SUMMARY
Transcriptional processes involved in the development of human cerebral neocortex are poorly understood. Here, we analyzed the temporal dynamics and laterality of gene expression in human and macaque monkey neocortex. We found that inter-areal differences exhibit a temporal hourglass pattern, dividing the human neocortical development into three major phases. The first phase, corresponding to prenatal development, is characterized by the highest number of differential expressed genes among areas and gradient-like expression patterns, including those that are different between human and macaque. The second, preadolescent phase, is characterized by lesser inter-areal expression differences and by an increased synchronization of areal transcriptomes. During the third phase, from adolescence onwards, differential expression among areas reappears driven predominantly by a subset of areas, without obvious gradient-like patterns. Analyses of left-right gene expression revealed population-level global symmetry throughout the fetal and postnatal timespan. Thus, human neocortical topographic gene expression is temporally specified and globally symmetric.
INTRODUCTION
The cerebral neocortex (NCX) is organized into functionally distinct sensory, motor, and association areas that provide the biological substrates underlying perception, behavior, and cognition (Brodmann, 1909; O’Leary and Sahara, 2008; Rakic, 1988; Rash and Grove, 2006; Sur and Rubenstein, 2005). While the basic architecture of this areal map is shared among mammals, important species-specific organizational differences have allowed for the elaboration of human-specific cognition and behavior (Hill et al., 2010; Judas et al., 2013; Kaas, 2012; Kennedy and Dehay, 2012; Lui et al., 2011; Molnar and Clowry, 2012; Preuss, 2011).
Another key feature of the human NCX is that it covers the surface of the left and right hemispheres, each comprising a topographically matched, though slightly structurally and functionally asymmetric areal map (Amunts et al., 2003; Gazzaniga et al., 1962; Geschwind and Levitsky, 1968). This asymmetric organization plays a crucial role in functional lateralization of many cognitive and motor functions, such as language and handedness, between the hemispheres. Several lines of evidence indicate that these asymmetries are reflected at the molecular (Sun et al., 2005) and cellular (Amunts et al., 2003; Hayes and Lewis, 1993) levels. Structural asymmetry first appears during the late mid-fetal period (Chi et al., 1977; Kasprian et al., 2011) and becomes more prominent during early postnatal development when functional asymmetries become noticeable (Amunts et al., 2003; Hill et al., 2010).
Multiple lines of evidence indicate that distinct human neocortical areas, and the hemispheres as a whole, mature at different rates (Flechsig, 1901; Giedd et al., 1999; Giedd and Rapoport, 2010; Huttenlocher and Dabholkar, 1997; Sowell et al., 2003). For example, axons in primary sensory-motor areas start to myelinate before those in the association areas (Flechsig, 1901). Other processes such as synaptogenesis also exhibit prominent inter-areal differences in their maturational trajectories (Huttenlocher and Dabholkar, 1997). Furthermore, the right hemisphere appears to mature faster than the left during late fetal and early postnatal development (Taylor, 1969; Thatcher et al., 1987).
There is increasing evidence to suggest that processes regulating areal patterning and asymmetry, as well as the maturational trajectories of these processes, are affected in major psychiatric and neurological disorders (Cullen et al., 2006; Faludi and Mirnics, 2011; Piao et al., 2004; Rapoport and Gogtay, 2007). Moreover, the progression of certain neuropathologies follows a stereotypic areal pattern (Braak et al., 1993), indicating that the mechanisms involved in patterning and asymmetry may play a role in the manifestation of disease. However, little is known about these developmental processes in normal or diseased human brains, or how they differ among mammals, especially closely related nonhuman primates (NHPs).
Gene expression has previously been profiled in the developing human NCX (Abrahams et al., 2007; Colantuoni et al., 2011; Ip et al., 2010; Johnson et al., 2009; Kang et al., 2011; Lambert et al., 2011; Sun et al., 2005). However, most of these studies were restricted to a small number of areas and time points. Furthermore, a number of genes was found to be expressed asymmetrically in the early fetal (Sun et al., 2005) NCX, but not in late, mid-fetal or adult NCX (Hawrylycz et al., 2012; Johnson et al., 2009; Lambert et al., 2011), suggesting that transcriptional asymmetry may be temporally regulated. In the present study, we analyzed the temporal dynamics and left-right asymmetry of NCX topographic gene expression across the full course of fetal and postnatal development, and adulthood.
RESULTS
Inter-areal transcriptional divergence exhibits a temporal hourglass pattern
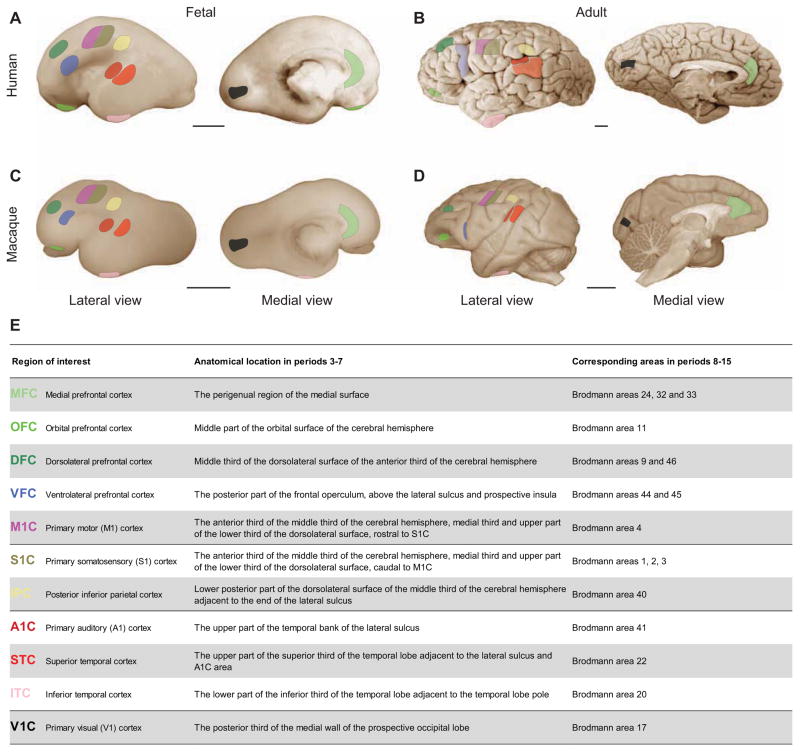
Our previous analyses of gene expression in the human brain revealed robust transcriptional differences among topographically defined areas of the fetal and, to a lesser extent, adult NCX (Johnson et al., 2009; Kang et al., 2011). To analyze temporal progression and left-right asymmetry of areal gene expression, we performed a secondary analysis of this dataset (Kang et al., 2011) that included 11 topographically defined NCX areas corresponding to the orbital (OFC), dorsolateral (DFC), ventrolateral (VFC), medial (MFC), and primary motor (M1C) cortices of the frontal lobe; the primary somatosensory (S1C) and posterior inferior (IPC) cortices of the parietal lobe; the primary auditory (A1C), posterior superior (STC) and anterior inferior (ITC) cortices of the temporal lobe; and the primary visual (V1C) cortex of the occipital lobe (Figure 1 and Supplemental Note 2 for topographic sampling). This dataset was generated using 886 tissue samples isolated from left and right hemispheres of 53 clinically unremarkable post-mortem human brain specimens spanning from early fetal development through old age (from 10 weeks of post-conception (PCW) to 82 years of age), which corresponded to periods 3–15, as previously designated, an interval during which all of the analyzed putative functional areas were represented (Kang et al., 2011) (Tables S1A and S2).
Figure 1. Topographic locations of NCX samples in the fetal and adult human and macaque brains.
(A–E) Schematic shows anatomical positions of dissections of NCX samples in (A) prenatal and (B) postnatal human, and (C) prenatal and (D) postnatal macaque brains, and their corresponding Brodmann areas in the postnatal NCX (E). Each NCX sample is colored by the same color in all figures of this article. Scale bar represents 1 cm. See also Tables S1 and S2.
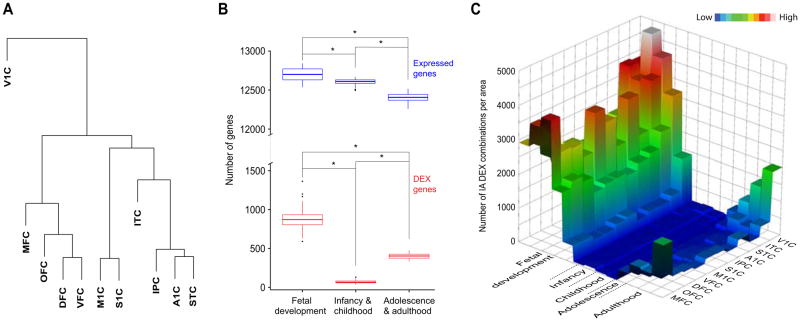
The areal localization of dissected NCX samples was previously verified by histology in the postnatal brains and matched across fetal periods using the same anatomical landmarks (Supplemental Note 2; see also Kang et al. 2011). A hierarchical clustering of both fetal and postnatal NCX samples confirmed their grouping by topographical proximity and functional overlap (Figure 2A). Principal component analysis also revealed that transcriptional differences across periods account for the majority of the variance among NCX samples (Figure S1A), indicating that NCX areal gene expression is strongly developmentally regulated.
Figure 2. Neocortical inter-areal differences in gene expression exhibit a temporal hourglass pattern.
(A) Unsupervised hierarchical clustering of the 11 NCX areas profiled in this study, based on the transcriptome of each area from the period of fetal development throughout life, showing relative transcriptional differences. (B) Boxplots of subsampling permutations show the number of expressed (blue) and differentially expressed (red) genes among neocortical areas across fetal development (periods 3–7), infancy (periods 8 and 9) childhood (periods 10 and 11), adolescence (period 12) and adulthood (periods 13–15). (C) Post-hoc Tukey tests were used to identify significant inter-areal (IA) DEX combinations. A 3D heatmap shows the number of IA DEX combinations per NCX area over time. See also Figure S1 and Table S3.
To investigate how putative areal transcriptional differences change over time, we used analysis of variance (ANOVA) to identify genes that exhibit differential expression (DEX) among areas in each period (henceforth referred to as the inter-areal divergence; Supplemental Note 3.3). This analysis revealed that inter-areal transcriptional divergence, but not the total number of expressed genes, exhibits a previously unrecognized temporal hourglass pattern, with robust and dynamic differences found prenatally and, to a lesser extent from adolescence onwards, with few in infancy and childhood (Figures 2B and S1B). In contrast, differential expression among other (non-neocortical) brain regions did not exhibit the same temporal hourglass pattern (Figure S1B).
We also analyzed the genotype of individuals using the data previously generated using Illumina HumanOmni 2.5 SNP arrays (Kang et al., 2011) to test if the hourglass pattern could be explained by a reduction in the genetic diversity among individuals in periods 8 to 11. We observed that the genetic diversity of samples varies throughout development in a random way, without any observable pattern for periods 8 to 11 compared with others. We also found no relation with the number of samples (Figure S1C).
To estimate the contribution of differences in specific putative areas to the overall inter-areal divergence, Tukey’s pairwise comparison was performed after ANOVA to determine the total number of significant (P < 0.01) DEX gene comparisons of each area with all the other areas for a given period. The relative contribution of areas to the overall hourglass shape varied across periods. During fetal periods, MFC, ITC, and the primary areas (V1C, A1C, S1C and M1C) exhibited the most prominent dissimilarity, whereas only MFC and V1C showed robust dissimilarities during adolescence and adulthood (Figure 2C). Together, these findings show that the pattern of inter-areal transcriptional divergence is specified over time and exhibits an hourglass pattern, with infancy and childhood representing a long phase of minimal divergence. Our results also show that the spatial pattern of inter-areal divergence is mainly driven by a subset of putative functional areas.
Temporal transcriptional hourglass pattern reflects putative areal and functional differences
We hypothesized that increased inter-areal transcriptional divergence during fetal development and from adolescence onwards reflects the differences in the underlying molecular and cellular processes between these two phases. Consistent with this hypothesis, only 848 of 3,125 (27%) inter-areal DEX genes were DEX both in fetal development and from adolescence onwards. To gain insights into the differential organization of the NCX transcriptomes during the two phases of increased inter-areal differences, we performed weighted gene co-expression network analysis (Supplemental Note 3.7) to identify modules of co-expressed genes with often-shared functional relevance. Within fetal development, we identified 122 modules (M1–M122: Table S4A), and from adolescence on we found 207 modules (M123–M329; Table S4B). Functional annotation of the modules revealed significant differences between the organization of fetal and adolescent/adult differential putative areal transcriptomes. Furthermore, the gene ontology (GO) enrichment analysis (Supplemental Note 3.8) revealed significant differences between fetal and adolescent/adult DEX and also between co-expression modules in the enrichment for GO categories (Tables S3 and S4). Among exclusively fetal GO categories were mostly categories related with developmental processes, such as phosphoprotein, neuron differentiation, cell cycle, neuron development, cell morphogenesis, mitosis, cell morphogenesis involved in neuron differentiation, cell adhesion, and cellular component morphogenesis (Bonferroni-adjusted P ≤ 6.61E-11 per category; Table S3A). In contrast, exclusively adolescent and adult GO categories were mostly related to processes associated with neuronal and synaptic function, including synaptic vesicle, transport, domain:C2 1 and 2, intrinsic to plasma membrane, intrinsic to membrane, clathrin-coated vesicle, neurological system process, neurotransmitter binding, coated vesicle, integral to membrane, and monovalent inorganic cation transport (Bonferroni-adjusted P ≤ 2.39E-06 per category; Table S3C).
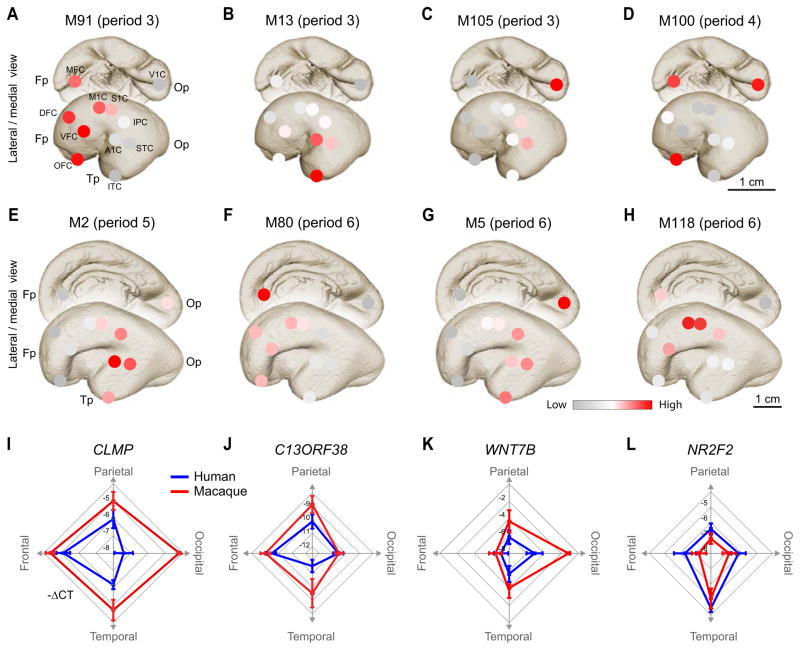
Many fetal co-expression modules had pronounced gradients and/or well-defined compartments with distinct boundaries (Figures 3A–H). In addition to expected gradients with prefrontal/frontal-enriched graded expression along the anterio-posterior axis (e.g., M54, M62, M80, M91), we found patterns including those with temporal (e.g., M2, M13), occipital (e.g., M105), occipito-temporal (e.g., M5), perisylvian (e.g., M118), and ventro-medial areas (MFC, OFC, V1C; e.g., M100). The majority of the modules exhibited temporally specified areal patterns that changed dramatically across fetal periods and lost their prominent areal differences postnatally (Figure S2A–H and also see Tables S4A and S4B with the description of spatiotemporal patterns). Areal expression patterns of 6 selected intra-modular hub genes were successfully validated by quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR; median correlation with the array data, r = 0.88; Figures 3I–L and S3A–L). CLMP, a gene encoding an adhesion molecule (Figures 3I, S3A, and S3B), and WNT7B, a gene involved in early brain development and dendritic arborization (Harrison-Uy and Pleasure, 2012) (Figures 3K, S3I, and S3J) displayed expression gradients along the rostro-caudal axis in anterior-posterior and posterior-anterior directions, respectively. We also validated genes with enriched expression in the temporal lobe areas, such as NR2F2 (COUP-TF2) (Figure 3L, S3C, and S3D), a gene enriched in the caudal ganglionic eminence and involved in migration of interneurons (Tripodi et al., 2004), or restricted to the putative temporal lobe areas, including the previously uncharacterized gene C13ORF38 (Figures 3J, S3E, and S3F). Immunohistochemical analysis validated regional differences in the expression of WNT7B and NR2F2 and revealed cell type specific differences in their expressions. Immunolabeling for WNT7B was enriched in future layer 5 pyramidal neurons of the occipito-temporal cortical plate, which were organized into columns (Figure S4A). NR2F2 was enriched in the upper part of the fetal temporal cortical plate including the islets in the entorhinal cortex (Figure S4B).
Figure 3. Fetal gene co-expression modules and hub genes in human and macaque monkey.
(A–H) Average scaled-expression of all genes in co-expression module (M) 91, M13, M105, M100, M2, M80, M5, and M118 show a gradient-like expression pattern in periods (A–C) 3, (D) 4, (E) 5, and (F–H) 6. I–L, Radar charts of qRT-PCR of intra-modular hub genes (I) CLMP (M91), (J) C13ORF38 (M80), (K) WNT7B (M6), AND (L) NF2R2 (M13) demonstrate a gradient-like expression pattern in human (blue) in directions of areas grouped by their corresponding lobes (e.g., frontal, parietal, occipital and temporal; versions of these radar charts with all 11 areas across different periods are shown in Figure S3A–L). Rhesus macaque (red) expression patterns are similar to those of human. See also Figures S2, S3, S4, and Table S4.
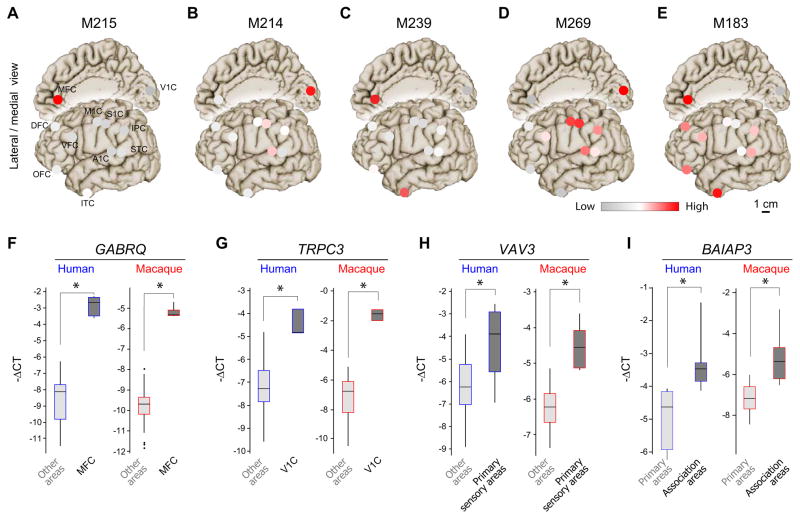
Adolescence and adulthood co-expression modules exhibited more stability over time and less complex spatial patterns (Figures 4A–E and S2I–L). Consistent with our previous finding of dramatic differences in the number of DEX genes among areas in these periods (Figure 2C), the most common areal co-expression patterns reflected enrichments in MFC (e.g., M215), V1C (e.g., M214), MFC and ITC (e.g., M239), and differences between the primary sensory-motor areas (e.g., M269), and association areas (e.g., M183). Expression patterns of several intra-modular hub genes were validated with qRT-PCR (median r = 0.82), such as GABRQ in MFC, TRPC3 in V1C, VAV3 in primary sensory areas, and BAIAP3 in all association areas (Figures 4F–I and S3M–T). Together, these findings indicate that the transcriptional hourglass pattern reflects differences over time in processes related to the construction and functional specializations of areas.
Figure 4. Adolescent/adult gene co-expression modules and hub genes in human and rhesus macaque.
(A–E) Average scaled-expression of all genes in M215, M214, M239, M269 and M183, in period 13, (A–C) shows area-specific or (D) primary sensory and (E) associative areas-specific patterns. (F–I) Box plots show qRT-PCR expression levels of area-enriched human (blue) and rhesus macaque genes (red): (F) GABRQ (M215), (G) TRPC3 (M214). Genes with patterns that discriminate primary sensory areas and associative areas are also shown: (H) VAV3 (M214) and (J) BAIAP3 (M183), respectively. Radar charts of qRT-PCR data for the four genes with all 11 areas across different periods are shown in Figure S3M–T. Error bars represent standard deviation. See also Figures S2 and S3.
Certain prenatal expression patterns differ between human and macaque
To investigate whether some of the observed temporally regulated inter-areal expression patterns exhibit inter-primate divergence, we performed qRT-PCR on the previously selected intra-modular hub genes (Figures 3I–L, 4F–I, and S3) in homologous regions of human and rhesus macaque neocortices at equivalent ages (Supplemental Note 5). In total, we analyzed all 11 NCX areal samples in 38 human and 21 macaque developing and adult brains (Supplemental Notes 4 and 5, and Table S1C). Of the hub genes that had prominent inter-areal differential expression prenatally but not postnatally (CLMP, C13ORF38, NR2F2, WNT7B, KCNK12, and WBSCR17), only NR2F2 and WNT7B had spatiotemporal areal expression profiles that were well correlated (r > 0.8) between human and macaque, indicating that their expression patterns are well conserved. In contrast, the inter-areal expression pattern of WBSCR17 was only conserved during late-fetal periods (r > 0.8), while KCNK12 had moderate correlation only during early fetal periods (r = 0.63), indicating that some of the spatiotemporal expression patterns are shaped by regulatory programs that differ between humans and NHPs (Figures 3I–L and S3A–L). Comparative analysis of adolescent and adult intra-modular hub genes (BAIAP3, GABRQ, TRPC3, and VAV3) revealed that the inter-areal expression patterns of BAIAP3 and VAV3 were conserved in both adolescent and adult human and macaque NCX (r > 0.8) (Figures 4F–I and S3M–T). In contrast, the spatial expression patterns of GABRQ and TRPC3 were only conserved in adulthood (r > 0.8) but not in adolescence (r ≤ 0.6). While limited in their scope, these findings show that certain intra-modular hub genes exhibit temporally regulated and species-differential inter-areal expression patterns, and may reflect broader differences in developmental gene expression programs between the two primates.
Increased synchronization of postnatal areal transcriptonal trajectories
Previous work has shown that NCX areas mature at different rates with primary sensory-motor areas maturing first, followed by association areas (Flechsig, 1901; Rapoport and Gogtay, 2007; Sowell et al., 2003). To investigate the transcriptional dynamics associated with areal maturation, we generated intra-areal global expression trajectories by correlating the transcriptome profile of each sample to the averaged transcriptome profile of the corresponding area in mid-adulthood (period 14; Supplemental Note 3.5.1). We also correlated the transcriptome profiles to the corresponding putative area in early fetal development (period 3), to control for putative bias of selecting a specific period (Figure 5A). These trajectories allowed us to detect large-scale changes in the transcriptome over time in individual putative functional areas. Surprisingly, all areal maturational trajectories had similar shapes, with steep increases during mid- and late fetal development and the major inflection point on the curve during late infancy (Figure 5A). However, the average deviation of the areal trajectory from the average overall maturational trajectory (maturational difference index; see Supplemental Note 3.5.2) varied more among putative areas during fetal than postnatal periods (Figure 5B). MFC and ITC appear to mature faster than other areas prenatally, while DFC and V1C mature slower. Thus, inter-areal differences in maturational rates did not follow the global antero-posterior or medio-lateral neurogenetic gradients observed in rodents (Bayer and Altman, 1987). Furthermore, these findings indicate that at the global level putative areal transcriptomes become more synchronized during postnatal development. Interestingly, we also observed a decline in some areal trajectories, most prominently in V1C, in period 15. Moreover, V1C changes in aging deviate in the same direction in both fetal and adult trajectories suggesting that the changes are not a reversal toward fetal expression as previously reported (Colantuoni et al., 2011), but instead are likely unique to the aging process. These transcriptome changes could not be explained by the difference in confounding factors between V1C and non-V1C areas (RIN, P = 0.41; PMI, P = 1; pH, P = 1), or between adult periods (RIN, P = 0.96; PMI, P = 0.84; pH, P = 0.13). Consistent with previous findings in aged prefrontal cortex (Colantuoni et al., 2011), this suggests that aging is associated with global transcriptomic changes that vary considerably among NCX areas.
Figure 5. Areal transcriptional trajectories become more synchronized during postnatal development.
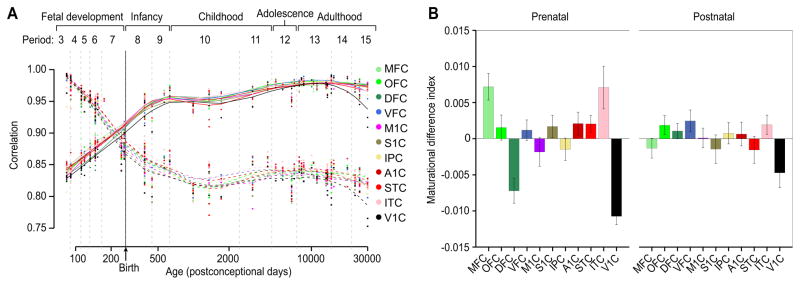
(A) A maturational trajectory plot shows the Pearson correlation of gene expression in each sample to the corresponding averaged gene expression in period 14 (solid line) or period 3 (dashed line). (B) Bar plots show the average deviation of the areal trajectory from the average overall maturational trajectory (maturational difference index). Prenatal (fetal) development exhibits more deviation than postnatal development. Error bars represent standard deviation. See also Figure S5 and S6.
Next, we followed the maturational trajectories of specific neurobiological processes and categories, using the first principal component to summarize the expression of genes associated with various developmental processes (Supplemental Note 3.6). Notable trajectories and differences in their onset times, rates of increase and decrease, and shapes were observed within and between putative areas. Many showed more prominent inter-areal variations prenatally than postnatally (Supplemental Note 3.6, and Figures S5 and S6). These included cell proliferation (which displayed antero-posterior and medio-lateral gradients), dendrite development, synapse development (which showed accelerated rate in M1C and S1C). Some trajectories, such as myelination, were variable both prenatally and postnatally (Figures S6E and S6F). These findings indicate that specific transcriptional trajectories, similar to inter-areal divergence, vary across the putative areas, with prenatal development being generally more variable.
Putative areal transcriptomes are globally symmetric across time
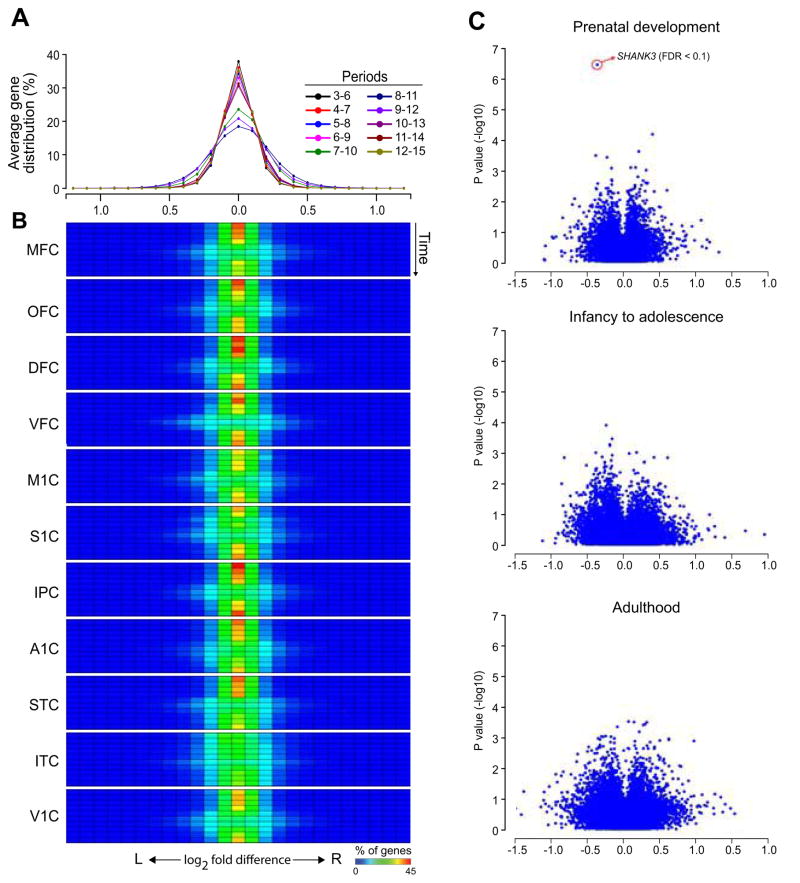
Since the initial discoveries of the functional lateralization of speech and language in the perisylvian areas of the NCX (Broca, 1861; Dax, 1865; Wernicke, 1874), an abundance of data has demonstrated functional and structural asymmetries between hemispheres (Amunts et al., 2003; Chi et al., 1977; Cullen et al., 2006; Geschwind and Levitsky, 1968; Hayes and Lewis, 1993; Kasprian et al., 2011). However, transcriptome studies of left-right differences in NCX gene expression are rare and limited by a small and temporally restricted dataset (Hawrylycz et al., 2012; Johnson et al., 2009; Lambert et al., 2011; Sun et al., 2005). Furthermore, previous findings are also inconsistent, as evidence for gene expression asymmetry has been found in early fetal (Sun et al., 2005), but not in mid-fetal or adult NCX (Hawrylycz et al., 2012; Johnson et al., 2009; Lambert et al., 2011). Since gene expression differences between hemispheres might be subtle, we relaxed our criteria, and used an FDR < 0.1 without fold change cut-off. Nevertheless, we failed to identify statistically significant inter-hemispheric differences across fetal and postnatal time span, using a paired t-test (Figure 7A and 7B).
Figure 7. Population-level global areal transcriptional expression trajectories are bilaterally symmetric.
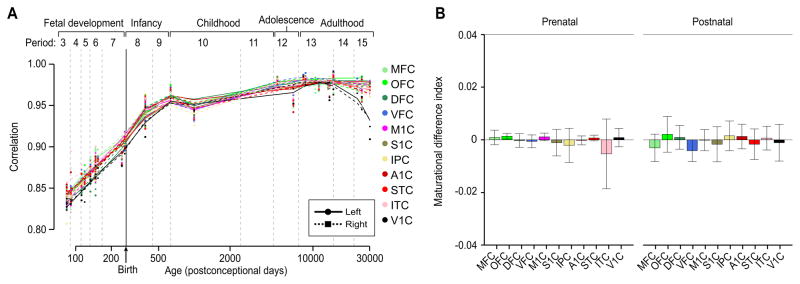
(A) Transcriptional maturational trajectories of left (solid line) and right (dashed line) hemispheres of each NCX area and (B) maturational difference index between hemispheres in prenatal (fetal) and postnatal development further demonstrate a symmetric gene expression profile. Error bars represent standard deviation.
Since microarray-based analyses are limited to those genes included in the platform, we further investigated the lateralization of gene expression using mRNA-sequencing (mRNA-seq). This technique allows for unbiased profiling of mRNA transcripts in a wider dynamic range than microarrays (Wang et al., 2009) and thus can detect rare and unknown transcripts as well as more subtle gene expression differences between samples. We performed mRNA-seq (see Figure S7 for quality control measures) on the left and right STC samples from 11 randomly selected brains covering the fetal and postnatal time span (periods 3–15; Table S1B). STC plays an important role in language lateralization (Geschwind and Levitsky, 1968; Wernicke, 1874) and is the earliest NCX area known to show signs of structural asymmetry in the fetal brain (Chi et al., 1977; Kasprian et al., 2011). mRNA-seq analysis detected only one gene (SHANK3) with a statistically significant (FDR<0.1) left-right bias in expression (Figure 7C). However, the right-biased SHANK3 expression was not detected in the larger number of samples analyzed by exon array extended probesets (data not shown). This indicates that the asymmetric SHANK3 expression observed with mRNA-seq was likely due to the sampling of a subset of analyzed fetal brains, as this bias disappeared when a larger number of brains were analyzed.
To test the concept of the arrival of corresponding developmental stages sooner in the right than in the left hemisphere (Taylor, 1969; Thatcher et al., 1987), we compared the individual areal maturational trajectories between hemispheres. This analysis revealed no trend in faster maturation of one hemisphere over the other (Figure 7). Our findings indicate that, at the population level, areal transcriptomes are globally symmetric across almost the full course of human neocortical development and adulthood.
DISCUSSION
Taken together, our analyses of the temporal dynamics and bilaterality of putative areal transcriptomes revealed several novel and unexpected aspects of human neocortical development. We uncovered that the degree of transcriptional differences between areas, as measured by the number of DEX genes, is regulated over time in an hourglass pattern. The greatest inter-areal differences were found prenatally, following by a phase of minimal inter-areal transcriptional divergence during infancy and childhood and the reappearance of inter-areal difference, to a slighter degree, from adolescence onwards.
We also provided evidence that these three major phases of inter-areal transcriptional divergence reflect underlying developmental and functional differences. Fetal DEX genes and co-expression modules had highly dynamic spatio-temporal expression patterns and tended to be related to early neurodevelopmental processes associated with cell generation and neural circuit construction. Consistent with this finding, previous neuroanatomical studies have shown that area-specific features of axonal projections and neuronal differentiation are already evident in the mid-fetal NCX (Johnson et al., 2009; Kwan et al., 2012; Vasung et al., 2010). Interestingly, the relative contribution of areas to the overall inter-areal transcriptional divergence varied across fetal development and may reflect the underlying processes involved in areal patterning. Additionally, our results indicate that fetal MFC and ITC, which have high numbers of DEX genes and global transcriptional trajectories that point to faster maturation, may have differentiated earlier than other analyzed areas. This developmental pattern brings to mind the evolutionary pattern of NCX differentiation arising from dual archicortical and paleocortical moieties (Sanides, 1969). Also, the increased transcriptional divergence of prospective primary motor-sensory areas (V1C, A1C, S1C, and M1C) is suggestive of their early transcriptional specifications.
The fetal phase of robust transcriptional differences is followed by a phase of sharply reduced inter-areal transcriptional divergence that spans late fetal development through the extended period of infancy and childhood, during which time areas become more transcriptionally similar. In addition, the global gene expression maturational trajectories become more synchronized over the course of early postnatal development. We hypothesize that this areal transition from high divergence to similarity as well as increased synchronization are related to coincidental changes in neurodevelopmental processes. Over one hundred years ago, Brodmann discovered (Brodmann, 1909) that the anatomy of the entire neocortical plate is transiently transformed by the appearance of an ontogenetic six-layered Grundtypus (i.e., the fetal equivalent of future layers 2–6) during late fetal and neonatal development (periods 7 and 8 in this study). During those periods layer 5 pyramidal neurons also transiently express nitric oxide synthase 1 protein throughout the human NCX (Kwan et al., 2012). In addition, the human NCX exhibits more widespread synchronized intra- and inter-hemispheric network activity around birth and in infancy (Fransson et al., 2007; Khazipov and Luhmann, 2006). Thus, these global molecular and functional changes in the late fetal and neonatal human NCX may provide the mechanisms by which areas become more similar in their expression patterns.
The drop in inter-areal transcriptional divergence occurs during early postnatal development when neocortical map undergoes reorganization largely through the influence of experience, and environmental and social inputs (Johnson, 2001). Furthermore, infancy and childhood are critical and sensitive periods for the acquisition and refinement of cognitive and motor functions characterized by substantial plasticity (Johnson, 2001; Rapoport and Gogtay, 2007). Thus, we hypothesize that following the initial establishment of area-specific subcortical and cortico-cortical projections during prenatal development, the robust inter-areal transcriptional differences are no longer needed and are superseded by more general neuronal and glial differentiation transcriptional programs involved in the maturation of neural circuits. Some of these more general programs, such as the transcription of myelination genes, also exhibit inter-areal difference in the neonatal and postnatal NCX, but they appear to be less robust or less dynamically regulated than many fetal programs. Specific transcriptional trajectories seem to be more synchronized between areas than expected from analyses of maturational trajectories of several phenotypic traits such as synaptogenesis (Huttenlocher and Dabholkar, 1997), cortical thickness (Giedd et al., 1999; Sowell et al., 2003), and myelination (Flechsig, 1901). This observation also suggests that some phenotypic inter-areal differences arise in part by post-transcriptional and activity-dependent mechanisms. Interestingly, in contrast to the classic hierarchical model of human cortical areal development, the peak of synaptogenesis and overproduction of neurotransmitter receptors occur concurrently among layers and areas of the developing macaque cerebral cortex (Rakic et al., 1986; Lidow et al., 1991).
Inter-areal transcriptional divergence increases again during adolescence, a period of great developmental changes in the human brain (Giedd et al., 1999; Johnson, 2001; Sowell et al., 2003), and throughout adulthood as the brain matures and ages. Unlike genes from the fetal phase, DEX genes and co-expressed genes during adolescence and adulthood tend to be related to categories such as synaptic transmission and cell-cell signaling. Thus, adolescence and adulthood, especially the aging process, are accompanied by specific changes in regional/areal transcriptomes.
Interestingly, the temporal pattern of inter-areal transcriptional divergence resembled the hourglass model of transcriptome divergence among various species of fruitfly (Domazet-Loso and Tautz, 2010; Kalinka et al., 2010). We show here that some of the genes that show inter-areal differential expression over time also display divergence between humans and rhesus macaques, suggesting a possible role for differences in gene expression patterns in human evolution.
The finding of dramatic temporal differences in inter-areal patterns of gene expression also points to even small differences in age as an important confound in the analysis of areal differences in gene expression across development in healthy and disease brains.
Our findings also show that putative areal transcriptomes are largely symmetrical at the population level and that left and right hemispheres mature at similar rates across the full course of human brain development and adulthood. Noteworthy, our study design does not allow us to exclude the possibility that there could be transcriptional asymmetry present in areas we did not profile, that we are overlooking asymmetry diluted at the population level by inter-individual variations, that asymmetry might be limited to the small subset of genes not present on the array or uniquely mapped by RNA-seq, or that it might be driven by more subtle changes in specific cellular components that we were unable to detect. Our findings are consistent with structural imaging studies showing strong bilateral symmetry of the neocortex (Chen et al., 2011) as well as reports of an unexpectedly high percentage of monozygotic twin pairs that are discordant for handedness (see McManus and Bryden, 1991). Together, these observations suggest that at least some of the aspects of neocortical asymmetry could be driven by stochastic, hormonal, epigenetic, functional connectivity-based, and/or experience-dependent mechanisms (see also Geschwind and Galaburda, 1985; Hobert et al., 2002; McManus and Bryden, 1991; Sun and Walsh, 2006; Toga and Thompson, 2003) that are not clearly reflected in the population-level hemispheric gene expression differences.
EXPERIMENTAL PROCEDURES
Samples and time periods
For this study, we used a subset of a previously reported dataset (Kang et al., 2011). In that dataset, the full course of human brain development and adulthood was divided into 15 periods. Neocortical areas from periods 3 to 15 were analyzed in the current study. Additionally, for validation of selected hub genes we used homologous areas from rhesus macaque, at equivalent developmental periods (Clancy et al., 2007, http://www.translatingtime.net/). The complete list of periods and NCX samples, including corresponding putative functional areas, analyzed in this study can be found in Table S2 and Figure 1, respectively.
Differential expression analyses
We used analysis of variance (ANOVA) to identify differentially expressed (DEX) genes in the NCX. P-values from ANOVA were corrected for multiple comparisons using the Benjamini and Hochberg false discovery rate (FDR) method (Benjamini and Hochberg, 1995), and a conservative statistical threshold (FDR <0.01 and minimum fold difference >2 between NCX areas) was used to identify DEX genes. To assess the influence of each area to DEX genes, a post-hoc Tukey’s HSD (honestly significant difference) test was used.
To control for differences in sample size, we used random subsampling to calculate the number of expressed and DEX genes. Samples were divided into three groups: prenatal, postnatal, and adult. The number of samples randomly chosen was calculated as ¾ of the lowest number of samples contained within any of the three groups. For 100 trials, this fixed number of samples was randomly drawn from each group and the number of expressed or DEX genes was calculated using the selected subset of samples.
Transcriptome maturation
We calculated the mean gene expression for every neocortical area for periods 14 and 3 as references for adulthood and early fetal development, respectively. Then, we compared each sample to their reference sample using Pearson correlation.
We also calculated the average maturational trajectory for all areas. The deviation from the average maturational trajectory for each area was used to calculate the maturational difference index.
Weighted gene co-expression network analysis (WGCNA)
WGCNA package (Langfelder and Horvath, 2008, 2012) was applied to samples from periods 3–6 and periods 12–15. Only genes with log2-expression value greater than 6 in any sample and coefficient of variation (CV) greater than 0.05 were used for analysis. Each module was summarized by an eigengene, which is the first principal component of the scaled module expression. Thus, the module eigengene explained the maximum amount of variation of the module expression levels.
Gene ontology (GO) enrichment analysis
Functional enrichment was assessed using DAVID Bioinformatics Resources 6.7 (Huang et al., 2009a, b).
Analysis of left-right bias in gene expression
Differences in gene expression between left and right hemispheres were analyzed by the combination of a sliding window algorithm and a paired t-test. The sliding-window algorithm was used to detect left/right gene expression differences within a group of sequential periods. The window size was set to span 4 periods.
Using the Illumina sequencing protocol, we also sequenced STC from both left and right hemispheres of 11 brains. After processing the data (see the supplemental experimental procedures for more details), a paired t-test was applied to determine if the expression level of a given gene in one hemisphere was significantly different from that in the other hemisphere, for fetal, postnatal, and adult individuals.
P-values from t-tests were transformed to FDR using the Benjamini and Hochberg method (Benjamini and Hochberg, 1995). For each gene, the fold difference (log2-transformed) between left and right hemisphere samples in each region was also calculated. An FDR of 0.1 was used as a cutoff to identify genes that are DEX between left and right hemispheres in each window.
Quantitative real-time RT-PCR
An aliquot of the total RNA that was previously extracted from each brain region was used for secondary validation through real-time PCR analysis. One μg of total RNA was used for cDNA synthesis using SuperScript III First-strand synthesis Supermix (Invitrogen) and subsequently diluted with nuclease-free water to 1 ng/μl cDNA. Gene-specific high-melting temperature primers for genes of interest were designed using NCBI/Primer-BLAST and expressed sequence information obtained from GenBank (NCBI). PCR reactions were conducted on an ABI 7900 Sequence Detection System (Applied Biosystems) using a hot start SYBR-green based method (Fast SYBR Green Master Mix, ABI) followed by melt curve analysis to verify specificity of the product. The Ct value (cycle number at threshold) was used to calculate the relative amount of mRNA molecules. The Ct value of each target gene was normalized by subtraction of the Ct value from housekeeping genes to obtain the ΔCt value. The relative gene expression level was shown as −ΔCt. All genes of interest were normalized to the housekeeping gene RPL32. For each qRT-PCR experiment, 3 technical replicates from 3 brains per stage (except in period 7, where we had 2) were used. Monkey brains used for qRT-PCR are listed in Table S1C.
Supplementary Material
Figure 6. Areal transcriptomes are globally symmetric at the population-level across fetal development and postnatal lifespan.
(A) Average distribution of hemispheric bias for all profiled neocortical areas through overlapping time points of development and adulthood. (B) A heat map shows the distribution of fold differences in expression of all analyzed genes between left and right hemispheres for each profiled neocortical area across time using the exon array technique. Each row corresponds to a 4-period sliding window (first row: 3–6; last row: 12–15). (C) RNA-seq analysis of inter-hemispheric bias. Volcano plots show the range of fold-differences and P-values of gene expression between left and right hemispheres in fetal, infancy to adolescence, and adulthood. SHANK3 is the only significantly DEX gene, during the fetal period (FDR threshold of 0.1), but with low fold ratio. See also Figure S6 and Table S5.
Acknowledgments
We thank M. Horn for helping with processing macaque tissue, Xuming Xu for help with data analyses, and the members of the Sestan laboratory for valuable comments. Human post-mortem tissue was obtained from sources listed in Supplemental Information. Support for predoctoral fellowships was provided by the Portuguese Foundation for Science and Technology (A.M.M.S.), the Croatian Science Foundation (G.S.), the National Science Foundation (DGE-1122492; K.A.M.), and the China Scholarship Council (Y.Z.). This work was supported by grants from the NIH (MH081896, MH089929, NS051869), the Kavli Foundation, NARSAD, the Foster-Davis Inc. Foundation, and by a James S. McDonnell Foundation Scholar Award (N.S.).
Footnotes
Supplemental information includes Supplemental Data, Supplemental Experimental Procedures, and Supplemental References, and can be found with the article online at www.cell.com/neuron.
Accession Numbers
Exon array and RNA-seq data have been deposited under NCBI BioProjects with accession numbers PRJNA134455 and PRJNA223168, respectively.
We declare that there is no conflict of interest that might be constructed to influence the results or their interpretation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahams B, Tentler D, Perederiy J, Oldham M, Coppola G, Geschwind D. Genome-wide analyses of human perisylvian cerebral cortical patterning. Proc Natl Acad Sci U S A. 2007;104:17849–17854. doi: 10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts K, Schleicher A, Ditterich A, Zilles K. Broca’s region: cytoarchitectonic asymmetry and developmental changes. J Comp Neurol. 2003;465:72–89. doi: 10.1002/cne.10829. [DOI] [PubMed] [Google Scholar]
- Bayer SA, Altman J. Directions in neurogenetic gradients and patterns of anatomical connections in the telencephalon. Prog Neurobiol. 1987;29:57–106. doi: 10.1016/0301-0082(87)90015-3. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Braak H, Braak E, Bohl J. Staging of Alzheimer-related cortical destruction. Eur Neurol. 1993;33:403–408. doi: 10.1159/000116984. [DOI] [PubMed] [Google Scholar]
- Broca P. Perte de la parole, ramollissement chronique et destruction partielle du lobe antérieur gauche. Bull Soc Anthropol. 1861;2:235–238. [Google Scholar]
- Brodmann K. Lokalisationslehre der Großhirnrinde in ihren Prinzipien dargestellt auf Grund des Zellenbaues. Leipzig: Barth; 1909. [Google Scholar]
- Chen CH, Panizzon MS, Eyler LT, Jernigan TL, Thompson W, Fennema-Notestine C, Jak AJ, Neale MC, Franz CE, Hamza S, et al. Genetic influences on cortical regionalization in the human brain. Neuron. 2011;72:537–544. doi: 10.1016/j.neuron.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi JG, Dooling EC, Gilles FH. Left-right asymmetries of the temporal speech areas of the human fetus. Arch Neurol. 1977;34:346–348. doi: 10.1001/archneur.1977.00500180040008. [DOI] [PubMed] [Google Scholar]
- Clancy B, Kersh B, Hyde J, Darlington RB, Anand KJ, Finlay BL. Web-based method for translating neurodevelopment from laboratory species to humans. Neuroinformatics. 2007;5:79–94. doi: 10.1385/ni:5:1:79. [DOI] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye TZ, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TJ, Walker MA, Eastwood SL, Esiri MM, Harrison PJ, Crow TJ. Anomalies of asymmetry of pyramidal cell density and structure in dorsolateral prefrontal cortex in schizophrenia. Brit J Psychiat. 2006;188:26–31. doi: 10.1192/bjp.bp.104.008169. [DOI] [PubMed] [Google Scholar]
- Dax M. Lésions de la moitié gauche de l’encéphale coincidant avec l’oubli des signes de la pensée: Lu au Congrès méridional tenu à Montpellier en 1836. Gaz Hebd Med Chir (Paris), 2me série. 1865;2:259–262. [Google Scholar]
- Domazet-Loso T, Tautz D. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature. 2010;468:815–818. doi: 10.1038/nature09632. [DOI] [PubMed] [Google Scholar]
- Faludi G, Mirnics K. Synaptic changes in the brain of subjects with schizophrenia. Int J Dev Neurosci. 2011;29:305–309. doi: 10.1016/j.ijdevneu.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flechsig P. Developmental (myelogenetic) localisation of the cerebral cortex in the human subject. Lancet. 1901;158:1027–1030. [Google Scholar]
- Fransson P, Skiöld B, Horsch S, Nordell A, Blennow M, Lagercrantz H, Åden U. Resting-state networks in the infant brain. Proc Natl Acad Sci U S A. 2007;104:15531–15536. doi: 10.1073/pnas.0704380104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzaniga MS, Sperry RW, Bogen JE. Some functional effects of sectioning cerebral commissures in man. Proc Natl Acad Sci U S A. 1962;48:1765–1769. doi: 10.1073/pnas.48.10.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind N, Galaburda AM. Cerebral lateralization - Biological mechanisms, associations, and pathology .3. A hypothesis and a program for research. Arch Neurol. 1985;42:634–654. doi: 10.1001/archneur.1985.04060070024012. [DOI] [PubMed] [Google Scholar]
- Geschwind N, Levitsky W. Human brain: left-right asymmetries in temporal speech region. Science. 1968;161:186–187. doi: 10.1126/science.161.3837.186. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Blumenthal J, Jeffries NO, Castellanos FX, Liu H, Zijdenbos A, Paus T, Evans AC, Rapoport JL. Brain development during childhood and adolescence: a longitudinal MRI study. Nat Neurosci. 1999;2:861–863. doi: 10.1038/13158. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Rapoport JL. Structural MRI of pediatric brain development: what have we learned and where are we going? Neuron. 2010;67:728–734. doi: 10.1016/j.neuron.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison-Uy SJ, Pleasure SJ. Wnt signaling and forebrain development. Cold Spring Harb Perspect Biol. 2012;4:a008094. doi: 10.1101/cshperspect.a008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes TL, Lewis DA. Hemispheric differences in layer III pyramidal neurons of the anterior language area. Arch Neurol. 1993;50:501–505. doi: 10.1001/archneur.1993.00540050053015. [DOI] [PubMed] [Google Scholar]
- Hill J, Inder T, Neil J, Dierker D, Harwell J, Van Essen D. Similar patterns of cortical expansion during human development and evolution. Proc Natl Acad Sci U S A. 2010;107:13135–13140. doi: 10.1073/pnas.1001229107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O, Johnston RJ, Jr, Chang S. Left-right asymmetry in the nervous system: the Caenorhabditis elegans model. Nature Rev Neurosci. 2002;3:629–640. doi: 10.1038/nrn897. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Prot. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huttenlocher PR, Dabholkar AS. Regional differences in synaptogenesis in human cerebral cortex. J Comp Neurol. 1997;387:167–178. doi: 10.1002/(sici)1096-9861(19971020)387:2<167::aid-cne1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Ip BK, Wappler I, Peters H, Lindsay S, Clowry GJ, Bayatti N. Investigating gradients of gene expression involved in early human cortical development. J Anat. 2010;217:300–311. doi: 10.1111/j.1469-7580.2010.01259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanovic D, Geschwind DH, Mane SM, State MW, Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MH. Functional brain development in humans. Nat Rev Neurosci. 2001;2:475–483. doi: 10.1038/35081509. [DOI] [PubMed] [Google Scholar]
- Judas M, Sedmak G, Kostovic I. The significance of the subplate for evolution and developmental plasticity of the human brain. Front Hum Neurosci. 2013;7:423. doi: 10.3389/fnhum.2013.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas JH. The evolution of neocortex in primates. Prog Brain Res. 2012;195:91–102. doi: 10.1016/B978-0-444-53860-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinka AT, Varga KM, Gerrard DT, Preibisch S, Corcoran DL, Jarrells J, Ohler U, Bergman CM, Tomancak P. Gene expression divergence recapitulates the developmental hourglass model. Nature. 2010;468:811–814. doi: 10.1038/nature09634. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprian G, Langs G, Brugger PC, Bittner M, Weber M, Arantes M, Prayer D. The prenatal origin of hemispheric asymmetry: an in utero neuroimaging study. Cereb Cortex. 2011;21:1076–1083. doi: 10.1093/cercor/bhq179. [DOI] [PubMed] [Google Scholar]
- Kennedy H, Dehay C. Self-organization and interareal networks in the primate cortex. Prog Brain Res. 2012;195:341–360. doi: 10.1016/B978-0-444-53860-4.00016-7. [DOI] [PubMed] [Google Scholar]
- Khazipov R, Luhmann HJ. Early patterns of electrical activity in the developing cerebral cortex of humans and rodents. Trends Neurosci. 2006;29:414–418. doi: 10.1016/j.tins.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Johnson MB, Dube U, Shim S, Rasin MR, Sousa AM, Fertuzinhos S, Chen JG, Arellano JI, et al. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell. 2012;149:899–911. doi: 10.1016/j.cell.2012.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert N, Lambot MA, Bilheu A, Albert V, Englert Y, Libert F, Noel JC, Sotiriou C, Holloway AK, Pollard KS, et al. Genes expressed in specific areas of the human fetal cerebral cortex display distinct patterns of evolution. PloS One. 2011;6:e17753. doi: 10.1371/journal.pone.0017753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. Fast R functions for robust correlations and hierarchical clustering. J Stat Softw. 2012;46:1–17. [PMC free article] [PubMed] [Google Scholar]
- Lidow MS, Goldman-Rakic PS, Rakic P. Synchronized overproduction ofneurotransmitter receptors in diverse regions of the primate cerebral cortex. Proc Natl Acad Sci U S A. 1991;88:10218–10221. doi: 10.1073/pnas.88.22.10218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus IC, Bryden MP. Geschwind’s theory of cerebral lateralization: developing a formal, causal model. Psychol Bull. 1991;110:237–253. doi: 10.1037/0033-2909.110.2.237. [DOI] [PubMed] [Google Scholar]
- Molnar Z, Clowry G. Cerebral cortical development in rodents and primates. Prog Brain Res. 2012;195:45–70. doi: 10.1016/B978-0-444-53860-4.00003-9. [DOI] [PubMed] [Google Scholar]
- O’Leary DD, Sahara S. Genetic regulation of arealization of the neocortex. Curr Opin Neurobiol. 2008;18:90–100. doi: 10.1016/j.conb.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao XH, Hill RS, Bodell A, Chang BS, Basel-Vanagaite L, Straussberg R, Dobyns WB, Qasrawi B, Winter RM, Innes AM, et al. G protein-coupled receptor-dependent development of human frontal cortex. Science. 2004;303:2033–2036. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- Preuss TM. The human brain: rewired and running hot. Ann N Y Acad Sci. 2011;1225(Suppl 1):E182–191. doi: 10.1111/j.1749-6632.2011.06001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- Rakic P, Bourgeois JP, Eckenhoff MF, Zecevic N, Goldman-Rakic PS. Concurrent overproduction of synapses in diverse regions of the primate cerebral cortex. Science. 1986;232:232–235. doi: 10.1126/science.3952506. [DOI] [PubMed] [Google Scholar]
- Rapoport JL, Gogtay N. Brain neuroplasticity in healthy, hyperactive and psychotic children: Insights from neuroimaging. Neuropsychopharmacology. 2007;33:181–197. doi: 10.1038/sj.npp.1301553. [DOI] [PubMed] [Google Scholar]
- Rash BG, Grove EA. Area and layer patterning in the developing cerebral cortex. Curr Opin Neurobiol. 2006;16:25–34. doi: 10.1016/j.conb.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Sanides F. Comparative architectonics of the neocortex of mammals and their evolutionary interpretation. Ann N Y Acad Sci. 1969;167:404–423. [Google Scholar]
- Sowell ER, Peterson BS, Thompson PM, Welcome SE, Henkenius AL, Toga AW. Mapping cortical change across the human life span. Nat Neurosci. 2003;6:309–315. doi: 10.1038/nn1008. [DOI] [PubMed] [Google Scholar]
- Sun T, Patoine C, Abu-Khalil A, Visvader J, Sum E, Cherry TJ, Orkin SH, Geschwind DH, Walsh CA. Early asymmetry of gene transcription in embryonic human left and right cerebral cortex. Science. 2005;308:1794–1798. doi: 10.1126/science.1110324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Walsh CA. Molecular approaches to brain asymmetry and handedness. Nat Rev Neurosci. 2006;7:655–662. doi: 10.1038/nrn1930. [DOI] [PubMed] [Google Scholar]
- Sur M, Rubenstein JL. Patterning and plasticity of the cerebral cortex. Science. 2005;310:805–810. doi: 10.1126/science.1112070. [DOI] [PubMed] [Google Scholar]
- Taylor DC. Differential rates of cerebral maturation between sexes and between hemispheres - evidence from epilepsy. Lancet. 1969;2:140–142. doi: 10.1016/s0140-6736(69)92445-3. [DOI] [PubMed] [Google Scholar]
- Thatcher RW, Walker RA, Giudice S. Human cerebral hemispheres develop at different rates and ages. Science. 1987;236:1110–1113. doi: 10.1126/science.3576224. [DOI] [PubMed] [Google Scholar]
- Toga AW, Thompson PM. Mapping brain asymmetry. Nat Rev Neurosci. 2003;4:37–48. doi: 10.1038/nrn1009. [DOI] [PubMed] [Google Scholar]
- Tripodi M, Filosa A, Armentano M, Studer M. The COUP-TF nuclear receptors regulate cell migration in the mammalian basal forebrain. Development. 2004;131:6119–6129. doi: 10.1242/dev.01530. [DOI] [PubMed] [Google Scholar]
- Vasung L, Huang H, Jovanov-Milosevic N, Pletikos M, Mori S, Kostovic I. Development of axonal pathways in the human fetal fronto-limbic brain: histochemical characterization and diffusion tensor imaging. J Anat. 2010;217:400–417. doi: 10.1111/j.1469-7580.2010.01260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernicke C. Der Aphasische Symptomenkomplex: Eine Psychologische Studie auf Anatomischer Basis. Breslau: Max Cohn & Weigert; 1874. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.