culture donors for proteomic analysis and validation studies is provided in Table E1 in the online supplement. Information on cell culture, transepithelial electrical resistance measurements, protein identification, and quantification are provided in the online supplement.
Collection of Apical and Basolateral Conditioned Media
Differentiated HBE cultures were gently washed four times apically and basolaterally with 0.5 and 1 ml of PBS, respectively, to remove residual proteins from previous media, floating cells, and debris. One milliliter per well of protein-free bronchial epithelial basal media (BEBM) (Lonza, Walkersville, MD) was added to the basolateral compartment, and the epithelia were incubated for another 24 hours. At 23 hours, 0.5 ml of PBS was added to the apical surface for 30 minutes, and the apical secretions were collected. Incubation with 0.5 ml of PBS for 30 minutes was repeated, and samples were pooled with the previous collection. A total of 12 ml of apical and 12 ml of basolateral conditioned media per donor were thus collected from a 12-well Transwell plate.
Sample Preparation and Mass Spectrometry Analysis
Samples were filtered and concentrated on Amicon Ultra-4 3K Centrifugal Filters (Millipore, Billerica, MA). Proteins were fractionated by gel electrophoresis; bands were excised and trypsin-digested. The resulting peptides were analyzed by LC-MS/MS as previously described (24–26). Further details are provided in the online supplement.
Protein Characterization
Proteins were included for final analysis only if they were identified by at least two unique peptides in each donor culture. Protein characterization was determined using Uniprot knowledge database (http://www.uniprot.org) and evaluated for conventional secretion using SignalP version 4.1 (http://www.cbs.dtu.dk/services/SignalP) and unconventional secretion using SecretomeP version 2.0 (http://www.cbs.dtu.dk/services/SecretomeP). Proteins were classified as exosomal based on their identification in apical HBE secretions (27). Molecular and cellular functions were determined using Ingenuity Pathways Analysis; biological functions were identified using the PANTHER database (http://pantherdb.org) (28).
Statistical Analysis
Directional secretion of a given protein was assessed on the basis of relative abundance in apical versus basolateral compartments. Unique apical proteins were defined as having two or more peptides in all three cell culture apical secretions and fewer than two peptides in one or more basolateral secretions. Unique basolateral proteins were defined as having at least two peptides in all three basolateral secretions and fewer than two peptides in one or more apical secretions. Significant differential secretion of proteins found in apical and basolateral secretions was calculated using the QSpec algorithm (29) based on the following parameters: false discovery rate < 0.05 and fold change ≥ 2. Nonparametric Wilcoxon signed-rank test (SAS 9.3; SAS Institute Inc., Cary, NC) was performed to validate significant differences in apical and basolateral Desmoglein-2 expression. Statistical significance was determined based on a P value < 0.05.
Validation of Secreted Proteins
Apical and basolateral secretions collected from three independent cell cultures (not used for proteomic analysis) were used to validate the directional secretion of Annexin 4 by Western blot using mouse monoclonal antibody to Annexin A4 (Santa Cruz Biotechnology, Santa Cruz, CA) and monoclonal antimouse IgG (BioRad, Hercules, CA) and Desmoglein-2 by ELISA using a Desmoglein-2 Elisa Kit (USCN Life Science, Inc., Houston, TX). Further details are provided in the online supplement.
Results
In Vitro Airway Epithelium Characteristics
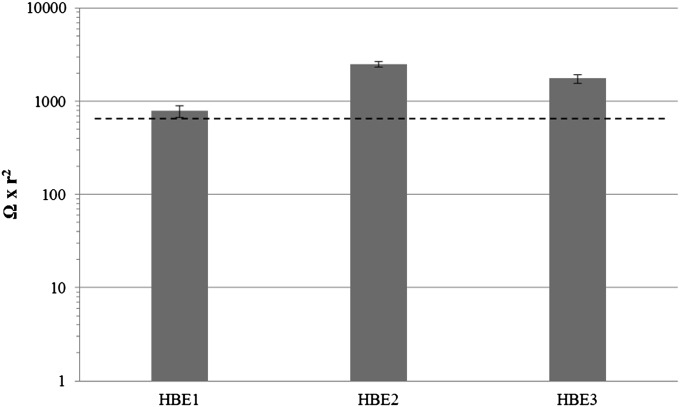
HBE cells from healthy donors differentiated to a mucociliary epithelium with apical polarization, as monitored by microscopy to observe cilia beating. Western blot analysis of apical secretions 3 days before initiating the proteomics study demonstrated the presence of MUC5B and/or MUC5AC mucins in secretions from cell cultures (data not shown). These mucins are considered markers of a polarized differentiated mucociliary airway epithelium (30). Figure 1 shows the transepithelial resistance measured between the upper and bottom chamber confirming the formation of a polarized epithelial layer of cells.
Figure 1.
Average transepithelial electrical resistance of human bronchial epithelial (HBE) cell cultures measured between Days 14 and 21 of air–liquid interface (ALI). Dashed line represents average TEER in normal HBE cultures at Days 14 and 21 of ALI as previously described (47).
Characterization of In Vitro HBE Directional Secretomes
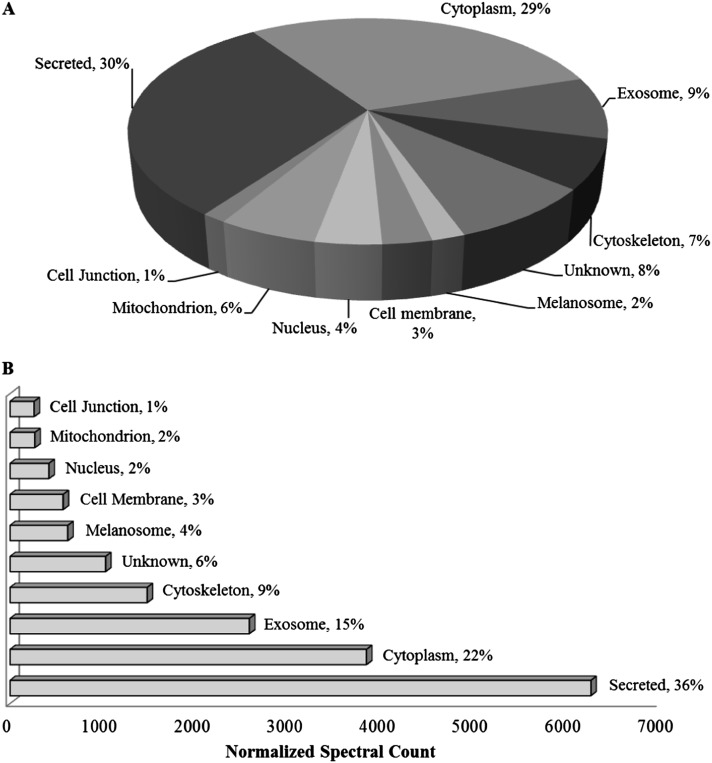
Proteome profiling of the apical and basolateral secretomes by LC-MS/MS analysis identified a total of 377 proteins (Table E2), of which 243 were detected across all cell cultures from three different donors. Ingenuity Pathways Analysis demonstrated that the major molecular and cellular functions of these 243 proteins were consistent with normal cellular development and functions (Table 1). Proteins were then classified by cellular location based on protein knowledge database and prediction tools (see Materials and Methods), with exosomal proteins classified based on a proteomics study of exosomes isolated from apical secretions of HBE cells (27). After excluding cytokeratins, 30% of the proteins were identified as true secreted, 9% as exosomal, 29% as cytosolic, and 8% with unknown subcellular localization; the remaining proteins (23%) were associated with organelles, cell junction, and cell membrane (Figure 2A). When normalized spectral counts were used to assess relative protein abundance, the majority of proteins (51.4%) were classified as secreted via classical pathway and/or exosomes (Figure 2B).
Table 1:
Ingenuity Pathways Analysis of Molecular and Cellular Functions
| Function* | P value | Number of Proteins |
|---|---|---|
| Cell death and survival | 4.55E-17–3.85E-03 | 136 |
| Cellular movement | 1.16E-11–3.62E-03 | 83 |
| Cellular growth and proliferation | 9.69E-11–4.78E-03 | 116 |
Top three functions are listed and based on the 243 proteins identified in all cell cultures.
Figure 2.
Characterization of proteins in apical and basal secretomes. Identified proteins (n = 243) in apical and basolateral secretions of all normal HBE cell cultures (n = 3) after incubating in protein-free media for 24 hours and excluding cytokeratins (n = 29). (A) Classification of protein location based on Uniprot and SecretomeP databases; exosome distinction based on previous study (27). (B) Adjusted protein characterization based on abundance determined by average normalized spectral count of detected peptides (from ProteoIQ).
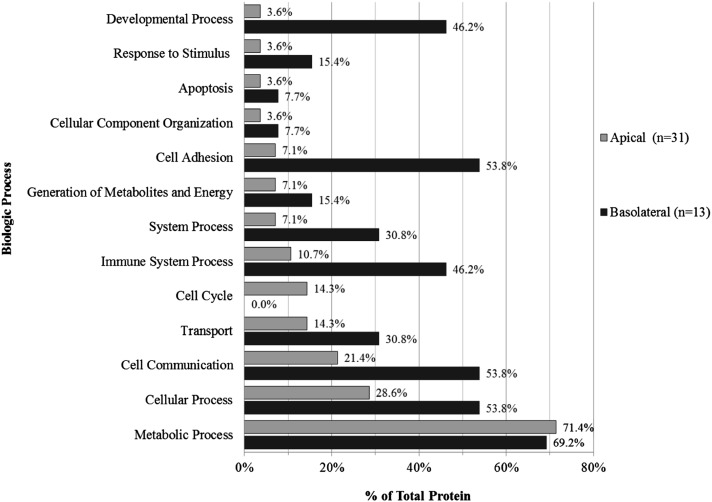
Of the 243 identified proteins, 69 were consistently detected in the apical compartment relative to the basolateral compartment. Thirty-one of these proteins had fewer than two peptides detected in the basolateral compartment of all cell cultures, of which 17 were detected only in the apical compartment in all three cultures; these are defined as “unique apical proteins” (Table 2). Additional data analysis of apically secreted proteins using protein knowledge database tools revealed that the proteins were associated with 55 biological processes defined by 12 categories, the most prevalent being metabolic process, cellular process, cellular communication, transport, and cell cycle (Figure 3).
Table 2:
Proteins Unique to the Apical Compartment
| Uniprot Identification Number | Protein* (n = 17) | Average Spectral Count† | Location‡ |
|---|---|---|---|
| O95436 | Sodium-dependent phosphate transport protein 2B | 24 | Membrane |
| P06576 | ATP synthase subunit β, mitochondrial | 19 | Mitochondion |
| P09525 | Annexin A4 | 17 | Unknown |
| P00558 | Phosphoglycerate kinase 1 | 17 | Cytoplasm |
| P08758 | Annexin A5 | 15 | Unknown |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | 13 | Cytoplasm |
| Q8WXI7 | Mucin-16§ | 13 | Secreted |
| P15941 | Mucin-1§ | 13 | Secreted |
| P00352 | Retinal dehydrogenase 1 | 12 | Cytoplasm |
| P40199 | Carcinoembryonic antigen-related cell adhesion molecule 6 | 12 | Secreted |
| P07237 | Protein disulfide-isomerase | 11 | Secreted |
| O14818 | Proteasome subunit α type-7 | 6 | Cytoplasm |
| P60660 | Myosin light polypeptide 6§ | 5 | Exosome |
| P27105 | Erythrocyte band 7 integral membrane protein§ | 5 | Exosome |
| P28072 | Proteasome subunit β type-6 | 4 | Cytoplasm |
| P49720 | Proteasome subunit β type-3 | 4 | Cytoplasm |
| P61160 | Actin-related protein 2 | 3 | Cytoplasm |
Listed proteins were present in all apical secretions and no basolateral secretions. Of the 69 proteins identified in all apical secretions but in fewer than three basal secretions, 31 proteins were associated with fewer than 2 peptides in basolateral secretions.
Spectral count calculated and normalized based on ProteoIQ; average based on values from all cell cultures.
Location based on Uniprot and SecretomeP databases.
Identified in exosomes from in vitro primary differentiated airway epithelial cell cultures in a previous study (27).
Figure 3.
Biological processes of uniquely secreted apical and basolateral proteins. Using the PANTHER database, a total of 55 processes were associated with the 31 proteins found in all apical secretions and with fewer than two peptides in basolateral secretions for all cell cultures. A total of 56 processes were identified for the 13 proteins present in all basolateral secretions and with fewer than two peptides in apical secretions.
Although 69 proteins were consistently expressed in the apical secretome, only 13 proteins were consistently expressed in the basolateral secretome (Table 3). Of these, seven had fewer than two peptides, and three (Desmoglein-2, Cathepsin B, and Carboxypeptidase A4) had zero peptides in apical secretomes. These 13 proteins were identified in 56 overlapping biological processes defined by 12 categories, including developmental process, cell adhesion, immune system process, and cell communication (Figure 3).
Table 3:
Proteins Unique to the Basolateral Compartment
| Uniprot Identification Number | Protein* (n = 13) | Average Spectral Count† | Location‡ | Molecular Weight (kD) |
|---|---|---|---|---|
| P29401 | Transketolase | 29 | Exosome | 67,878 |
| P24821 | Tenascin§ | 28 | Secreted | 240,853 |
| P31431 | Syndecan-4§ | 25 | Secreted | 21,642 |
| Q13740 | CD166 antigen§ | 16 | Secreted | 65,102 |
| O00391 | Sulfhydryl oxidase 1 | 14 | Secreted | 82,578 |
| P01034 | Cystatin-C | 11 | Secreted | 15,799 |
| P98160 | Basement membrane-specific heparan sulfate proteoglycan§ | 11 | Secreted | 468,830 |
| Q13753 | Laminin subunit γ-2 | 9 | Secreted | 130,976 |
| Q16787 | Laminin subunit α-3 | 8 | Secreted | 366,649 |
| P99999 | Cytochrome c | 7 | Mitochondrion | 11,749 |
| Q14126 | Desmoglein-2|| | 6 | Secreted | 122,294 |
| P07858 | Cathepsin B|| | 5 | Secreted | 37,822 |
| Q9UI42 | Carboxypeptidase A4|| | 5 | Secreted | 47,351 |
Listed proteins were identified in all basal secretions but fewer than three apical secretions in all three cell cultures.
Spectral count calculated and normalized based on ProteoIQ; average based on values from all cell cultures.
Location based on Uniprot Database and SecretomeP databases; exosome distinction based on previous study (27).
Present in all basal secretions and fewer than two peptides in all apical secretions.
Present in all basal secretions but no peptides in all apical secretions.
Other proteins exhibited significant directional secretion between apical and basolateral compartments (Table 4). Five of these were highly abundant in basolateral secretions, whereas the remaining 20 proteins were more highly abundant in apical secretions.
Table 4:
Differentially Expressed Proteins in Apical Versus Basolateral Compartments
| Protein | Protein* (n = 25) | Location† | Log Fold Change‡ | FDR |
|---|---|---|---|---|
| P04083 | Annexin A1 | Exosome | −4.58 | 0.00 |
| P07355 | Annexin A2 | Exosome | −3.45 | 0.00 |
| A6NMY6 | Putative annexin A2-like protein | Secreted | −3.35 | 0.00 |
| P08729 | Keratin, type II cytoskeletal 7 | Cytoplasm | −2.35 | 0.00 |
| P02768 | Serum albumin | Secreted | −2.19 | 0.00 |
| P08238 | Heat shock protein HSP 90-β | Cytoplasm | −1.88 | 0.01 |
| P07900 | Heat shock protein HSP 90-α | Cytoplasm | −1.67 | 0.00 |
| P11021 | 78 kDa glucose-regulated protein | Cytoplasm | −1.64 | 0.00 |
| P00751 | Complement factor B | Secreted | −1.47 | 0.00 |
| P30838 | Aldehyde dehydrogenase 3 | Cytoplasm | −1.22 | 0.01 |
| O43707 | α-actinin-4 | Nucleus | −1.15 | 0.00 |
| P09211 | Glutathione S-transferase P | Cytoplasm | −1.14 | 0.01 |
| P01024 | Complement C3 | Secreted | −1.12 | 0.00 |
| P19013 | Keratin, type II cytoskeletal 4 | Unknown | −1.12 | 0.00 |
| P12814 | α-actinin-1 | Cytoskeleton | −1.09 | 0.02 |
| P15924 | Desmoplakin | Cell junction | −1.07 | 0.03 |
| P68133 | Actin, α skeletal muscle | Cytoskeleton | −0.90 | 0.00 |
| P63261 | Actin, cytoplasmic 2 | Cytoskeleton | −0.83 | 0.00 |
| P60709 | Actin, cytoplasmic 1 | Cytoskeleton | −0.82 | 0.00 |
| P48594 | Serpin B4 | Cytoplasm | −0.75 | 0.04 |
| Q14525 | Myosin-4 | Cytoplasm | 0.75 | 0.00 |
| P81605 | Dermcidin | Secreted | 1.00 | 0.00 |
| P12830 | Cadherin-1 (E-cadherin) | Cell junction | 1.52 | 0.00 |
| P36955 | Pigment epithelium-derived factor | Secreted | 2.26 | 0.01 |
| O94985 | Calsyntenin-1 | Cell junction | 3.19 | 0.01 |
Definition of abbreviation: FDR, false discovery rate.
Identified in all apical and basolateral secretions with differential expression based on Student t test.
Location based on Uniprot and SecretomeP databases; exosome distinction based on previous study (27).
Significant differential expression of relative protein abundance calculated using the QSpec algorithm (29). Values are relative to the basolateral secretome. Negative value denotes increased expression in the apical secretome and positive value denotes increased expression in the basolateral secretome.
Validation of Directional Protein Secretions
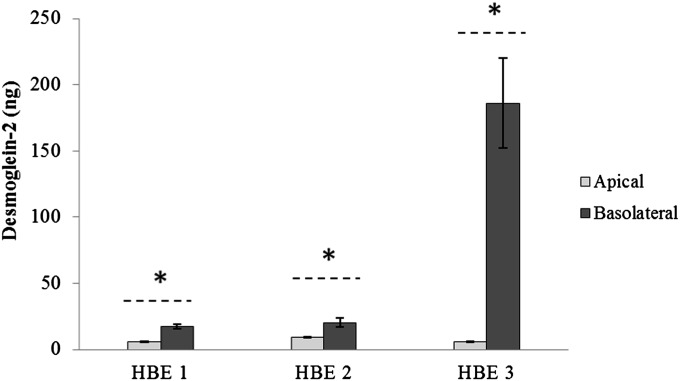
Proteomic analysis showed that 17 proteins were uniquely secreted apically (Table 2) and three basolaterally (Table 3). We interrogated the localization of one protein from each compartment (e.g., apical and basolateral) in three independent cell cultures using Western blot or ELISA. Annexin A4 was chosen to validate the proteomic findings for unique apical secretion because of its function in two highly prevalent biological processes identified by pathway analysis: cellular processing and communication. Desmoglein-2 was chosen to validate uniquely secreted basolateral protein because it is a junctional protein involved in cell adhesion—a biological process that is specific to the basolateral side of the epithelium—and because it was the most abundant basolateral protein that was absent in all three apical secretions. Western blot analysis demonstrated that Annexin A4 was present only in apical secretions and not detectable in basal secretions (Figure 4). ELISA, a more sensitive method of detecting proteins compared with Western blot analysis, was chosen to validate basolateral proteins because these proteins were in low amounts and biologic samples were limited. ELISA demonstrated that Desmoglein-2 was present at much higher levels in basolateral than apical secretions across all cell cultures (Figure 5).
Figure 4.
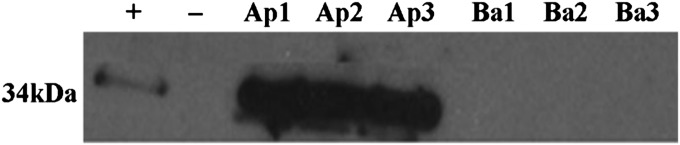
Western blot analysis of Annexin A4. A total of 20 μg of protein from apical (Ap) and basal (Ba) samples for all three cultures (1–3) were loaded. Results confirm proteomic findings of Annexin A4 only in apical, and not basolateral, secretions. Positive control (+): cell lysate; negative control (−): loading buffer.
Figure 5.
Expression of Desmoglein-2 in HBE secretions. Average protein concentration (± SEM) for each HBE cell culture was measured using ELISA. Statistical differences between amount of Desmoglein-2 in apical and basolateral secretions for each cell culture were based on Wilcoxon signed-rank test. *P < 0.05.
Discussion
Although airway epithelium communicates simultaneously with the environment via the apical membrane and secretions and with underlying tissues via basolateral secretions, only apical secretions can be obtained in vivo using minimally invasive procedures. In contrast, an in vitro approach is likely to prove informative when evaluating how apical challenges affect basolateral signaling to subepithelial structures (5). Our study demonstrates that a polarized, differentiated airway epithelium has distinct apical and basolateral secretomes.
Molecular function analysis (Ingenuity Pathways Analysis) identified basic homeostatic processes, including cellular growth, cellular proliferation, cellular movement, cell death, and cell survival in these secretomes. Bioinformatic analysis assigned unique apical and basolateral secretome proteins to biological processes that are homeostatic in nature. This analysis also identified differences and similarities in the distribution of processes between apical and basolateral secreted proteins (Figure 3). Proteins associated with metabolic processes comprised the most abundant category in both secretomes, and the distribution was similar between the two compartments (72.4% apical vs. 69.2% basolateral). The greatest distribution difference between apical and basolateral proteins demonstrated that the polarized epithelium directionally secretes unique proteins at baseline (e.g., unchallenged conditions), predictably for functional purposes. Basolateral secretomes displayed a notable increase in cell adhesion (7.1% apical vs. 53.8% basolateral), developmental process (3.6% vs. 46.2%), and immune system process (10.7% vs. 46.2%).
Overall, our analysis demonstrated that approximately 51% of the identified proteins are classified as conventional secreted proteins (e.g., secretory signal) or as unconventional secreted proteins, and 15% have previously been identified in HBE exosomes. Of the remaining proteins, 13% were associated with the epithelial membrane (cell junction, cell membrane, cytoskeleton), and 33% were classified as intracellular (nucleus, mitochondria, melanosome, and cytoplasm) proteins. Although cell leakage and cell death may be responsible for the presence of proteins classified as intracellular (despite efforts to minimize contamination by washing with PBS before secretion collection), intracellular proteins are characteristically found in secretomes (31). This raises the possibility that some of these proteins may be secreted despite their current characterization as intracellular proteins. Future studies are necessary to resolve this issue.
We evaluated Annexin A4, unique to our apical secretomes, and Desmoglein-2, unique to the basal secretomes, as markers of directional secretion by the human bronchial epithelium. Annexin A4 is vital to cellular processing and cellular communication, two of the three most significant biological processes identified for apical proteins. It is a member of an evolutionarily conserved multigene superfamily, whose members are characterized by their ability to interact with biological membranes in a calcium-dependent manner (32). Although its specific function in human airway epithelium has not been fully studied, it is known to promote membrane fusion and is involved in exocytosis (33). In rabbit trachea and alveoli, Annexin A4 is expressed only in differentiated cells (34), although its specific location appears unclear, and it has been suggested to play a role in cell proliferation (35). Annexin A2, a member of the same superfamily, is vital for intestinal epithelial differentiation and polarization, specifically affecting sorting of hydrolases and general function at the apical border (36). Although its subcellular location is classified as unknown by bioinformatic databases, Annexin A4 has been identified as an exosomal protein (37–42) in many cells (parotid gland, bladder, colon, B lymphocytes) and bodily fluids (urine). Although this suggests that Annexin A4 may play a secretory role as an exosomal protein in airway epithelium, it is not reported in a study that isolated and identified HBE apical exosome proteins (27) and has not been reported in apical secretions from HBE cultures (22, 43) or induced sputum in a comparison study (22). However, it is present in BALF of normal patients (11), and we validated apical secretion of Annexin A4 in an independent set of normal HBE secretions (n = 3). In addition, we have observed that this protein is secreted only apically in other airway cell cultures, including normal differentiated nasal epithelium and asthmatic nasal and bronchial epithelium (unpublished data). These results suggest that unidirectional expression of this protein may contribute to as yet unrecognized homeostatic functions in apical secretions of airway epithelium.
Desmoglein-2, a unique basolateral protein in our system, is a secreted intercellular desmosome junction protein that functions in cellular adhesion (44). Although Desmoglein-2 is a structural protein, it contains a 23-peptide signaling region that is responsible for its classification as a secreted protein. Cell adhesion was the second most prominent biological process identified in uniquely expressed basal proteins and was markedly higher than in apical secretions. The role of Desmoglein-2 in airway epithelium is poorly understood. However, in intestinal epithelium, another polarized system, Desmoglein-2 was found to be essential to maintaining barrier integrity and function as disrupting its secretory (extracellular) domain dissociates cells by rupturing tight junctions (45). In addition, mutations in Desmoglein-2 have been implicated in arrhythmogenic right ventricular cardiomyopathy, a potentially fatal heart condition. In transgenic mice with mutant Desmoglein-2, cellular dissociation at the desmosome junction was noted, followed by necrosis and fibrosis (46). This led to alterations in myocardial polarity, decreasing the action potential responsible for normal cardiac rhythm and the subsequent fatal abnormalities found in this disease. In our model, basolateral expression of Desmoglein-2 was validated, indicating it may be vital to maintaining the homeostatic barrier function of airway epithelium as required for myocardial tissue. It will be interesting to see whether basolateral expression and secretion of this protein is altered in diseases such as asthma, where the barrier function of epithelium is intrinsically disrupted (47).
We compared our apical secretome with a similar in vitro primary differentiated bronchial apical secretome established in a study that compared pooled apical secretions from HBE cell cultures with pooled induced sputum samples (22). When we applied the protein selection criteria used in that study (i.e., pooled apical proteins from all cultures including cytokeratins and the presence of at least one peptide), we detected 377 proteins from our three HBE cell cultures, compared with the 134 proteins in the pooled secretome. The disparity in the number of proteins in apical secretions between the two studies may reflect differences in how samples were processed and methods of proteomic analysis. Kesimer and colleagues isolated mucin-rich and protein fractions for proteomic analyses, whereas we electrophoresed unfractionated samples. They also used Q-TOF Micro mass spectrometry whereas our study used an Orbitap XL, which is considered much more sensitive in detecting ions than Q-TOF. In addition, there were some differences between cell cultures that may have affected apical secretion. We replaced the bronchial epithelial cell growth medium (typically used in basolateral compartment to effect and maintain differentiation) with protein-free media (BEBM) 24 hours before to collecting basolateral secretions because our proteomic analyses had shown that bronchial epithelial cell growth medium contains numerous proteins that would interfere with our ability to detect truly secreted proteins.
Another apparent difference between our study and that of Kesimer and colleagues (22) is the lack of MUC5AC and MUC5B peptides in our proteomic data. They are the major secretory mucins in lung mucus/sputum (48) and in apical secretions from HBE differentiated cells (49). MUC5B is typically seen in ALI secretions, whereas MUC5AC is not always detectable (43). With this in mind, we evaluated MUC5AC and MUC5B mucins by Western blot analysis on 1% agarose gels before collecting secretions for proteomic analysis. Consistent with current literature, MUC5B was well expressed in all three HBE samples used for proteomics and MUC5AC in HBE1 and HBE2 (data not shown). Separation of samples into high- and low-molecular-weight fractions has been used before proteomic analysis to target identification of mucins, which are typically underrepresented because of their large size and physical properties (22, 43). The PAGE fractionation and LC-MS/MS proteomics approach used in our study have previously identified mucin peptides near the top of polyacrylamide gels (50), suggesting that mucins in otitis media secretions are endogenously fragmented by the high level of proteases in pathological samples. Because MUC5B and MUC5AC were detected by Western blot data, the absence of the corresponding mucin peptides in our proteomic data indicates that HBE secretory mucins were trapped in the polyacrylamide gel wells, precluding their electrophoresis and subsequent detection by proteomic analysis.
Overall there was a substantially higher amount of protein in our apical secretions compared with our basolateral secretions. Apical secretions were collected starting at 23 hours using PBS in two successive 30-minute incubation periods, whereas basolateral secretions were collected after incubation in BEBM for 24 hours. The basolateral compartment is separated from epithelial cells by a collagen-coated Transwell membrane (0.4 μm pore size). Although this may limit the size and subsequent amount of proteins in basal secretions, the proteins that were uniquely secreted basolaterally had an average molecular weight of 129.2 kD, and the largest, basement membrane-specific heparan sulfate proteoglycan has a molecular weight of 468.8 kD (Table 3).
We also compared our data with that of BALF (n = 4) from healthy control subjects (11) because apical secretions in vivo are removed by bronchial washing of the airways. Although the BALF study used a different proteomic approach (shotgun proteomics with two-dimensional liquid chromatography–electrospray ionization–tandem mass spectrometry of suspended trypsin digested samples), the authors identified all proteins present in lavage samples from subjects without illness or known disease. We compared our proteins of interest—those consistently present in one secretome but with fewer than two peptides in the opposite secretome (31 apical and 13 basolateral proteins) and those differentially expressed between the two compartments (n = 25)—with the list of proteins found in healthy BALF. Overall, we identified 51% (35/69) of proteins in normal BALF. Specifically, 45% (14/31) of apical, 23% (3/13) of basolateral, and 72% (18/26) of differentially expressed proteins of interest were identified in normal BALF (Table E3). In addition, 55% (21/38) of our proteins of interest associated with the basolateral compartment (basolateral and differentially expressed) were found in normal BALF. Studying ex vivo airway samples alone or in comparison to in vitro apical secretions would minimize the impact of these biologically relevant proteins for a given disease state because basolateral proteins and their interaction with the apical secretome would not be evaluated. To this end, a primary differentiated bronchial epithelial model should be used for mechanistic lung disease studies to investigate the effect of ex vivo BAL proteins on basolateral processes and structures through directional epithelial signaling. This would allow for true translational studies evaluating mechanisms of lung disease for a targeted disease process.
In summary, to our knowledge this is the first comparison of in vitro apical and basolateral secretomes in a homeostatic environment and is the first proteomic description of basolateral secretions from primary differentiated bronchial epithelium. Specifically, we identified the normal apical and basolateral secretomes and proteins differentially expressed between compartments in cell cultures from multiple donors in an in vitro system that morphologically mimics the ex vivo bronchial epithelium and is routinely used for studying mechanisms of airway diseases. This analysis sets the stage for future comparative studies through global analysis of apical and basolateral secreted proteins for a more comprehensive understanding of mechanisms underlying diseases affecting the lung. Future validation of this in vitro secretome model in ex vivo specimens will enhance our understanding of lung diseases, suggesting that targeted therapy toward specific proteins may be a future step for airway secretome studies.
Acknowledgments
Acknowledgments
The authors thank Amarel Saieg, M.S., for assistance with the development and processing of Western blots.
Footnotes
This work was supported by National Institutes of Health grants K12HL090020 and NICHD 5K12HD001399 with partial support from R01 HL033152 (M.C.R.) and by National Institutes of Health Integrated Molecular Core for Rehabilitation Medicine grant NCMRR/NINDS 5R24 HD050846.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2013-0188OC on September 6, 2013
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Simons K, Wandinger-Ness A. Polarized sorting in epithelia. Cell. 1990;62:207–210. doi: 10.1016/0092-8674(90)90357-k. [DOI] [PubMed] [Google Scholar]
- 2.Feigin ME, Muthuswamy SK. Polarity proteins regulate mammalian cell-cell junctions and cancer pathogenesis. Curr Opin Cell Biol. 2009;21:694–700. doi: 10.1016/j.ceb.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feigin ME, Muthuswamy SK. ErbB receptors and cell polarity: new pathways and paradigms for understanding cell migration and invasion. Exp Cell Res. 2009;315:707–716. doi: 10.1016/j.yexcr.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 4.Homolya L, Steinberg TH, Boucher RC. Cell to cell communication in response to mechanical stress via bilateral release of ATP and UTP in polarized epithelia. J Cell Biol. 2000;150:1349–1360. doi: 10.1083/jcb.150.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malavia NK, Raub CB, Mahon SB, Brenner M, Panettieri RA, Jr, George SC. Airway epithelium stimulates smooth muscle proliferation. Am J Respir Cell Mol Biol. 2009;41:297–304. doi: 10.1165/rcmb.2008-0358OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong AM, Chow AW, Au SC, Wong CC, Ko WH. Apical versus basolateral P2Y(6) receptor-mediated Cl(-) secretion in immortalized bronchial epithelia. Am J Respir Cell Mol Biol. 2009;40:733–745. doi: 10.1165/rcmb.2008-0020OC. [DOI] [PubMed] [Google Scholar]
- 7.Holgate ST, Lackie PM, Davies DE, Roche WR, Walls AF. The bronchial epithelium as a key regulator of airway inflammation and remodelling in asthma. Clin Exp Allergy. 1999;29:90–95. doi: 10.1046/j.1365-2222.1999.00016.x. [DOI] [PubMed] [Google Scholar]
- 8.Davies DE. The role of the epithelium in airway remodeling in asthma. Proc Am Thorac Soc. 2009;6:678–682. doi: 10.1513/pats.200907-067DP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Regamey N, Jeffery PK, Alton EW, Bush A, Davies JC. Airway remodelling and its relationship to inflammation in cystic fibrosis. Thorax. 2011;66:624–629. doi: 10.1136/thx.2009.134106. [DOI] [PubMed] [Google Scholar]
- 10.Burgel PR, Bourdin A, Chanez P, Chabot F, Chaouat A, Chinet T, de Blic J, Devillier P, Deschildre A, Didier A, et al. Update on the roles of distal airways in COPD. Eur Respir Rev. 2011;20:7–22. doi: 10.1183/09059180.10010610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gharib SA, Vaisar T, Aitken ML, Park DR, Heinecke JW, Fu X. Mapping the lung proteome in cystic fibrosis. J Proteome Res. 2009;8:3020–3028. doi: 10.1021/pr900093j. [DOI] [PubMed] [Google Scholar]
- 12.McMorran BJ, Patat SA, Carlin JB, Grimwood K, Jones A, Armstrong DS, Galati JC, Cooper PJ, Byrnes CA, Francis PW, et al. Novel neutrophil-derived proteins in bronchoalveolar lavage fluid indicate an exaggerated inflammatory response in pediatric cystic fibrosis patients. Clin Chem. 2007;53:1782–1791. doi: 10.1373/clinchem.2007.087650. [DOI] [PubMed] [Google Scholar]
- 13.Sloane AJ, Lindner RA, Prasad SS, Sebastian LT, Pedersen SK, Robinson M, Bye PT, Nielson DW, Harry JL. Proteomic analysis of sputum from adults and children with cystic fibrosis and from control subjects. Am J Respir Crit Care Med. 2005;172:1416–1426. doi: 10.1164/rccm.200409-1215OC. [DOI] [PubMed] [Google Scholar]
- 14.Gray RD, MacGregor G, Noble D, Imrie M, Dewar M, Boyd AC, Innes JA, Porteous DJ, Greening AP. Sputum proteomics in inflammatory and suppurative respiratory diseases. Am J Respir Crit Care Med. 2008;178:444–452. doi: 10.1164/rccm.200703-409OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, Kobayashi M, Sousa EA, Liu W, Cai J, Goldman SJ, Dorner AJ, Projan SJ, Kavuru MS, Qiu Y, et al. Differential proteomic analysis of bronchoalveolar lavage fluid in asthmatics following segmental antigen challenge. Mol Cell Proteomics. 2005;4:1251–1264. doi: 10.1074/mcp.M500041-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Wu R, Yankaskas J, Cheng E, Knowles MR, Boucher R. Growth and differentiation of human nasal epithelial cells in culture: serum-free, hormone-supplemented medium and proteoglycan synthesis. Am Rev Respir Dis. 1985;132:311–320. doi: 10.1164/arrd.1985.132.2.311. [DOI] [PubMed] [Google Scholar]
- 17.Kondo M, Finkbeiner WE, Widdicombe JH. Cultures of bovine tracheal epithelium with differentiated ultrastructure and ion transport. In Vitro Cell Dev Biol. 1993;29A:19–24. doi: 10.1007/BF02634367. [DOI] [PubMed] [Google Scholar]
- 18.Fulcher ML, Gabriel S, Burns KA, Yankaskas J, Randell SH. Human cell culture protocols. Totowa, NJ: Humana Press; 2005. [Google Scholar]
- 19.Gruenert DC, Finkbeiner WE, Widdicombe JH. Culture and transformation of human airway epithelial cells. Am J Physiol. 1995;268:L347–L360. doi: 10.1152/ajplung.1995.268.3.L347. [DOI] [PubMed] [Google Scholar]
- 20.Matsui H, Grubb BR, Tarran R, Randell SH, Gatzy JT, Davis CW, Boucher RC. Evidence for periciliary liquid layer depletion, not abnormal ion composition, in the pathogenesis of cystic fibrosis airways disease. Cell. 1998;95:1005–1015. doi: 10.1016/s0092-8674(00)81724-9. [DOI] [PubMed] [Google Scholar]
- 21.Davies DE, Wicks J, Powell RM, Puddicombe SM, Holgate ST. Airway remodeling in asthma: new insights. J Allergy Clin Immunol. 2003;111:215–225, quiz 226. doi: 10.1067/mai.2003.128. [DOI] [PubMed] [Google Scholar]
- 22.Kesimer M, Kirkham S, Pickles RJ, Henderson AG, Alexis NE, Demaria G, Knight D, Thornton DJ, Sheehan JK. Tracheobronchial air-liquid interface cell culture: a model for innate mucosal defense of the upper airways? Am J Physiol Lung Cell Mol Physiol. 2009;296:L92–L100. doi: 10.1152/ajplung.90388.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freishtat RJ, Watson AM, Benton AS, Iqbal SF, Pillai DK, Rose MC, Hoffman EP. Asthmatic airway epithelium is intrinsically inflammatory and mitotically dyssynchronous. Am J Respir Cell Mol Biol. 2011;44:863–869. doi: 10.1165/rcmb.2010-0029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.An E, Lu X, Flippin J, Devaney JM, Halligan B, Hoffman EP, Strunnikova N, Csaky K, Hathout Y. Secreted proteome profiling in human RPE cell cultures derived from donors with age related macular degeneration and age matched healthy donors. J Proteome Res. 2006;5:2599–2610. doi: 10.1021/pr060121j. [DOI] [PubMed] [Google Scholar]
- 25.Zhang A, Williamson CD, Wong DS, Bullough MD, Brown KJ, Hathout Y, Colberg-Poley AM. Quantitative proteomic analyses of human cytomegalovirus-induced restructuring of endoplasmic reticulum-mitochondrial contacts at late times of infection. Mol Cell Proteomics. 2011;10:M111 009936. doi: 10.1074/mcp.M111.009936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hathout Y, Flippin J, Fan C, Liu P, Csaky K. Metabolic labeling of human primary retinal pigment epithelial cells for accurate comparative proteomics. J Proteome Res. 2005;4:620–627. doi: 10.1021/pr049749p. [DOI] [PubMed] [Google Scholar]
- 27.Kesimer M, Scull M, Brighton B, DeMaria G, Burns K, O'Neal W, Pickles RJ, Sheehan JK. Characterization of exosome-like vesicles released from human tracheobronchial ciliated epithelium: a possible role in innate defense. FASEB J. 2009;23:1858–1868. doi: 10.1096/fj.08-119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, et al. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi H, Fermin D, Nesvizhskii AI. Significance analysis of spectral count data in label-free shotgun proteomics. Mol Cell Proteomics. 2008;7:2373–2385. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernacki SH, Nelson AL, Abdullah L, Sheehan JK, Harris A, Davis CW, Randell SH. Mucin gene expression during differentiation of human airway epithelia in vitro: muc4 and muc5b are strongly induced. Am J Respir Cell Mol Biol. 1999;20:595–604. doi: 10.1165/ajrcmb.20.4.3442. [DOI] [PubMed] [Google Scholar]
- 31.Brown KJ, Formolo CA, Seol H, Marathi RL, Duguez S, An E, Pillai D, Nazarian J, Rood BR, Hathout Y. Advances in the proteomic investigation of the cell secretome. Expert Rev Proteomics. 2012;9:337–345. doi: 10.1586/epr.12.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lizarbe MA, Barrasa JI, Olmo N, Gavilanes F, Turnay J. Annexin-phospholipid interactions: functional implications. Int J Mol Sci. 2013;14:2652–2683. doi: 10.3390/ijms14022652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piljic A, Schultz C. Annexin A4 self-association modulates general membrane protein mobility in living cells. Mol Biol Cell. 2006;17:3318–3328. doi: 10.1091/mbc.E06-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayran N, Traverso V, Maroux S, Massey-Harroche D. Cellular and subcellular localizations of annexins I, IV, and VI in lung epithelia. Am J Physiol. 1996;270:L863–L871. doi: 10.1152/ajplung.1996.270.5.L863. [DOI] [PubMed] [Google Scholar]
- 35.Lin LL, Huang HC, Juan HF. Revealing the molecular mechanism of gastric cancer marker annexin A4 in cancer cell proliferation using exon arrays. PLoS ONE. 2012;7:e44615. doi: 10.1371/journal.pone.0044615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hein Z, Schmidt S, Zimmer KP, Naim HY. The dual role of annexin ii in targeting of brush border proteins and in intestinal cell polarity. Differentiation. 2011;81:243–252. doi: 10.1016/j.diff.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez-Begne M, Lu B, Han X, Hagen FK, Hand AR, Melvin JE, Yates JR. Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT) J Proteome Res. 2009;8:1304–1314. doi: 10.1021/pr800658c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buschow SI, van Balkom BW, Aalberts M, Heck AJ, Wauben M, Stoorvogel W. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunol Cell Biol. 2010;88:851–856. doi: 10.1038/icb.2010.64. [DOI] [PubMed] [Google Scholar]
- 39.Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, Mason MD, Clayton A. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9:1324–1338. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi DS, Lee JM, Park GW, Lim HW, Bang JY, Kim YK, Kwon KH, Kwon HJ, Kim KP, Gho YS. Proteomic analysis of microvesicles derived from human colorectal cancer cells. J Proteome Res. 2007;6:4646–4655. doi: 10.1021/pr070192y. [DOI] [PubMed] [Google Scholar]
- 41.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, Kleta R, Wang NS, Knepper MA. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol. 2009;20:363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ali M, Lillehoj EP, Park Y, Kyo Y, Kim KC. Analysis of the proteome of human airway epithelial secretions. Proteome Sci. 2011;9:4. doi: 10.1186/1477-5956-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cirillo N, Lanza M, De Rosa A, Cammarota M, La Gatta A, Gombos F, Lanza A. The most widespread desmosomal cadherin, desmoglein 2, is a novel target of caspase 3-mediated apoptotic machinery. J Cell Biochem. 2008;103:598–606. doi: 10.1002/jcb.21431. [DOI] [PubMed] [Google Scholar]
- 45.Schlegel N, Meir M, Heupel WM, Holthöfer B, Leube RE, Waschke J. Desmoglein 2-mediated adhesion is required for intestinal epithelial barrier integrity. Am J Physiol Gastrointest Liver Physiol. 2010;298:G774–G783. doi: 10.1152/ajpgi.00239.2009. [DOI] [PubMed] [Google Scholar]
- 46.Rizzo S, Lodder EM, Verkerk AO, Wolswinkel R, Beekman L, Pilichou K, Basso C, Remme CA, Thiene G, Bezzina CR. Intercalated disc abnormalities, reduced Na(+) current density, and conduction slowing in desmoglein-2 mutant mice prior to cardiomyopathic changes. Cardiovasc Res. 2012;95:409–418. doi: 10.1093/cvr/cvs219. [DOI] [PubMed] [Google Scholar]
- 47.Xiao C, Puddicombe SM, Field S, Haywood J, Broughton-Head V, Puxeddu I, Haitchi HM, Vernon-Wilson E, Sammut D, Bedke N, et al. Defective epithelial barrier function in asthma. J Allergy Clin Immunol. 2011;128:549–556. doi: 10.1016/j.jaci.2011.05.038. e1–12. [DOI] [PubMed] [Google Scholar]
- 48.Kirkham S, Sheehan JK, Knight D, Richardson PS, Thornton DJ. Heterogeneity of airways mucus: variations in the amounts and glycoforms of the major oligomeric mucins MUC5AC and MUC5B. Biochem J. 2002;361:537–546. doi: 10.1042/0264-6021:3610537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmén JM, Karlsson NG, Abdullah LH, Randell SH, Sheehan JK, Hansson GC, Davis CW. Mucins and their O-glycans from human bronchial epithelial cell cultures. Am J Physiol Lung Cell Mol Physiol. 2004;287:L824–L834. doi: 10.1152/ajplung.00108.2004. [DOI] [PubMed] [Google Scholar]
- 50.Preciado D, Goyal S, Rahimi M, Watson AM, Brown KJ, Hathout Y, Rose MC. MUC5B Is the predominant mucin glycoprotein in chronic otitis media fluid. Pediatr Res. 2010;68:231–236. doi: 10.1203/PDR.0b013e3181eb2ecc. [DOI] [PMC free article] [PubMed] [Google Scholar]