Abstract
Purpose
Small cell neuroendocrine carcinoma of the prostate is likely to become increasingly common with recent advances in pharmacologic androgen suppression. Thus, developing molecular markers of small cell differentiation in prostate cancer will be important to guide diagnosis and therapy of this aggressive tumor.
Experimental Design
We examined the status of RB1, TP53 and PTEN in prostatic small cell and acinar carcinomas via immunohistochemistry (IHC), copy number alteration analysis and sequencing of formalin fixed paraffin-embedded specimens.
Results
We found Rb protein loss in 90% (26/29) of small cell carcinoma cases with RB1 allelic loss in 85% (11/13) of cases. Of acinar tumors occurring concurrently with prostatic small cell carcinoma, 43% (3/7) showed Rb protein loss. In contrast, only 7% (10/150) of primary high grade acinar carcinomas, 11% (4/35) of primary acinar carcinomas with neuroendocrine differentiation, and 15% (2/13) of metastatic castrate resistant acinar carcinomas showed Rb protein loss. Loss of PTEN protein was seen in 63% (17/27) of small cell carcinomas, with 38% (5/13) showing allelic loss. By IHC, accumulation of p53 was observed in 56% (14/25) of small cell carcinomas, with 60% (6/10) of cases showing TP53 mutation.
Conclusions
Loss of RB1 by deletion is a common event in prostatic small cell carcinoma and can be detected by validated IHC assay. As Rb protein loss rarely occurs in high grade acinar tumors, these data suggest that Rb loss is a critical event in the development of small cell carcinomas and may be a useful diagnostic and potential therapeutic target.
Keywords: Prostatic adenocarcinoma, small cell carcinoma, tumor suppressor, RB1, TP53, PTEN
Introduction
Small cell neuroendocrine carcinoma is an unusual and aggressive subtype of prostate cancer, accounting for 0.5 to 2% of all untreated primary tumors (1, 2). Small cell carcinoma represents an extreme example of neuroendocrine differentiation in prostate cancer. The other end of the spectrum encompasses more typical acinar primary tumors, where scattered rare cells expressing neuroendocrine markers can be found in 10–100% of adenocarcinomas of the prostate (3, 4). Interestingly, autopsy studies have suggested that more extensive neuroendocrine differentiation (including small cell carcinoma) is present in up to 25% of lethal, widely metastatic cases (3–5). This apparent selection for the neuroendocrine phenotype in end-stage disease is likely due to the fact that tumors with extensive neuroendocrine differentiation, like small cell carcinoma, are almost invariably castration-resistant and typically lack expression of both androgen receptor and downstream targets of the androgen signaling axis (2, 4, 6–12). Indeed, it is anticipated that recent improvements in pharmacologic options for androgen suppression therapy such as abiraterone and enzalutamide, that are even more effective at blocking androgen receptor signaling than traditional methods, will lead to a relative upsurge in small cell carcinoma cases in the near future (13), making it especially important to fully understand the molecular and immunophenotypic underpinnings of this tumor type.
Both de novo and treatment-related small cell carcinoma cases commonly occur concurrently with typical acinar adenocarcinoma (1, 2). We and others have shown limited molecular evidence of a clonal relationship between the two components based on the concordant presence of ERG gene rearrangements or TP53 mutations (12, 14–17), however it remains unclear how small cell carcinoma differs at a molecular level from its acinar counterpart. Indeed, delineating the unique molecular features of small cell carcinoma that distinguish it from acinar carcinoma would be useful for both diagnosis and potential therapy of this aggressive subtype. Accurately distinguishing small cell carcinoma from high grade (Gleason pattern 4 or 5) acinar prostate carcinoma can be difficult based on morphology alone, and current immunohistochemical markers for neuroendocrine differentiation may be non-specific (18). However, this distinction is extremely consequential for clinical patient management since small cell carcinomas are often resistant to androgen deprivation therapy at the outset, and are at least temporarily sensitive to cisplatin-based chemotherapeutic regimens in contrast to acinar carcinoma (2, 6–11). Thus, identifying additional molecular differences between small cell carcinoma and acinar carcinoma would not only aid in accurate diagnosis of these lesions, but also provide additional potential therapeutic targets for this currently uniformly lethal tumor type.
Although prior molecular studies of prostatic small cell carcinomas have been limited, a number of studies of small cell carcinomas arising in other organs have suggested that loss of the RB1 and TP53 genes are extremely common events in this tumor type in humans, and these changes are sufficient to generate small cell carcinomas in the lung of mice (19–21). Intriguingly, prostate-specific inactivation or deletion of these two genes in the mouse is sufficient to result in highly penetrant metastatic carcinoma with neuroendocrine features (22, 23). In prostatic acinar carcinomas, loss of the RB1 and TP53 tumor suppressors does occur in a subset of primary tumors, however PTEN is typically the most commonly lost tumor suppressor in this setting (24–26). Here, we comprehensively examined the protein and genomic status of RB1, PTEN and TP53 in a series of small cell and acinar carcinomas. We report that RB1 loss occurs virtually uniformly in small cell carcinoma of the prostate, a finding that may be useful in the future for both diagnosis and therapy of this tumor.
Materials and Methods
Tissue Selection
A total of 29 specimens from patients with prostatic small cell carcinoma were retrieved from the surgical pathology and consultation files of the Johns Hopkins Hospitals from 1994–2008 as previously reported (12). 19 cases (66%) were transurethral resections of the prostate (TURP), 8 (27%) were bladder, prostate or rectal biopsies, and 1 (3%) was a radical prostatectomy and 1 (3%) was a liver metastasis. (Table 1). Prostatic origin was documented for the majority of cases using the following criteria: in 76% (22/29) of cases, a concurrent or prior history of prostatic acinar carcinoma was established, while 7% (2/29) of cases were diagnosed by positive PSA (prostate specific antigen) immunostaining and 3% (1/29) had a documented negative cystoscopy. Thus, in the vast majority of cases, the small cell carcinoma arose in the setting of a current or prior acinar prostatic adenocarcinoma. Although 97% (28/29) of the tissue samples were from the prostate or contiguous sites (ie, most likely non-metastatic specimens), due to the fact that the majority of these cases were consultations, adequate clinical history was not available in 16 (55%) cases to determine which small cell carcinomas arose in the context of prior hormonal therapy (t-NEPC). Overall, of the cases in which we did have clinical data, 7 (23%) patients had no prior history of therapy for prostate cancer and represented new diagnoses, 3 (10%) had a history of prior radiation treatment for prostate cancer, and 3 (10%) of patients had a documented history of prior treatment with hormonal therapies.
Table 1.
RB, PTEN and p53 immunohistochemistry results in small cell carcinoma (SCC) and associated acinar carcinoma (ACa) component where present. n=negative, p=positive. TURP = transurethral resection of prostate; RP=radical prostatectomy.
| Case #: |
Specimen type: |
Prostatic SCC confirmed by: |
RB | PTEN | P53 | |||
|---|---|---|---|---|---|---|---|---|
| SCC | ACa | SCC | ACa | SCC | ACa | |||
| 1 | TURP | concurrent ACa | n | p | n | n | n | p |
| 4 | TURP | PSA | n | n | p | |||
| 5 | TURP | concurrent ACa | n | n | p | |||
| 6 | rectal biopsy | history of ACa | n | n | p | |||
| 7 | TURP | unknown | n | p | n | |||
| 8 | TURP | concurrent ACa | n | p | p | p | p | p |
| 9 | rectal biopsy | history of ACa | n | |||||
| 10 | TURP | concurrent ACa | n | p | p | |||
| 11 | prostate biopsy | unknown | n | n | p | |||
| 13 | TURP | unknown | n | n | p | |||
| 14 | TURP | concurrent ACa | n | n | n | |||
| 15 | TURP | PSA | p | n | n | |||
| 16 | TURP | history of ACa | n | p | n | |||
| 17 | RP | concurrent ACa | n | p | p | n | p | p |
| 18 | prostate biopsy | concurrent ACa | n | n | ||||
| 19 | TURP | negative cystoscopy | n | n | n | |||
| 20 | bladder biopsy | concurrent ACa | p | p | n | |||
| 21 | TURP | concurrent ACa | n | p | n | |||
| 23 | TURP | unknown | p | p | ||||
| 24 | bladder biopsy | concurrent ACa | n | n | n | n | n | n |
| 25 | TURP | concurrent ACa | n | p | n | |||
| 26 | TURP | concurrent ACa | n | p | n | n | p | p |
| 28 | TURP | concurrent ACa | n | n | p | p | p | |
| 29 | TURP | concurrent ACa | n | n | n | n | p | n |
| 30 | bladder biopsy | history of ACa | n | n | p | |||
| 31 | bladder biopsy | history of ACa | n | n | p | |||
| 32 | TURP | history of ACa | n | n | n | |||
| 33 | TURP | history of ACa | n | n | p | |||
| 34 | liver | history of ACa | n | |||||
A tissue microarray (TMA) was manually constructed from these cases as previously described (12). In each case, a minimum of three 1.0 mm cores were punched from the small cell carcinoma component, the acinar carcinoma component (when present) and the paired benign prostatic tissue, with 3 to 18 cores from each patient represented on the array. 24% (7/29) of cases had a concurrent acinar carcinoma component present on the TMA.
For assessment of Rb protein expression in high grade acinar prostatic carcinoma cases unassociated with a small cell carcinoma component, 170 cases from previously described TMAs comprising several cohorts of high grade, high stage prostate tumors were utilized (27–29). Rb protein expression was also assessed on standard sections of an additional 15 cases of non-small cell primary prostatic carcinoma with neuroendocrine differentiation (including 6 cases of large cell neuroendocrine carcinoma) from the consultation files of the Johns Hopkins Hospital. Finally, Rb immunostaining was performed on standard sections of 13 samples of metastatic castrate resistant prostate cancer that were previously characterized for RB1 copy number alteration using a high density single nucleotide polymorphism array (Affymetrix 6.0 SNP microarray) (Supplementary Table 1) (30). For validation of the nCounter Cancer Copy Number Variation panel at the PTEN and RB1 loci (described below), DNA derived from frozen tissue samples from 5 of these cases of metastatic acinar prostatic carcinoma in this series was utilized (30).
Immunohistochemistry
Tissue sections were deparaffinized, rehydrated, and briefly equilibrated in water. Antigen unmasking was performed by either steaming in HTTR (Target Retrieval Solution; DAKO, Glostrup, Denmark) for 50 minutes (p53, Rb) or in EDTA for 45 minutes (PTEN). Endogenous peroxidase activity was quenched by incubation with peroxidase block for 5 minutes at RT. Slides were incubated with a mouse monoclonal anti-human p53 antibody (Clone DO-7; Dako, Glostrup, Denmark;1:400 dilution), a mouse monoclonal anti-human Rb antibody (Cell Signaling, Danvers, MA; 1: 100 dilution), or a rabbit anti-human PTEN antibody (Clone D4.3 XP; Cell Signaling, Danvers, MA; 1: 100 dilution). A horseradish peroxidase-labeled polymer (PowerVision, Leica Microsystems, Bannockburnm IL) was applied for 30 minutes at RT. Signal detection was performed using 3,3'-diaminobenzidine tetrahydrochloride (DAB) as the chromagen. Slides were counterstained with hematoxylin, dehydrated, and mounted.
Validation and Scoring of Immunohistochemistry
Rb, p53 and PTEN immunostains were scored in small cell carcinoma and the associated acinar carcinoma component by a urologic pathologist (TLL). Based on controls described in the Results section below, a case was considered to have lost Rb protein if any TMA spot showed loss (0+ staining) in >95% of tumor nuclei. Positive nuclear staining in surrounding endothelial cells provided an internal control in most cases and a case was excluded if it lacked this endothelial staining.
The immunohistochemical protocol and interpretation of staining for cytoplasmic PTEN protein detection were extensively validated using multiple genetic controls (cell lines and tissues) as described previously (31). A case was considered to have lost PTEN protein if the intensity of staining in tumor cell cytoplasm was markedly decreased or entirely negative across all tumor cells compared with the surrounding benign glands and/or stroma in any given TMA spot. A given spot was dropped from the analysis if these benign areas lacked PTEN staining (internal positive control).
Accumulation of p53 protein is frequently associated with deleterious mutations in this tumor suppressor gene which extend the protein’s half-life. The immunohistochemical staining protocol for p53 was validated using prostate cancer cell lines with known p53 mutations or lines that are known to be null for p53 (32). Using this protocol, DU145 cells which have two TP53 mutations stain positively, and PC3 cells which are TP53-null and stain negatively. While there is no strict cut-off for the amount of p53 immunostaining that provides definitive evidence of an underlying mutation in TP53, a number of studies, predominantly in bladder cancer, used 10% as a cut-off. In the current study, we used an even more stringent cut-off: positive p53 overexpression was scored if any spot in a case showed strong expression (3–4+) in >50% of tumor cells.
DNA preparation
For 13 of the 29 prostatic small cell carcinoma samples there was adequate tissue available for DNA preparation. Between one and three 10 µm sections of formalin-fixed paraffin-embedded tumor tissue were de-paraffinized and briefly rehydrated. Eleven tumor samples (11/13) were enriched for tumor by macrodissection using a 26 gauge needle under stereomicroscopic visualization guided by a continguous H&E section. Two tumor samples (2/13) were subjected to laser capture microdissection (LCM) using a Leica P.A.L.M. laser microdissection system (Leica Microsystems, Wetzlar, Germany) according to the manufacturer’s directions. All tumor samples were at least 80% pure by visual estimation using a contiguous H&E section. For each sample, genomic DNA was extracted by using QIAamp DNA Micro kit (Qiagen) following manufacturer’s directions.
Copy Number Alteration Analysis
300–500 ng of genomic DNA from each small cell carcinoma sample was used according to the manufacturer’s directions for nanoString nCounter® Cancer Copy Number Assay (nanoString, Seattle, WA). This assay, optimized for DNA prepared from formalin-fixed paraffin embedded tissue samples, detects copy number changes in 86 genes which are commonly amplified or deleted in cancer using 255 gene-specific probes and an additional 54 probes for invariant regions of the genome. DNA from 5 additional samples of metastatic acinar prostatic carcinoma (for which copy number alterations have previously been reported by high density SNP microarray) was also used for a validation set. Data analysis was performed using the nSolver analysis software (nanoString, Seattle, WA). Raw counts were first normalized to the average counts across probes to invariant regions to normalize for any differences in DNA input. The copy number for each probe was then normalized to the reference sample (Human blood genomic DNA; Roche) and the average copy number call reported for each gene with a standard deviation (0–0.5 = 0 copies; 0.6–1.4 = 1 copy, 1.6–2.4 = 2 copies; etc). Probes for TP53 were unintentionally omitted from the version 1 nanoString assay, thus TP53 copy number was not available for the present analyses.
Next Generation Sequencing (NGS)
Ion Torrent Ion AmpliSeq™ Cancer Hotspot Panel V2 (Life Technologies, Carlsbad, CA) was used for sequencing hotspot regions in 50 frequently mutated tumor suppressor and oncogenes, covering approximately 2,800 COSMIC mutations in total. For this manuscript, we focused on exonic regions of RB1 (the panel included 10 amplicons covering roughly 10 out of 27 exons: exon 4, 6, 10, 11, 14, 17, 18, 20, 21, 22), TP53 (8 amplicons covering roughly 7 out of 11 exons: exon 2, 4, 5, 6, 7, 8, 10), and PTEN (8 amplicons covering roughly 6 out of 9 exons: exon 1, 3, 5, 6, 7, 8). Genomic DNA (10 ng input) from 13 out of the 30 prostatic small cell carcinoma cases was selected for the assay and Ion 318™ chips were used for greater sample multiplexing. Ion Torrent Variant Caller, Ion Reporter, and Integrated Genomic Viewer (IGV, Broad Institute) were used for analyzing target mutations. Variant frequency was determined by the ratio of variant allele reads to total reads.
Sanger Sequencing
Direct sequencing for exons 4, 5, 7 and 8 of TP53 was performed on PCR-amplified DNA from 13 paraffin embedded prostatic small cell carcinoma samples using established primers and protocols available at the IARC p53 Database Website (http://p53.iarc.fr/Download/TP53_DirectSequencing_IARC.pdf). Both forward and reverse strands of the PCR product were subjected to sequencing. Sequence variants were verified on both strands as well as by sequencing an independent PCR reaction from the same sample. Sequence analysis was conducted using Mutation Surveyor® (SoftGenetics, State College, PA). The predicted effect of the mutation (deleterious, non-deleterious) was determined using the IARC p53 Database (http://p53.iarc.fr/TP53GeneVariations.aspx).
Results
Validation of Rb immunohistochemistry assay
Immunohistochemical staining for Rb protein was validated by blindly scoring Rb protein status on a TMA constructed from 72 formalin-fixed paraffin-embedded cell lines, predominantly from the NCI-60 panel (31). Cell lines and samples were scored as negative for Rb protein if >95% of cells in all spots lacked Rb (0+ staining). In all, 5 (7%) cell lines completely lacked Rb protein by immunohistochemistry (Figure 1). These included the only two cell lines on the TMA known to have complete homozygous deletions involving RB1: BT549 cells have a homozygous 343 base pair deletion in the gene, and SF-539 cells have a 4 base pair deletion resulting in a frameshift mutation (33). Of the 3 other cell lines that lacked Rb protein by immunohistochemical assay (SNB-75, OVCAR-8 and NCI-adr-Res), none have known mutations in the RB1 gene, however all have previously been shown to be negative for Rb protein by western blotting or reverse phase protein arrays (33–35). In addition, 7 cell lines showed reduced, but not absent, immunohistochemical staining for Rb protein (CAKI-1, DU-145, HCC-2998, Hep3B, MDA-PCA2b, NCI-H522, PrSc, and RWPE cells) defined as 1+ weak staining in >5% of cells. Of these, DU-145 cells are known to have homozygous deletion resulting in a truncation of the protein at exon 21, and HCC-2998 cells have a heterozygous nonsense mutation in the gene (32, 33).
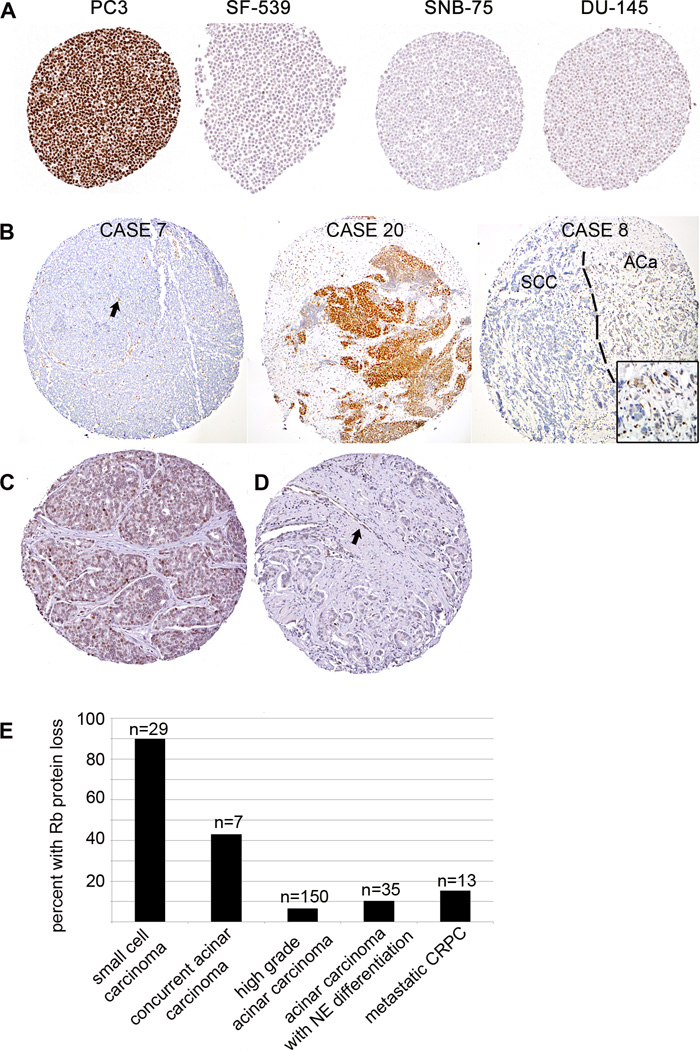
Figure 1.
(A) Immunostaining for Rb was validated using a tissue microarray made from the NCI-60 cell line panel. PC3 cells show high Rb expression with wildtype RB1. SF-539 cells are negative for Rb protein and are known have a 4 base pair deletion resulting in a frameshift mutation. SNB-75 cells are not known to have a homozygous deletion in RB1, however numerous previous studies have shown absence of the Rb protein by western blotting in this cell line. DU-145 cells have a homozygous deletion resulting in a truncation of the protein at exon 21 and show very low Rb staining by immunohistochemistry. (B) Representative Rb immunostaining results from Case 7 shows negative staining in tumor cells with retained endothelial staining (arrow) as an internal positive control. Case 20 shows strongly positive staining. Case 8 shows negative staining in the small cell carcinoma (SCC) component and positive staining in the adjacent acinar carcinoma component (ACa). Images in this panel were assembled from a mosaic of higher power images. (C) Representative positive Rb immunostaining results in a high grade acinar carcinoma unassociated with small cell carcinoma. (D) Representative negative Rb immunostaining in a high grade acinar carcinoma unassociated with small cell carcinoma. Endothelial cells (arrow) provide an internal positive control. (E) Graphical representation of percent of cases with Rb protein loss comparing small cell carcinomas, acinar carcinomas associated with small cell carcinoma and high grade acinar carcinomas unassociated with small cell carcinoma. Rb loss is most frequent in small cell carcinomas and rarely seen in high grade acinar carcinomas unassociated with small cell carcinoma, while acinar carcinomas occurring concurrently with small cell carcinoma show an intermediate rate of Rb protein loss.
Rb protein expression in small cell carcinoma and high grade acinar carcinoma cases
90% (26/29) of prostatic small cell carcinoma cases showed Rb protein loss, compared to only 43% (3/7) of concurrent acinar carcinoma cases (Figure 1, 2, Table 1). In cases with concurrent small cell and acinar carcinoma components, 57% (4/7) showed concordance of the small cell carcinoma and acinar carcinoma components for Rb status, while 43% (3/7) showed presence of Rb protein in the acinar carcinoma component with loss of Rb in the small cell carcinoma component. Interestingly, where present, the acinar component typically showed weak or reduced staining similar to DU-145 cells (Figure 1). Cases where the acinar component had loss of Rb protein and the small cell carcinoma component retained the protein were not identified.
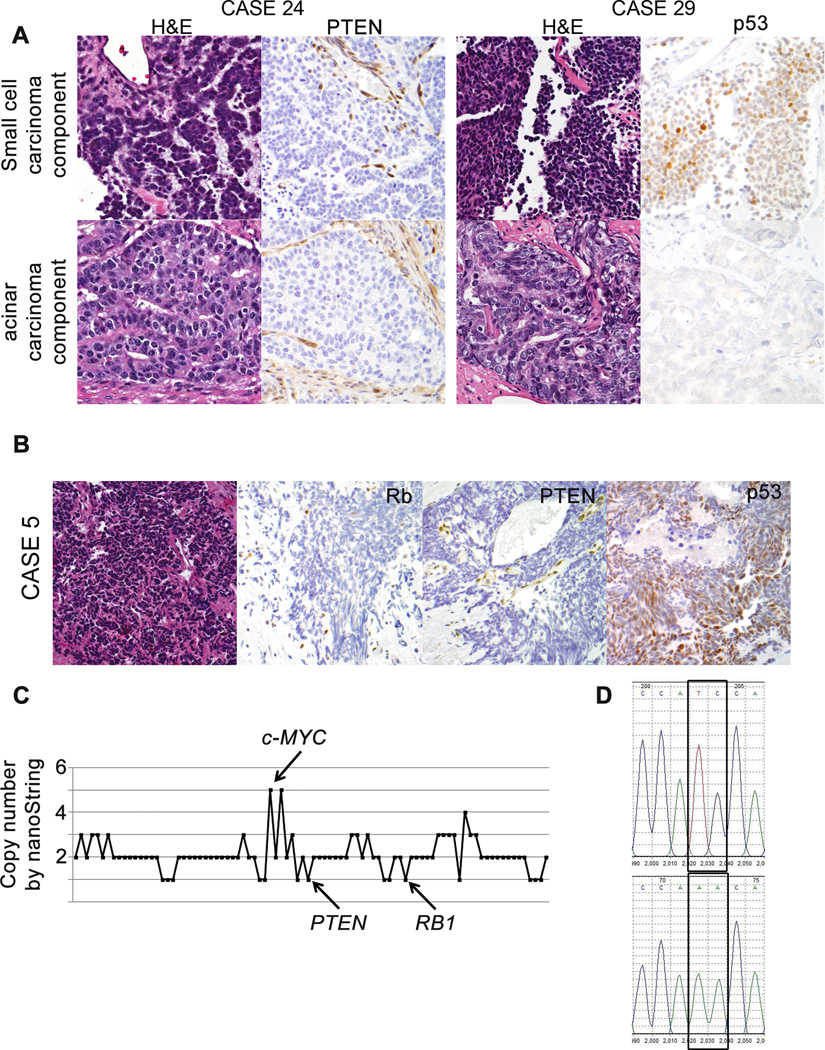
Figure 2.
Immunohistochemical assessment of PTEN and p53 tumor suppressor loss in small cell carcinomas and their associated acinar carcinoma component. (A) Case 24 shows PTEN loss in both the small cell and associated acinar carcinoma, with retained endothelial/stromal staining for PTEN providing an internal positive control. Case 29 shows p53 accumulation in the small cell component only. (B) Immunohistochemistry for Rb, PTEN and p53 in Case 5 demonstrates loss of Rb and PTEN, with over-expression for p53. (C) Copy number by nanoString nCounter Cancer CNV Panel across 86 probed genes for Case 5. c-MYC amplification (5 copies), and hemizygous PTEN and RB1 loss are present. (D) Sanger sequencing chromatogram for TP53 from Case 5 shows a two base pair missense mutation (exon 7: c.695_696TC>AA) predicted to deleteriously affect the DNA binding domain.
To assess whether the very high frequency of Rb protein loss is unique to small cell differentiation in prostate cancer, we assessed Rb expression in a spectrum of high grade primary acinar carcinomas unassociated with small cell carcinoma, primary carcinomas with neuroendocrine differentiation and metastatic castrate resistant acinar carcinomas. First, we scored Rb expression in an additional 150 acinar carcinoma cases enriched for high grade disease and occurring without concurrent small cell carcinoma that have been described elsewhere (27–29). Gleason score was available in 108 (72%) of these cases: 11% (n=12) Gleason 6, 26% (n=28) Gleason 7, 32% (n=35) Gleason 8, 29% (n=31) Gleason 9 and 2% (n=2) Gleason 10. Of these acinar tumors, only 7% (10/150) showed Rb protein loss (Figure 1).
In addition, we examined primary non-small cell acinar carcinomas with evidence of neuroendocrine differentiation. Since the pathologic classification of non-small cell neuroendocrine differentiation in prostate carcinoma was the topic of a recent consensus meeting and is currently undergoing revision (Mahul Amin, Mark Rubin, Himisha Beltran, Tamara Lotan, Juan-Miguel Mosquera, Victor Reuter, Brian Robinson, Patricia Troncoso and Jonathan Epstein, in preparation), we utilized several definitions for this entity. First, we examined the frequency of Rb protein loss in primary prostate tumors diagnosed as “high grade prostatic adenocarcinoma with neuroendocrine differentiation.” Morphologically, these cases fell short of a diagnosis of small cell carcinoma, but showed focal architectural or cytological features suggestive of neuroendocrine differentiation, combined with non-focal (generally >20%) expression of either chromogranin or synaptophysin. Of these cases, 22% (2/9) showed Rb protein loss (Supplementary Figure S1). Next we examined cases of large cell neuroendocrine prostate carcinoma, characterized by large nests of tumor cells with peripheral palisading and often geographic necrosis, where the cytology is that of non-small cell carcinoma (prominent nucleoli, vesicular clumpy chromatin and/or large cell size and abundant cytoplasm). These tumors generally express at least one neuroendocrine marker and frequently lack prostatic/AR axis signaling markers. Of these rare cases, 17% (1/6) showed Rb protein loss (Supplementary Figure S1).
Finally, we examined Rb protein status in usual prostatic adenocarcinomas with neuroendocrine differentiation established solely by non-focal immunohistochemical expression of chromogranin and/or synaptophysin. Since as many as 100% of prostate acrinar carcinomas can show focal expression of neuroendocrine markers, we arbitrarily defined this category as cases where >20% of the cells expressed chromogranin and/or synaptophysin. Of the 170 high grade acinar prostatic adenocarcinomas assayed for non-focal chromogranin and synaptophysin positivity, 12% (20/170) met the criterion of neuroendocrine marker staining in >20% of the cells in at least one TMA spot. Of these cases, only 5% (1/20) showed Rb protein loss.
To determine whether Rb protein loss is specific to small cell carcinoma or simply a result of progression of disease, we also examined Rb expression in a series of metastatic castrate resistant acinar prostatic carcinoma samples. Multiple metastases from these patients have been previously characterized for copy number alterations at the RB1 locus using high density SNP microarray (30). Importantly, however, for Rb IHC studies, we did not have access to the identical tissue specimen characterized by SNP microarray, although in 7 cases (54%) we were able to analyze tissues from the same metastatic site as had been characterized by SNP microarray. Overall, 15% (2/13) of cases showed Rb protein loss (Supplementary Table 1). By SNP microarray, 23% (3/13) of the castrate resistant cases had homozygous loss of RB1, although in two out of three of these cases, homozygous RB1 loss was heterogeneous among the 5 metastatic sites examined, strongly suggesting that complete loss of RB1 is a late event in metastatic progression. Concordant with this, Rb protein was lost only in the single case with homozygous deletion in all 4 metastases examined, and was actually only focally lost in this metastasis, suggesting both inter- and intra-metastatic heterogeneity in Rb status is common in advanced prostate cancer (Supplementary Figure S1). An additional 38% (5/13) of the castrate resistant metastases had hemizygous deletion of RB1. Of these cases, 100% (5/5) had hemizygous loss in all metastatic sites examined by SNP array and 20% (1/5) had Rb protein loss, albeit in a metastasis from a separate site as the ones characterized by SNP microarray. The remaining 5 cases (38%) were copy-number neutral for RB1, including 2 cases that had deletion of one allele with amplification of the remaining allele (loss of heterozygosity). Of these, all were positive for Rb protein.
PTEN and p53 protein expression in small cell carcinoma cases
63% (17/27) of small cell carcinoma cases and 71% (5/7) of concurrent acinar carcinoma cases displayed PTEN protein loss (Figure 2, Table 1). 86% (6/7) cases showed concordance in small cell carcinoma and acinar carcinoma components for PTEN status. One case showed retained PTEN protein in the small cell component and loss in the acinar component. 56% (14/25) of small cell carcinoma cases and 66% (4/6) of concurrent acinar carcinoma cases showed p53 accumulation (Figure 2, Table 1). 66% (4/6) of cases showed concordance in the small cell carcinoma and acinar carcinoma components for p53 protein status.
Assessment of RB1 and PTEN copy number alteration
To determine the molecular mechanism of the high rate of Rb and PTEN protein loss among small cell carcinoma cases, we next assessed RB1 and PTEN gene copy number in 13 of the 29 small cell carcinoma cases with adequate tissue available for DNA isolation. In order to validate the nanoString nCounter® Cancer Copy Number Assay for this purpose, we first tested the assay on DNA purified from 5 previously described samples of metastatic prostatic adenocarcinoma for which copy number alterations across the genome have been reported by high density SNP microarray (Affymetrix Genome Wide Human SNP Array 6.0; 30). For 4 of these samples, the correlation between the PTEN copy number as assessed by nanoString assay and the Affymetrix SNP array was high (R2=0.85, Supplementary Figure S2). For the fifth sample, a partial homozygous deletion encompassing exons 2–9 of the gene was detected by both assays, with only the most 5’ of the 3 nanoString probes to PTEN hybridizing to this sample (data not shown). Similarly, at the RB1 locus, the copy number data from the nanoString CNV assay and the Affymetrix SNP array was highly correlated across all 5 samples (R2=0.92).
Of the prostatic small cell carcinoma samples, RB1 allelic loss was detected in 85% (11/13), of which 73% (8/11) showed hemizygous loss and 27% (3/11) of cases showed homozygous loss at RB1 (Table 2). Of the cases with RB1 allelic loss, 100% (11/11) showed Rb protein loss by immunohistochemistry (Table 3A, Figure 2). Of the cases without RB1 loss, 50% (1/2) showed Rb protein loss. At the PTEN locus, 38% (5/13) of prostatic small cell carcinoma cases showed allelic loss, with 80% (4/5) showing hemizygous loss and 20% (1/5) showing homozygous loss. Of the cases with allelic loss, 80% (4/5) showed PTEN protein loss, while only 50% (4/8) of cases without PTEN allelic loss showed PTEN protein loss (Table 3B).
Table 2.
Association of protein status, copy number status and mutation status for Rb, PTEN and p53, by case.
| Case #: |
Dissection method: |
Rb IHC |
RB1 Copy # |
RB1 sequence |
PTEN IHC |
PTEN copy # |
PTEN sequence |
mutation type |
mutation effect |
p53 IHC |
TP53 sequence |
mutation type |
mutation effect |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | LCM | n | 1 | NA | n | 1 | NA | p | ex 7: c.695_696T C>AA (SS) | missense; DNA binding | predicted deleterious | ||
| 7 | LCM | n | 1 | WT (NGS) | p | 2 | WT (NGS) | n | ex 5:c.497C>A (NGS only) | nonsense | deleterious | ||
| 10 | MACRO | n | 1 | WT (NGS) | p | 3 | WT (NGS) | p | ex 5: c.524G>A (NGS, SS) | missense; DNA binding | deleterious | ||
| 14 | MACRO | n | 0 | WT (NGS) | n | 0 | WT (NGS) | n | in 8: c.919+1G>A (NGS, SS) | splice site broken | deleterious | ||
| 15 | MACRO | p | 2 | NA | n | 2 | NA | n | NA | ||||
| 17 | MACRO | n | 0 | WT (NGS) | p | 1 | WT (NGS) | p | ex 8: c.817C>T (NGS) | missense; | deleterious | ||
| 19 | MACRO | n | 0 | WT (NGS) | n | 2 | WT (NGS) | n | WT (NGS) | ||||
| 21 | MACRO | n | 1 | WT (NGS) | p | 2 | WT (NGS) | n | WT (NGS) | ||||
| 24 | MACRO | n | 1 | NA | n | 1 | NA | n | NA | ||||
| 28 | MACRO | n | 1 | NA | p | 2 | NA | p | NA | ||||
| 31 | MACRO | n | 2 | WT (NGS) | n | 2 | ex8: c.863delA (NGS) | frameshift; C2 domain | predicted deleterious | p | WT (NGS, SS) | ||
| 32 | MACRO | n | 1 | WT (NGS) | n | 1 | WT (NGS) | n | WT (NGS, SS) | ||||
| 33 | MACRO | n | 1 | WT (NGS) | n | 2 | WT (NGS) | p | ex 5: c.524G>A (NGS, SS) | missense; DNA binding | deleterious |
LCM = laser capture microdissection; MACRO= macrodissection; WT = wild-type; NGS = Ampliseq Hotspot next generation sequencing assay; SS = Sanger sequencing; NA = not assessed.
Table 3.
|
A: Association between Rb protein levels by immunohistochemistry (IHC) and RB1 copy number by nanoString. | |||
|---|---|---|---|
|
RB1 Copy # |
Rb IHC | ||
| present | absent | ||
| normal | 1 | 1 | |
| LOH | 0 | 11 | |
|
B: Association between PTEN protein levels by immunohistochemistry (IHC) and PTEN copy number by nanoString. | |||
|---|---|---|---|
|
PTEN Copy # |
PTEN IHC | ||
| present | absent | ||
| normal | 4 | 4 | |
| LOH | 1 | 4 | |
|
C: Association between p53 protein levels by immunohistochemistry (IHC) and TP53 copy number by nanoString | |||
|---|---|---|---|
|
TP53 status |
p53 IHC | ||
| absent | present | ||
| wt | 3 | 1 | |
| mut | 2 | 4 | |
Sequencing of TP53, PTEN and RB1
Slightly more than half of the small cell carcinoma cases showed accumulation of p53 protein, indicating the possible presence of an inactivating mutation in TP53. To assess for TP53 mutations in a subset of 10 small cell carcinoma cases with adequate DNA available for analysis, we used a combination of next generation sequencing via the AmpliSeq™ Cancer Hotspot Panel on the Ion Torrent (which includes 8 amplicons covering the most commonly mutated exons in the gene: exons 2, 4, 5, 6, 7, 8, 10) and direct Sanger sequencing of exons 4, 5, 7 and 8. Due to limited DNA availability for some samples, six samples had both Ampliseq and Sanger sequencing data available, while 3 samples had only Ampliseq data available and one sample had only Sanger sequencing data available. Overall, 60% (6/10) small cell carcinoma samples showed mutations in TP53 (Table 2), including one deleterious missense mutation in the DNA binding domain in exon 5 (c.524G>A) that was seen in two separate samples and has been reported in multiple other tumor types and several families with Li-Fraumeni syndrome; one nonsense mutation in exon 5 (c.497C>A) that has been reported in a number of other tumors as well as tumor cell lines; a deleterious splice site mutation in intron 8 (c.919+1G>A) and a missense mutation in exon 8 (c.817C>T), both of which have been reported in other tumors and in families with Li-Fraumeni syndrome. In addition, we found a novel missense mutation in exon 7 (c.695_696TC>AA) predicted to deleteriously affect the DNA binding domain. Of the 6 samples subjected to both next generation and Sanger sequencing, 5 (83%) showed identical results by both assays, while one (17%) showed an additional mutation detected by next generation sequencing that was not detected in the Sanger sequence of the same sample (the nonsense mutation in exon 5 of sample 7: c.497C>A) (Table 2). Of the small cell carcinoma samples with a TP53 mutation detected, 66% (4/6) showed accumulation of p53 protein, compared to 25% (1/4) of the cases with wild-type TP53 (Table 3C).
As part of the AmpliSeq™ Cancer Hotspot Panel, we also examined the sequence of 6 of the 9 exons in PTEN which are most frequently mutated (exons 1, 3, 5, 6, 7, 8). Out of 9 small cell carcinoma samples with available DNA, we found only one frameshift mutation in the C2 domain of exon 8 of PTEN (c.863delA) in a single sample (Table 2). This mutation has been previously reported in several endometrial carcinoma samples as well as in a colon carcinoma sample, however the functional significance is unclear (36, 37). Interestingly, this sample had no detectable PTEN protein by immunohistochemical assay, despite two intact copies of the PTEN gene by nanoString analysis. Finally, we examined the sequence of 10 out of 26 exons of RB1 (exons 4, 6, 10, 11, 14, 17, 18, 20, 21, 22), and did not find any mutations in the 9 samples analyzed (Table 2).
Discussion
Although de novo small cell carcinoma of the prostate is a rare disease, treatment-related neuroendocrine prostate cancer (t-NEPC) is expected to become increasingly common with the recent availability of more potent anti-androgen therapeutics for prostate cancer treatment (aberaterone, enzalutamide, TAK700) (13). Because both de novo small cell carcinomas and t-NEPCs are resistant to androgen deprivation therapies and transiently responsive to platinum-based chemotherapeutics (2, 6–11), accurate diagnosis of these entities and distinction from ordinary high grade acinar prostate cancer is essential. Currently, the mainstay of pathologic diagnosis of small cell carcinoma is morphology (1, 18). Small cell carcinomas typically possess the classic “oat-cell” morphology first described for their counterparts in the lung, accompanied by a high mitotic index and frequent apoptosis. However, diagnosis by hematoxylin and eosin stain can be challenging and given the clinical consequences, use of an immunohistochemical panel to identify small cell carcinoma differentiation is frequently helpful. Unfortunately, neuroendocrine markers such as synaptophysin, chromogranin and CD56 do not comprise an optimal marker panel for small cell differentiation in prostate cancer, as they can be seen in up to 100% of acinar carcinomas in some series (38). Other markers that are useful in clinical practice include absence of PSA or other prostatic marker positivity in ~80% of cases (due to absence of androgen receptor expression), high ki-67 labeling index and TTF-1 expression (18, 39, 40). Recently CD44 was also proposed as a potential marker (41). However, given the rising prevalence of the disease and the lack of sensitivity and specificity of currently used IHC panels, identifying additional markers of small cell carcinoma and t-NEPC currently represents an important area of unmet clinical need.
A number of studies in human samples as well as transgenic mouse models have suggested that loss of the RB1 and TP53 tumor suppressors may be critical for the development of neuroendocrine carcinoma in multiple organ systems. Human lung small cell carcinomas were first shown to have frequent TP53 mutations as well as nearly universal loss of RB1 (19, 20). Subsequent studies in mouse models showed that germline RB1 heterozygosity is associated with the development of neuroendocrine tumors in multiple organ systems (42). Further, conditional RB1 and TP53 loss are sufficient to cause highly penetrant small cell carcinoma in the mouse lung (21). Recent studies have suggested that the cell of origin is likely neuroendocrine in this tumor model (43). Similarly, in the prostate, activation of the SV40 large T cell antigen (which inactivates all Rb family proteins and p53) has long been known to result in aggressive tumors with neuroendocrine features (22). More recent studies have shown that while conditional RB1 loss alone is not sufficient to cause invasive prostatic carcinoma in the mouse (44,45), concurrent loss of RB1 and TP53 leads to highly metastatic neuroendocrine carcinomas with a small cell-like morphology, likely originating from the proximal prostatic ducts (23, 46).
Despite the wealth of data for human lung and murine prostate tumors, ours is the first study to comprehensively evaluate the status of the RB1, TP53 and PTEN tumor suppressors in a large series of human prostatic small cell carcinomas. This is likely in part because these specimens are quite rare and almost uniformly formalin fixed and paraffin embedded (FFPE). Most small cell carcinomas are first discovered in biopsies or transurethral resection of the prostate (TURP) specimens (due to the aggressive nature of the disease, patients commonly present with urinary retention), and radical prostatectomy is contraindicated in these patients, making it difficult to collect frozen tissue specimens for analysis. However, recent improvements in genomic analysis from FFPE specimens have expanded our ability to cull genomic information from these small samples.
In the current studies, we took advantage of both nanoString and targeted next generation sequencing technologies to examine the status of the RB1, TP53 and PTEN tumor suppressors. We found that Rb loss occurs nearly universally at the protein level in small cell carcinomas, suggesting that absence of this protein may provide a useful marker of small cell differentiation in the prostate. In contrast, in high grade primary or metastatic acinar carcinomas, even those with immunohistochemical or morphologic evidence of (non-small cell) neuroendocrine differentiation, Rb protein loss occurred in only a minority of cases. Because negative markers always run the risk of producing false negative results due to technical failures, and because it is not 100% sensitive for detecting small cell carcinomas, Rb immunohistochemistry will likely be most useful as part of a larger panel of immunohistochemical markers for neuroendocrine differentiation, including synaptophysin, chromogranin, and markers of the AR signaling axis. As such, this panel of immunohistochemical markers would provide additional verification of small cell differentiation in histologically indefinite cases. In addition, it is tempting to speculate that inclusion of Rb in this panel may also help to identify a clinically important subgroup of high grade metastatic prostate tumors that show clinical resemblance to small cell carcinomas (including response to platinum-based chemotherapeutics), but lack apparent small cell or, in some cases, even neuroendocrine differentiation by conventional markers (47). Although studies are currently underway to test this hypothesis, the results of our current work suggest the possibility that Rb loss in the context of an otherwise unremarkable non-small cell adenocarcinoma may presage the later development of small cell carcinoma or castrate resistant prostate carcinoma with neuroendocrine differentiation, thus serving as a predictive biomarker.
Our finding that Rb loss is common in prostatic small cell carcinomas is in agreement with a recent study that examined prostatic xenografts with varying degrees of neuroendocrine differentiation (48). In this study, Rb protein loss was almost invariably seen in xenografts with evidence of high grade neuroendocrine differentiation, and microdeletions at the RB1 locus were detected by aCGH in many of these specimens. In 14 additional small cell carcinomas of the prostate, Rb protein levels were also markedly decreased. Adding to this study, we found that in contrast to small cell carcinomas, Rb loss at the protein level occurs in only 10% of high risk acinar primary carcinomas, despite the fact that RB1 allelic loss occurs in 18–40% of these cases (24–26, 49). Even in prostatic primaries with evidence of neuroendocrine differentiation (based on morphologic features and/or immunohistochemical expression of neuroendocrine markers), Rb protein was only lost in 11% of cases overall. In part, this is likely because homozygous deletion is extremely uncommon in the setting of acinar primary tumors. In a recent study of high risk primary tumors, only 1.6% or 2/125 of primary acinar cases showed homozygous deletion compared to 41.6% or 52/125 with hemizygous deletion (49 and Wennuan Liu, unpublished data).
Indeed, complete inactivation of RB1 generally occurs quite late in acinar tumor progression. In castrate resistant prostate tumors, the gene expression signature for Rb functional loss is relatively enriched compared to that in primary tumors (50) and up to 60% show RB1 hemizygous deletion (Supplementary Figure S1, 24, 26, 30). However the rate of homozygous RB1 deletion in castrate resistant tumors does not exceed 20% in the current series (Supplementary Figure S1, 24, 26, 30) and we confirmed that Rb protein loss is seen in a similar minority of cases. Interestingly, in our series, of the cases with homozygous RB1 loss, only 33% (1/3) had homozygous loss documented in more than one of the multiple metastatic sites sampled within the case. This provides further support for the concept that homozygous deletion of RB1 is a relatively late stage genomic alteration in acinar prostate cancer progression.
Though Rb protein loss is far more common in small cell carcinomas than acinar castrate resistant prostate tumors, the mechanism of Rb protein loss in small cell carcinomas remains unclear. Despite complete loss of the protein in almost every case, homozygous loss of RB1 was also relatively rare in our series of small cell carcinomas, suggesting that alternative mechanisms of RB1 loss may be common in this tumor type. Importantly, the relatively large amounts of DNA required for the nanoString assay (500 ng) preclude microdissection of tumor samples for analysis, thus we cannot entirely rule out the possibility that contamination by surrounding benign glands and stroma confounded the copy number call by the nanoString assay. However, all tumor samples were estimated to be >80% pure by macrodissection and complete loss of Rb protein expression was also frequently seen with low transcript levels despite the presence of one intact RB1 allele by CGH in the LuCaP castrate resistant prostate xenograft series (50). Another possible explanation for the apparent discrepancy between protein and copy number assays for Rb in our study and others could be that the second allele is frequently inactivated by mutation in prostate tumors. Yet, in primary and metastatic acinar prostate tumors, RB1 is inactivated by mutation in less than 5% of cases in most series (24, 25). In agreement with this data, we performed limited sequencing of RB1 using the Ampliseq Cancer HotSpot panel and did not find any mutations in the gene in small cell carcinomas. However, it is important to note that this panel covers only roughly 10 out 27 exons from this large gene (exons 4, 6, 10, 11, 14, 17, 18, 20, 21, 22) and it remains possible that we missed significant mutations in the second allele because of this limitation. Another alternative mechanism for RB1 inactivation could be epigenetic gene silencing. While differential deoxycytidine methylation within CpG dinucleotides near the regulatory regions of the RB1 locus has been reported previously in castration resistant prostate tumors, a recent study of neuroendocrine xenografts did not confirm this finding (48, 51). Thus, it remains unclear how the second allele of RB1 is inactivated in small cell carcinomas.
In addition to RB1 loss, we also found that TP53 loss is common in small cell carcinomas, with protein accumulation and underlying detrimental mutation of the gene occurring in well over half of all specimens examined. Overall, our immunohistochemistry results for p53 protein over-expression correlated quite well with the mutational status of the gene, as would be expected given that TP53 mutations commonly extend the half-life of the protein. In some cases, TP53 mutations can also decrease the expression of the protein (52), and this may account for a few cases where we observed a mutation in the absence of protein accumulation. In the only previous study of p53 in a series of prostatic small cell carcinomas, Chen et al found that p53 protein was over-expressed in 74% (23/31) of prostatic small cell carcinomas (53). Interestingly, on sequencing, 5/7 of these tumors showed the same novel G747A missense mutation in the gene. Similarly, a case study of small cell carcinoma and associated acinar carcinoma showed an identical TP53 mutation in both components, supporting a common origin for the two tumors (17). TP53 mutation can be seen in acinar prostate carcinomas, but it is less frequently present in primary tumors, occurring in less than 10% of specimens examined in recent reports (24–26). However, in castrate resistant tumors, it is substantially more common and can be seen in 30–40% of metastases (24). Thus, p53 accumulation at the protein level or evidence of TP53 mutations are not specific to small cell carcinoma, do not correlate with AR or neuroendocrine marker IHC and occur relatively commonly in advanced prostate cancer. Accordingly, it is unclear whether evidence of p53 inactivation or PTEN loss adds much information about potential small cell differentiation compared to Rb loss, which is seen nearly universally in this tumor type. Future studies will specifically examine whether p53 and PTEN are useful adjunct markers in combination with Rb.
A high proliferative rate is a key feature of small cell carcinoma in any organ, thus it may not be surprising that loss of critical cell cycle regulators such as RB1 and TP53 is common in this tumor type in multiple organ systems. Active, hypophosphorylated Rb binds to and inhibits the function of E2F family transcription factors which promote S-phase entry. p53 similarly inhibits the G1/S cell cycle transition upon DNA damage recognition. Along these lines, recent studies have suggested that other cell cycle regulators, such as Aurora A kinase and UBE2C are frequently amplified or over-expressed in small cell neuroendocrine tumors (16, 48). Of note, however, we did not find evidence of Aurora kinase A amplifications in our samples (n=13) by nanoString assay (data not shown). It is also important to point out that Rb protein can be inactivated by a number of non-genomic means, such as hyperphosphorylation following over-expression of cyclin D1, CDK4/6 or loss of p16, events that occur commonly in other tumor types (54). Though we could not reliably examine Rb phosphorylation status in our FFPE samples, hyperphosphorylation of Rb most commonly results in over-expression of the protein, a finding we did not see in the small cell carcinoma cases (55). Accordingly, we did not see evidence of high level CDK4/6 or cyclin D1 amplification in our small cell carcinoma cases by nanoString assay, with only a few samples showing 3 copies of these genes (data not shown). This finding was not mutually exclusive with homozygous RB1 loss, suggesting it is of unclear significance (data not shown). Additionally, recent reports suggest that cyclin D1 protein levels are low, rather than high, in this tumor type (48). We did find hemizygous copy loss of CDKN2A (p16) in 4 small cell carcinoma samples, none of which showed homozygous RB1 loss, however all of these cases were negative for Rb protein (data not shown), again making the significance of this finding unclear. Overall, our data suggest that Rb protein loss is overwhelmingly the preferential mechanism of Rb inactivation in these tumors.
The extraordinarily high frequency of Rb loss in prostatic small cell carcinomas suggests that loss of this tumor suppressor, perhaps in combination with inactivation of p53, may be an essential event in the development of this tumor type. However, other than increasing the proliferative potential of the tumor cells, it remains unclear what signals downstream of Rb and p53 loss mediate small cell differentiation. Since Rb protein loss occurs in the overwhelming majority of small cell carcinomas and is rarely seen in primary acinar carcinomas unassociated with a small cell tumor, it is tempting to speculate that Rb loss itself may be a critical mediator of neuroendocrine transdifferentiation, as has been suggested by some studies in the lung (56). However, it remains equally plausible that Rb is simply required to prevent the proliferative expansion of neuroendocrine cells in the prostate, and its functions in this capacity are less critical for non-neuroendocrine epithelial cells. If Rb loss is a prerequisite for the development of small cell carcinoma which is frequently AR-negative, this also raises the interesting question of how Rb function may interface with AR expression. Interestingly, studies performed in usual-type prostatic adenocarcinomas without small cell differentiation have supported the idea that Rb loss may actually increase AR levels and AR axis activity (50). The fact that small cell carcinomas are most commonly AR- and Rb-negative strongly suggests that the effects of Rb loss may be highly context dependent, with differing outcomes in tumors with and without small cell neuroendocrine differentiation, as has been suggested by others (54).
In sum, our data suggest that loss of the RB1 and TP53 tumor suppressors are common in prostatic small cell carcinomas, similar to their counterparts in the lung and other organs. Overall, genetic inactivation of these loci by allelic loss or mutation, respectively, was well-correlated with validated immunohistochemical assays for these tumor suppressor proteins. Indeed, we found total loss of Rb in the vast majority of prostatic small cell carcinomas, with only rare loss in high grade acinar carcinoma primary tumors, suggesting that as part of a panel, Rb protein loss is a potentially clinically useful marker of small cell differentiation in the prostate.
Supplementary Material
Translational Relevance.
Small cell carcinoma of the prostate represents an extreme example of neuroendocrine differentiation in prostate cancer. However, with the advent of potent androgen suppressive therapies such as abiraterone and enzalutamide, neuroendocrine differentiation is likely to become more common, representing an important mode of drug resistance. Thus, defining molecular and immunohistochemical markers for prostatic small cell carcinoma could be helpful in predicting treatment response in this setting. We report that loss of the retinoblastoma (Rb) tumor suppressor occurs nearly universally in prostatic small cell carcinomas. In contrast, loss of this tumor suppressor occurs rarely in conventional high grade acinar prostate tumors, acinar carcinomas with neuroendocrine differentiation and castrate resistant prostate carcinomas. These data suggest that Rb loss is a critical event in the development of small cell carcinomas of the prostate and may be a useful diagnostic and potential therapeutic target in the setting of neuroendocrine differentiation in prostate cancer.
Acknowledgments
The authors wish to thank Jianfeng Xu for SNP array copy number data used to validate the nanoString assay.
Financial Support: Funding for this research was provided in part by a Prostate Cancer Foundation Young Investigator Award (TLL) and a grant by the Patrick C. Walsh Prostate Cancer Research Fund (TLL).
Footnotes
Conflicts of Interest: None.
References
- 1.Epstein JI, Netto GN. Biopsy interpretation of the prostate. 2nd ed. Philadelphia, PA: Lippincott, Williams & Wilkins; 2008. [Google Scholar]
- 2.Palmgren JS, Karavadia SS, Wakefield MR. Unusual and underappreciated: Small cell carcinoma of the prostate. Semin Oncol. 2007;34(1):22–29. doi: 10.1053/j.seminoncol.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 3.Mucci NR, Akdas G, Manely S, Rubin MA. Neuroendocrine expression in metastatic prostate cancer: Evaluation of high throughput tissue microarrays to detect heterogeneous protein expression. Hum Pathol. 2000;31(4):406–414. doi: 10.1053/hp.2000.7295. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Coleman I, Coleman R, Doan K, Roudier M, Chéry L, et al. Characterizing the molecular features of the neuroendocrine phenotype in castration resistant prostate cancer; Proceedings of the 104th Annual Meeting of the American Association for Cancer Research; 2013. Apr 6–10, Abstract nr 406. [Google Scholar]
- 5.Shah RB, Mehra R, Chinnaiyan AM, Shen R, Ghosh D, Zhou M, et al. Androgen-independent prostate cancer is a heterogeneous group of diseases: Lessons from a rapid autopsy program. Cancer Res. 2004;64(24):9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- 6.Hindson DA, Knight LL, Ocker JM. Small-cell carcinoma of prostate. transient complete remission with chemotherapy. Urology. 1985;26(2):182–184. doi: 10.1016/0090-4295(85)90060-3. [DOI] [PubMed] [Google Scholar]
- 7.Amato RJ, Logothetis CJ, Hallinan R, Ro JY, Sella A, Dexeus FH. Chemotherapy for small cell carcinoma of prostatic origin. J Urol. 1992;147(3 Pt 2):935–937. doi: 10.1016/s0022-5347(17)37427-x. [DOI] [PubMed] [Google Scholar]
- 8.Moore SR, Reinberg Y, Zhang G. Small cell carcinoma of prostate: Effectiveness of hormonal versus chemotherapy. Urology. 1992;39(5):411–416. doi: 10.1016/0090-4295(92)90235-o. [DOI] [PubMed] [Google Scholar]
- 9.Rubenstein JH, Katin MJ, Mangano MM, Dauphin J, Salenius SA, Dosoretz DE, et al. Small cell anaplastic carcinoma of the prostate: Seven new cases, review of the literature, and discussion of a therapeutic strategy. Am J Clin Oncol. 1997;20(4):376–380. doi: 10.1097/00000421-199708000-00011. [DOI] [PubMed] [Google Scholar]
- 10.Papandreou CN, Daliani DD, Thall PF, Tu SM, Wang X, Reyes A, et al. Results of a phase II study with doxorubicin, etoposide, and cisplatin in patients with fully characterized small-cell carcinoma of the prostate. J Clin Oncol. 2002;20(14):3072–3080. doi: 10.1200/JCO.2002.12.065. [DOI] [PubMed] [Google Scholar]
- 11.Spiess PE, Pettaway CA, Vakar-Lopez F, Kassouf W, Wang X, Busby JE, et al. Treatment outcomes of small cell carcinoma of the prostate: A single-center study. Cancer. 2007;110(8):1729–1737. doi: 10.1002/cncr.22971. [DOI] [PubMed] [Google Scholar]
- 12.Lotan TL, Gupta NS, Wang W, Toubaji A, Haffner MC, Chaux A, et al. ERG gene rearrangements are common in prostatic small cell carcinomas. Mod Pathol. 2011;24(6):820–828. doi: 10.1038/modpathol.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mosquera JM, Beltran H, Park K, MacDonald TY, Robinson BD, Tagawa ST, et al. Concurrent AURKA and MYCN gene amplifications are harbingers of lethal treatment-related neuroendocrine prostate cancer. Neoplasia. 2013;15(1):1–10. doi: 10.1593/neo.121550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williamson SR, Zhang S, Yao JL, Huang J, Lopez-Beltran A, Shen S, et al. ERG-TMPRSS2 rearrangement is shared by concurrent prostatic adenocarcinoma and prostatic small cell carcinoma and absent in small cell carcinoma of the urinary bladder: Evidence supporting monoclonal origin. Mod Pathol. 2011;24(8):1120–1127. doi: 10.1038/modpathol.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo CC, Dancer JY, Wang Y, Aparicio A, Navone NM, Troncoso P, et al. TMPRSS2-ERG gene fusion in small cell carcinoma of the prostate. Hum Pathol. 2011;42(1):11–17. doi: 10.1016/j.humpath.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1(6):487–495. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansel DE, Nakayama M, Luo J, Abukhdeir AM, Park BH, Bieberich CJ, et al. Shared TP53 gene mutation in morphologically and phenotypically distinct concurrent primary small cell neuroendocrine carcinoma and adenocarcinoma of the prostate. Prostate. 2009;69(6):603–609. doi: 10.1002/pros.20910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Epstein JI. Small cell carcinoma of the prostate. A morphologic and immunohistochemical study of 95 cases. Am J Surg Pathol. 2008;32(1):65–71. doi: 10.1097/PAS.0b013e318058a96b. [DOI] [PubMed] [Google Scholar]
- 19.Yuan J, Knorr J, Altmannsberger M, Goeckenjan G, Ahr A, Scharl A, et al. Expression of p16 and lack of pRB in primary small cell lung cancer. J Pathol. 1999;189(3):358–362. doi: 10.1002/(SICI)1096-9896(199911)189:3<358::AID-PATH452>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 20.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44(10):1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4(3):181–189. doi: 10.1016/s1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]
- 22.Greenberg NM, DeMayo F, Finegold MJ, Medina D, Tilley WD, Aspinall JO, et al. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci U S A. 1995;92(8):3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Z, Flesken-Nikitin A, Corney DC, Wang W, Goodrich DW, Roy-Burman P, et al. Synergy of p53 and rb deficiency in a conditional mouse model for metastatic prostate cancer. Cancer Res. 2006;66(16):7889–7898. doi: 10.1158/0008-5472.CAN-06-0486. [DOI] [PubMed] [Google Scholar]
- 24.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beltran H, Yelensky R, Frampton GM, Park K, Downing SR, MacDonald TY, et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol. 2013;63(5):920–926. doi: 10.1016/j.eururo.2012.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antonarakis ES, Keizman D, Zhang Z, Gurel B, Lotan TL, Hicks JL, et al. An immunohistochemical signature comprising PTEN, MYC, and Ki67 predicts progression in prostate cancer patients receiving adjuvant docetaxel after prostatectomy. Cancer. 2012;118(24):6063–6071. doi: 10.1002/cncr.27689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chuang AY, DeMarzo AM, Veltri RW, Sharma RB, Bieberich CJ, Epstein JI. Immunohistochemical differentiation of high-grade prostate carcinoma from urothelial carcinoma. Am J Surg Pathol. 2007;31(8):1246–1255. doi: 10.1097/PAS.0b013e31802f5d33. [DOI] [PubMed] [Google Scholar]
- 29.Herawi M, Epstein JI. Immunohistochemical antibody cocktail staining (p63/HMWCK/AMACR) of ductal adenocarcinoma and Gleason pattern 4 cribriform and noncribriform acinar adenocarcinomas of the prostate. Am J Surg Pathol. 2007;31(6):889–894. doi: 10.1097/01.pas.0000213447.16526.7f. [DOI] [PubMed] [Google Scholar]
- 30.Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15(5):559–565. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lotan TL, Gurel B, Sutcliffe S, Esopi D, Liu W, Xu J, et al. PTEN protein loss by immunostaining: Analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res. 2011;17(20):6563–6573. doi: 10.1158/1078-0432.CCR-11-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubin SJ, Hallahan DE, Ashman CR, Brachman DG, Beckett MA, Virudachalam S, et al. Two prostate carcinoma cell lines demonstrate abnormalities in tumor suppressor genes. J Surg Oncol. 1991;46(1):31–36. doi: 10.1002/jso.2930460108. [DOI] [PubMed] [Google Scholar]
- 33.Ikediobi ON, Davies H, Bignell G, Edkins S, Stevens C, O'Meara S, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5(11):2606–2612. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ha TU, Segev DL, Barbie D, Masiakos PT, Tran TT, Dombkowski D, et al. Mullerian inhibiting substance inhibits ovarian cell growth through an rb-independent mechanism. J Biol Chem. 2000;275(47):37101–37109. doi: 10.1074/jbc.M005701200. [DOI] [PubMed] [Google Scholar]
- 35.Nishizuka S, Charboneau L, Young L, Major S, Reinhold WC, Waltham M, et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci U S A. 2003;100(24):14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rudd ML, Price JC, Fogoros S, Godwin AK, Sgroi DC, Merino MJ, et al. A unique spectrum of somatic PIK3CA (p110alpha) mutations within primary endometrial carcinomas. Clin Cancer Res. 2011;17(6):1331–1340. doi: 10.1158/1078-0432.CCR-10-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berg M, Danielsen SA, Ahlquist T, Merok MA, Agesen TH, Vatn MH, et al. DNA sequence profiles of the colorectal cancer critical gene set KRAS-BRAF-PIK3CA-PTEN-TP53 related to age at disease onset. PLoS One. 2010;5(11):e13978. doi: 10.1371/journal.pone.0013978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Sant Agnese PA, Cockett AT. The prostatic endocrine-paracrine (neuroendocrine) regulatory system and neuroendocrine differentiation in prostatic carcinoma: A review and future directions in basic research. J Urol. 1994;152(5 Pt 2):1927–1931. doi: 10.1016/s0022-5347(17)32417-5. [DOI] [PubMed] [Google Scholar]
- 39.Helpap B, Kollermann J. Undifferentiated carcinoma of the prostate with small cell features: Immunohistochemical subtyping and reflections on histogenesis. Virchows Arch. 1999;434(5):385–391. doi: 10.1007/s004280050357. [DOI] [PubMed] [Google Scholar]
- 40.Yao JL, Madeb R, Bourne P, Lei J, Yang X, Tickoo S, et al. Small cell carcinoma of the prostate: An immunohistochemical study. Am J Surg Pathol. 2006;30(6):705–712. doi: 10.1097/00000478-200606000-00005. [DOI] [PubMed] [Google Scholar]
- 41.Simon RA, di Sant'Agnese PA, Huang LS, Xu H, Yao JL, Yang Q, et al. CD44 expression is a feature of prostatic small cell carcinoma and distinguishes it from its mimickers. Hum Pathol. 2009;40(2):252–258. doi: 10.1016/j.humpath.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 42.Nikitin AY, Juarez-Perez MI, Li S, Huang L, Lee WH. RB-mediated suppression of spontaneous multiple neuroendocrine neoplasia and lung metastases in rb+/− mice. Proc Natl Acad Sci U S A. 1999;96(7):3916–3921. doi: 10.1073/pnas.96.7.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: Inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19(6):754–764. doi: 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Hayward SW, Donjacour AA, Young P, Jacks T, Sage J, et al. Sex hormone-induced carcinogenesis in rb-deficient prostate tissue. Cancer Res. 2000;60(21):6008–6017. [PubMed] [Google Scholar]
- 45.Hill R, Song Y, Cardiff RD, Van Dyke T. Heterogeneous tumor evolution initiated by loss of pRb function in a preclinical prostate cancer model. Cancer Res. 2005;65(22):10243–10254. doi: 10.1158/0008-5472.CAN-05-1579. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Z, Flesken-Nikitin A, Nikitin AY. Prostate cancer associated with p53 and rb deficiency arises from the stem/progenitor cell-enriched proximal region of prostatic ducts. Cancer Res. 2007;67(12):5683–5690. doi: 10.1158/0008-5472.CAN-07-0768. [DOI] [PubMed] [Google Scholar]
- 47.Aparicio AM, Harzstark AL, Corn PG, Wen S, Araujo JC, Tu SM, et al. Platinum-based chemotherapy for variant castrate-resistant prostate cancer. Clin Cancer Res. 2013;19(13):3621–3630. doi: 10.1158/1078-0432.CCR-12-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tzelepi V, Zhang J, Lu JF, Kleb B, Wu G, Wan X, et al. Modeling a lethal prostate cancer variant with small-cell carcinoma features. Clin Cancer Res. 2012;18(3):666–677. doi: 10.1158/1078-0432.CCR-11-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu W, Xie CC, Thomas CY, Kim ST, Lindberg J, Egevad L, et al. Genetic markers associated with early cancer-specific mortality following prostatectomy. Cancer. 2013;119(13):2405–2412. doi: 10.1002/cncr.27954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, et al. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest. 2010;120(12):4478–4492. doi: 10.1172/JCI44239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedlander TW, Roy R, Tomlins SA, Ngo VT, Kobayashi Y, Azameera A, et al. Common structural and epigenetic changes in the genome of castration-resistant prostate cancer. Cancer Res. 2012;72(3):616–625. doi: 10.1158/0008-5472.CAN-11-2079. [DOI] [PubMed] [Google Scholar]
- 52.Kuhn E, Kurman RJ, Vang R, Sehdev AS, Han G, Soslow R, et al. TP53 mutations in serous tubal intraepithelial carcinoma and concurrent pelvic high-grade serous carcinoma--evidence supporting the clonal relationship of the two lesions. J Pathol. 2012;226(3):421–426. doi: 10.1002/path.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen H, Sun Y, Wu C, Magyar CE, Li X, Cheng L, et al. Pathogenesis of prostatic small cell carcinoma involves the inactivation of the P53 pathway. Endocr Relat Cancer. 2012;19(3):321–331. doi: 10.1530/ERC-11-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aparicio A, Den RB, Knudsen KE. Time to stratify? the retinoblastoma protein in castrate-resistant prostate cancer. Nat Rev Urol. 2011;8(10):562–568. doi: 10.1038/nrurol.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chatterjee SJ, George B, Goebell PJ, Alavi-Tafreshi M, Shi SR, Fung YK, et al. Hyperphosphorylation of pRb: A mechanism for RB tumour suppressor pathway inactivation in bladder cancer. J Pathol. 2004;203(3):762–770. doi: 10.1002/path.1567. [DOI] [PubMed] [Google Scholar]
- 56.Wikenheiser-Brokamp KA. Rb family proteins differentially regulate distinct cell lineages during epithelial development. Development. 2004;131(17):4299–4310. doi: 10.1242/dev.01232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.