Abstract
A bacterial cell takes on the challenge to preserve and reproduce its shape at every generation against a substantial internal pressure by surrounding itself with a mechanical support, a peptidoglycan cell wall. The enlargement of the cell wall via net incorporation of precursors into the pre-existing wall conditions bacterial growth and morphology. However, generation, reproduction and/or modification of a specific shape requires that the incorporation take place at precise locations for a defined time period. Much has been learnt in the past few years about the biochemistry of the peptidoglycan synthesis process, but topological approaches to the understanding of shape generation have been hindered by a lack of appropriate techniques. Recent technological advances are paving the way for substantial progress in understanding the mechanisms of bacterial morphogenesis. Here we review the latest developments, focusing on the impact of new techniques on the precise mapping of cell wall growth sites.
Keywords: Peptidoglycan, PBPs, division, elongation
Introduction
Bacterial proliferation depends on the ability of each individual cell to periodically enlarge and divide in a way compatible with the regeneration of the initial shape and size in daughter cells. Conceptually the easiest way to achieve this goal is volume doubling followed by symmetric fission, as exemplified by many of the more commonly used model bacterial species (Escherichia coli, Bacillus subtilis, Staphylococcus aureus, Streptococcus pneumoniae, etc). However, this is by no means the only possible way. Indeed, Caulobacter crescentus is a well-known example of a species in which division results in unequal offspring [1].
The cell wall (peptidoglycan layer, abbreviated PG, or murein sacculus) is a major element to consider for an appropriate understanding of bacterial growth. Because the sacculus is a covalently closed, stress-bearing, net–like giant molecule coating the cytoplasmic membrane, cell shape and growth are constrained by coordinated changes in the physical dimensions of the sacculus. Although the sacculus is elastic and can be considerably stretched [2], continuous cell growth, division, and generation of shape and cell appendages requires incorporation of new precursors into the pre-existing cell wall fabric. Incorporation by itself would not lead to expansion of the sacculus unless some of the existing bonds were cleaved concomitantly with the insertion of new PG subunits, a function performed by specialized PG modifying enzymes called endopeptidases [3,4]. Otherwise, sacculi would simply become thicker. Furthermore, in order to generate a precise shape, both incorporation and cleavage must take place at specific locations, rates, and times, that is, with a defined topology. Unfortunately, appropriate experimental tools to explore the delicate balance of cell wall removal and incorporation have been in short supply, and only very recently efficient methods to either label [5–8] or visualize growth sites [9,10] have become available. Here we provide a survey of the state of research on cell wall growth topology and dynamics in rod-like bacteria. For the case of round and ovoid bacteria (Figure 1), the reader is referred to excellent recent reviews [11,12].
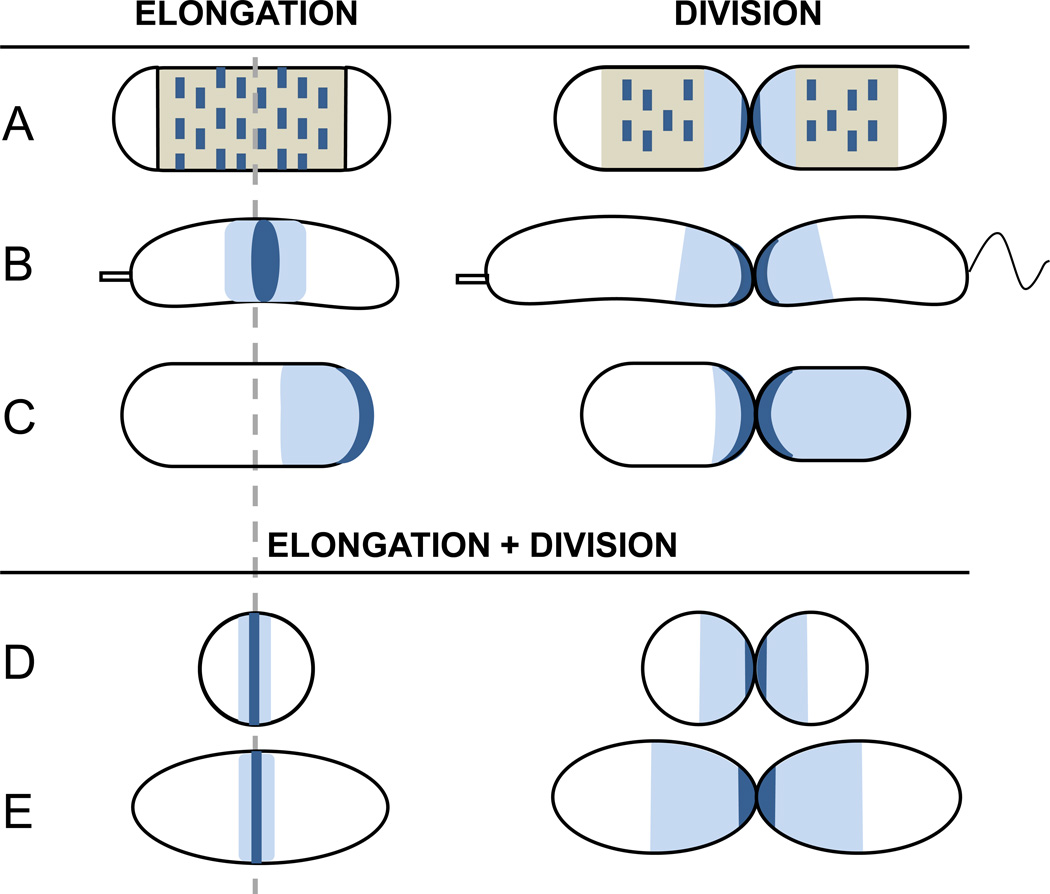
Figure 1. Schematics of different modes of cell wall growth in bacteria.
A) E. coli (or B. subtilis) follow a disperse mode of growth during elongation followed by activation of zonal synthesis at the time of septation. Short MreB filaments direct local incorporation of nascent PG by the elongation biosynthetic complexes. B) C. crescentus elongates and divides by zonal insertion of nascent PG at a location slightly offset from the axis of symmetry. Subsequent septation will lead to distinct offspring cells. C) A. tumefaciens elongates by zonal-apical growth, followed by activation of a central site of zonal synthesis and symmetric septation. D,E) In cocci (i.e. S. aureus) and ovococci (i.e. S. pneumoniae) elongation and septation are essentially a single process mediated by zonal central PG incorporation. Dark blue, active PG biosynthetic complexes (regions); light blue, PG produced by zonal insertion; beige, PG produced by disperse insertion.
Septal PG synthesis
Not surprisingly, research on E. coli has remained at the forefront of the bacterial cell wall biosynthesis field. The topology of PG synthesis in this species was first addressed 40 years ago in the pioneering work of Ryter et al. [13] using radioactive PG precursors and autoradiography. Further refinement of the methodology combined with the mathematical modeling of the data by Verwer and Nanninga led to the proposal of a dispersed mode of cell wall growth with a higher probability of incorporation at mid-cell [14]. These studies also lead to the idea that septal PG synthesis was inhibited at the mid-cell prior to nucleoid segregation, or “nucleoid occlusion” [15], much later substantiated by the discovery of the FtsZ-ring inhibitor proteins SlmA in E. coli [16] and Noc in B. subtilis [17]. Although the utilization of radioactive PG precursors is perhaps the least growth perturbing method, the imaging via autoradiography was intrinsically limited in both spatial and temporal resolution, forbidding high-resolution mapping. The introduction of the “D-cysteine (D-Cys) labeling technique” permitted higher resolution and facilitated systematic analysis of cell wall growth in E. coli strains under a variety of conditions [18]. Taking advantage of the inherent promiscuity of the cell wall machinery for D-amino acid incorporation, this technique showed a more complex pattern. Elongation of the cell wall is primarily performed by disperse incorporation of precursors on the cylindrical wall from cell birth until termination of the DNA replication round. At this time a zonal, annular incorporation region becomes active at mid-cell, starts a progressive reduction in diameter (invagination), and produces the new polar caps of the daughter cells sacculi. Zonal PG synthesis is then switched off at the new poles and, for still unknown reasons, no further PG synthesis nor removal seems to ever happen again at the new poles, generating what has been called inert PG regions [18,19]. Therefore, the cell wall growth cycle in E. coli encompasses two overlapping biosynthetic processes: An essentially continuous, dispersed mode along the cell cylinder driving elongation, and a periodic, zonal mode associated with septum formation (Figure 1).
Of course, a critical remaining question has been which factors direct and control the activation and location of the biosynthetic complexes involved in each insertion mode. The onset of midcell PG incorporation was soon shown to depend upon ring formation by the tubulin-like protein FtsZ, but incorporation was first observed prior to constriction and was independent of the division-specific penicillin binding protein (PBP) 3. This PBP3-independent PG synthesis (PIPS) is followed by PBP3-dependent incorporation concomitant with constriction [18]. FtsZ and its ancillary protein ZipA are the only cell division proteins required for PIPS [20], whereas none of the LMW or high molecular weight (HMW) class B PBPs seem to be required for this process [20,21]. Whether PIPS is a transition stage between elongation and division or what PG synthesizing enzymes are required for PIPS are still not known.
Completion of septation does require the participation of the whole plethora of division proteins as well as LMW-PBPs and N-acetylmuramyl-L-ala amidases. The former are important to ensure the new poles are correctly shaped [19,20], whereas the later are required for separation of the new poles at the end of division (see review [22]). These proteins associate with the FtsZ ring to form the septosome or divisome. Only five out of the more than 20 proteins involved in septal PG incorporation have recognized enzymatic activities in PG metabolism: PBP1B, monofunctional PG glycosyltransferase MtgA and FtsI (PBP3) as PG synthetases for septal growth [23], amidases required for separation of the inwards growing septum at the end of division, and DacA (PBP5), a D-alanyl-D-alanine carboxypeptidase necessary to avoid irregularities at cell poles [19]. Interestingly, no specific regulatory elements have been identified so far to control activation of PBP3 and PBP5, but amidases are under the control of a particularly sophisticated mechanism based on the relief of auto-inhibition by regulatory elements related to ABC-transporters in the cytokinetic ring [24,25]. In some species, septal PG synthesis is also the target for external modulators of cell division such as BslO, a component of the S-layer of Bacillus anthracis [26]. Interestingly when division is inhibited, the septation-associated zonal PG synthesis remains active for a limited period of time, resulting in the synthesis of a ring of new PG which only depends on FtsZ and ZipA and apparently shares the same properties as polar PG [19,20]. How and when zonal PG synthesis is switched off under these circumstances is still unknown.
Cell elongation by dispersed PG synthesis
In E. coli and other species like Bacillus subtilis, growth in length is not predominantly dependent on the zonal system, but rather on the dispersed incorporation of precursors at discrete sites over the cylindrical cell body (Figure 1). As is the case for septal PG synthesis, the elongation PG biosynthetic complexes are made of a large number of molecules [27]. Biosynthetic complexes include not only the polymerizing machinery (PBPs and ancillary proteins) but also (some) precursor-synthesizing enzymes [28,29], hydrolases [4] and outer-membrane lipoprotein cofactors [30,31].
The functional interactions between the enzymatic components of the biosynthetic complexes is also an area of active research. Results indicate the existence of cooperativity between Class A and Class B PBPs, proposed for a long time but never confirmed [32]. High-resolution techniques support the idea of “mobile” biosynthetic complexes. Early deconvolution fluorescent microscopy of the actin homologue MreB implied that the protein polymerized as helical, dynamic filaments along the lateral walls of rod-shaped bacteria [33]. This led to the idea that MreB serves as the motor to drive biosynthetic complexes around the cell body as incorporation of precursors proceeds. Although these early predictions were accurate about the essential role of MreB and associated proteins in defining the topology of the biosynthetic complexes, high resolution studies using total internal reflection fluorescence microscopy (TIRFM) revealed that MreB and paralogues (Mbl and MreBH) in B. subtilis are not polymerized into continuous helical fibers, but are organized in variable-length patches with a bidirectional movement [9,10,34] (Figure 1). The PG polymerization process itself would then be the movement generating force and MreB would work as a scaffold-tracking structure [9,10]. Analysis of the distribution of new and old PG in the lateral cell wall is consistent with a patchy organization of the biosynthetic complexes, and indicates a mosaic-like structure with isolated regions of new and old PG [35]. Clearly, the assembly of MreB into filaments regulates the subcellular arrangement of biosynthetic complexes. However, it is still unknown how polymerization of MreB determines the rod shape of bacterial cells. Other factor(s) are likely to be involved in controlling the function and dynamics of MreB. For instance FtsZ seems to be involved in the dynamics of lateral wall synthesis [36,37], as well as RodZ, PBP2 and RodA [38]. Furthermore, local architecture of the PG sacculus or the polymerized nature of the PG subunits could play a critical role on the differentiation or activation of new biosynthetic complexes [35,39,40].
The coexistence of two significantly different mechanisms for cell wall elongation and division brings up an essential question; How are they coordinated? As a simple model, it is possible to think of a continuous elongation process in which new PG biosynthetic complexes are generated according to surface availability and metabolic state of the cell, and a superposed discontinuous zonal synthesis subordinated to DNA replication. Whether both elongation and division mechanisms are simultaneously active or alternate is another open question, although short labeling pulses with fluorescent D-amino acids resulted in simultaneous labeling of the septum and lateral wall in E. coli and B. subtilis, suggesting that they might be simultaneous [6]. Simple superposition would look simpler, but not necessarily right. In E. coli the transition from elongation to division apparently requires the interaction of elongation associated (PBP2) and division specific (PBP3) proteins suggesting a close coordination between both phenomena [28]. Additional support to this idea is provided experiments which suggest that bacterial cell division is coupled to cell elongation via a direct and essential interaction between FtsZ and MreB [41]. A possible self-control mechanism could be the demand for precursors, in particular lipid II, which is present at a low concentration [42]. Sudden activation of extra biosynthetic complexes for septal PG synthesis could restrict lipid II availability for the dispersed PG synthesis complexes, therefore temporally reducing the elongation rate.
Cell elongation by zonal growth
Not all rod-shaped bacteria elongate by dispersed incorporation of precursors; indeed many do so by zonal incorporation. In such a situation, both elongation and division occur at the same cellular location, but not at the same time. Newborn cells differentiate a zonal growth site which first promotes cell elongation and then is modified to promote inwards growth of a septum (see review [43]). Division is coupled to inactivation of cell elongation PG synthesis at the new poles, and a new cycle requires differentiation of a new zonal region [44]. A good example is the α-proteobacterium C. crescentus (Figure 1). Exhaustive studies on the growth of this bacterium revealed significant differences from the E. coli model in the timing of localization of cell-division proteins and the genetic requirements for localization [45]. FtsZ localization early in the cell cycle drives a switch from dispersed elongation to a septal elongation mode responsible for a significant amount of growth [36,37]. In addition, environmental cues can modulate the localization of several elements of the biosynthetic complexes in C. crescentus. In this regard, it has been shown that PBP1A, PBP2 and RodA localization to the septa responds to osmotic variations [46].
Recent studies with another rod-shaped α-proteobacterium, Agrobacterium tumefaciens, revealed an unsuspected mode of bacterial growth and cell division [6,47]. In this species, as well as in other Rhizobiales, cell elongation happens from the new poles created by division until the cell doubles in length (Figure 1). A shallow constriction forms close to the growing pole in very young cells, but further constriction does not occur until late stages of cell elongation. Elongation is then turned off at the old pole as the new poles are created by division, and growth switches to these new poles after division, suggesting that the old biosynthetic complexes are either inactivated or relocated to the new poles [6,47] (Figure 1). Studies on the dynamics of FtsA and FtsZ suggest that both are required for continuous activity of the biosynthetic complexes irrespective of their polar (elongation) or central (division) location [47,48].
Polar cell wall growth is also characteristic of the hyphae of Streptomycetes, where it was described based on the binding of Vancomycin-FL[49,50], and the incorporation of fluorescent derivatives of D-amino acids [6]. In these organisms, polar growth is ultimately determined by the recruitment of PG biosynthetic enzymes to the hyphal tips by the “tip organizing center” composed of the polar determinant DivIVA, and the cytoskeletal element Scy [51]. New branch sites along the lateral wall are formed by DivIVA foci that break off from existing polar foci [52]. DivIVA focus splitting is regulated by its phosphorylation by the Ser/Thr protein kinase AfsK, which itself is activated by the inhibition of cell wall synthesis [53]. This may provide a mechanism for redirecting growth when a hyphae encounters an obstacle. A peculiarity of Streptomycetes is that neither MreB nor FtsZ homologues are involved in apical hyphal growth. Both cytoskeletal proteins are instead required for proper sporulation, and the later also for vegetative septation [54,55]. Mycobacteria are similarly characterized by polar cell wall growth driven by the DivIVA homologue, Wag31. In addition CwsA, a small membrane protein, plays a critical role in septal and polar PG synthesis modulating FtsZ ring assembly [56]. As in the case of E. coli, resolution of septa at the end of division requires a sophisticated regulation based on protein-protein interactions. In this instance a potential hydrolase, RipA, requires proteolytic activation to cleave septa [57], which happens through a progressive, inwards directed, degradation of the transversal cell wall starting once the septum is fully closed [58].
Conclusion
In the previous sections we described how current research is revealing the diversity of mechanisms underlying bacterial cell wall growth. As is often the case, the abundance of information on a few species is largely out-weighted by the restricted diversity of subjects. Nevertheless, it is quite evident that bacteria have found diverse ways to coordinate longitudinal growth with transverse division ensuring conservation of shape. The recently developed methodologies to visualize new PG in vivo, and specific components of the biosynthetic complexes involved in elongation and division at higher than ever resolution is producing a resurgence of interest in the field. However, we are still in the very early days of understanding the complexity of bacterial shape and growth patterns. Bacterial morphology can certainly be far more complex than a rod cell. Bacteria with polymorphic cycles, resistance forms, and specialized cell types are only starting to be studied in any detail. The case of C. crescentus and the surprising discovery of polar growth in the Rhizobiaceae provide a good indication of the complexities awaiting to be discovered. For example, while the model rod-shaped species E. coli and B. subtilis grow using a dispersed mode, labeling of natural populations of bacteria in situ using fluorescent D-amino acids indicates that zonal growth is likely a frequent mode of growth in the bacterial world [6,59] (Figure 2).
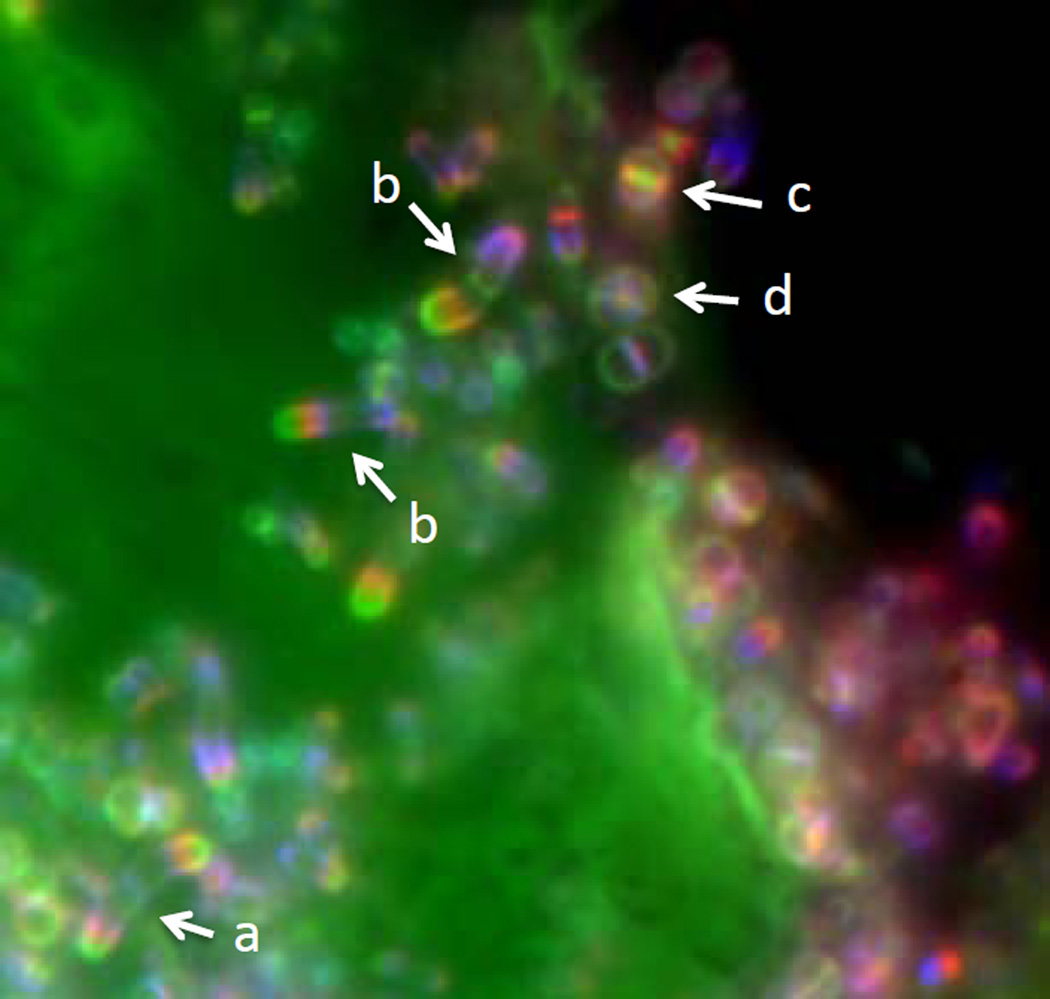
Figure 2. Different modes of growth in a natural population.
A saliva sample was pulse-labeled successively with a blue, a red, and a green Fluorescent-D-amino acid (FDAA) with washing of excess dye between each labeling. The large area labeled in green is a eukaryotic cell that was fortuitously labeled by a degradation product of the green FDAA (FDAAs do not label eukaryotic cells). The labeling patterns on this image provides a chronological account of the areas of PG synthesis during each pulse-labeling. Cells (a) and (b) grow polarly but cell (a) grows at the same rate from each pole, while cells (b) probably switched growth from the blue (or blue-red) pole to the red-green pole during the experiment. Cells (c) and (d) grow by septal incorporation. Alternating planes of growth are evident in cell (d).
Finally, how the assembly of new biosynthetic complexes is directed to generate specific morphological alterations is “terra ignota”. Bacterial shapes are precisely reproduced at every generation, are genetically encoded, and can be modulated by environmental conditions and developmental processes, suggesting that the generation of different shapes confers specific, selectable advantages [60]. Asymmetry in growth and division can generate phenotypic variation and provide a bet-hedging strategy to increase fitness in the face of rapid and/or unpredictable environmental shifts [61]. For example, a consequence of Mycobacterial polar growth is the generation of heterogeneity in elongation rate in the population and the concomitant decreased susceptibility of the slow growing cells to antibiotics that target PG synthesis [62]. Asymmetric growth may also facilitate the unequal inheritance of damage in daughter cells, generating an aging cell and a relatively damage-free sibling with higher fitness [61,63]. It is also important to have in mind that invasion of host cells by bacteria and the generation of biofilms or resistant forms are all processes involving shape alterations. Therefore, a precise knowledge of the mechanisms responsible for shape generation should have a strong incidence in the exploitation of bacteria.
Highlights.
Bacterial cell wall growth is conducted by an unsuspected diversity of mechanisms.
High-resolution techniques allow visualization of PG-biosynthetic complex dynamics.
Labeling with D-amino acid derivatives permits real-time, in vivo, tracking of PG-synthesis.
Division is universally committed to zonal (septal) PG synthesis.
Cell wall elongation follows a dispersed pattern in E. coli while in C. crescentus PG-synthesis concentrates at mid-cell section.
Rhizobiales species exhibit cell elongation by polar cell wall growth.
Acknowledgements
Research in the Cava lab is supported by The Laboratory for Molecular Infection Medicine Sweden (MIMS) and by the Knut and Alice Wallenberg Foundation (KAW). Y.V.B. was supported by Grant GM51986 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Of outstanding interest (**)
Of special interest (*)
- 1.Curtis PD, Brun YV. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev. 2010;74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao X, Jericho M, Pink D, Beveridge T. Thickness and elasticity of gram-negative murein sacculi measured by atomic force microscopy. J Bacteriol. 1999;181:6865–6875. doi: 10.1128/jb.181.22.6865-6875.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorr T, Cava F, Lam H, Davis BM, Waldor MK. Substrate specificity of an elongation-specific peptidoglycan endopeptidase and its implications for cell wall architecture and growth of Vibrio cholerae. Mol Microbiol. 2013 doi: 10.1111/mmi.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh SK, SaiSree L, Amrutha RN, Reddy M. Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol Microbiol. 2012;86:1036–1051. doi: 10.1111/mmi.12058. [DOI] [PubMed] [Google Scholar]
- 5.Siegrist MS, Whiteside S, Jewett JC, Aditham A, Cava F, Bertozzi CR. (D)-amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem Biol. 2013;8:500–505. doi: 10.1021/cb3004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, VanNieuwenhze MS. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl. 2012;51:12519–12523. doi: 10.1002/anie.201206749. ** Provides a revolutionary universal method for real-time tracking of peptidoglycan synthesis.
- 7.Olrichs NK, Aarsman ME, Verheul J, Arnusch CJ, Martin NI, Herve M, Vollmer W, de Kruijff B, Breukink E, den Blaauwen T. A novel in vivo cell-wall labeling approach sheds new light on peptidoglycan synthesis in Escherichia coli. Chembiochem. 2011;12:1124–1133. doi: 10.1002/cbic.201000552. [DOI] [PubMed] [Google Scholar]
- 8.Kocaoglu O, Calvo RA, Sham LT, Cozy LM, Lanning BR, Francis S, Winkler ME, Kearns DB, Carlson EE. Selective penicillin-binding protein imaging probes reveal substructure in bacterial cell division. ACS Chem Biol. 2012;7:1746–1753. doi: 10.1021/cb300329r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, Carballido-Lopez R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. ** These two concurrent reports provide high resolution microscopy evidence for the bi-directional movement of short patches of MreB and its dependence on PG synthesis in B. subtilis.
- 10. Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. ** These two concurrent reports provide high resolution microscopy evidence for the bi-directional movement of short patches of MreB and its dependence on PG synthesis in B. subtilis.
- 11.Pinho MG, Kjos M, Veening J-W. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nature Reviews Microbiology. 2013 doi: 10.1038/nrmicro3088. in press. [DOI] [PubMed] [Google Scholar]
- 12.Sham LT, Tsui HC, Land AD, Barendt SM, Winkler ME. Recent advances in pneumococcal peptidoglycan biosynthesis suggest new vaccine and antimicrobial targets. Curr Opin Microbiol. 2012;15:194–203. doi: 10.1016/j.mib.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryter A, Hirota Y, Schwarz U. Process of cellular division in Escherichia coli growth pattern of E. coli murein. J Mol Biol. 1973;78:185–195. doi: 10.1016/0022-2836(73)90437-3. [DOI] [PubMed] [Google Scholar]
- 14.Verwer RW, Nanninga N. Pattern of meso-dl-2,6-diaminopimelic acid incorporation during the division cycle of Escherichia coli. J Bacteriol. 1980;144:327–336. doi: 10.1128/jb.144.1.327-336.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mulder E, Woldringh CL. Autoradiographic analysis of diaminopimelic acid incorporation in filamentous cells of Escherichia coli: repression of peptidoglycan synthesis around the nucleoid. J Bacteriol. 1991;173:4751–4756. doi: 10.1128/jb.173.15.4751-4756.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernhardt TG, de Boer PA. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E coli. Mol Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 18.de Pedro MA, Quintela JC, Holtje JV, Schwarz H. Murein segregation in Escherichia coli. J Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Potluri LP, de Pedro MA, Young KD. Escherichia coli low-molecular-weight penicillin-binding proteins help orient septal FtsZ, and their absence leads to asymmetric cell division and branching. Mol Microbiol. 2012;84:203–224. doi: 10.1111/j.1365-2958.2012.08023.x. * Shows that low molecular weight penicillin binding proteins, particularly PBP5, have a role in maintaining the perpendicular geometry of the Z ring and subsequent septum in E. coli
- 20.Potluri LP, Kannan S, Young KD. ZipA is required for FtsZ-dependent preseptal peptidoglycan synthesis prior to invagination during cell division. J Bacteriol. 2012;194:5334–5342. doi: 10.1128/JB.00859-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Pedro MA, Donachie WD, Holtje JV, Schwarz H. Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J Bacteriol. 2001;183:4115–4126. doi: 10.1128/JB.183.14.4115-4126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Heijenoort J. Peptidoglycan hydrolases of Escherichia coli. Microbiol Mol Biol Rev. 2011;75:636–663. doi: 10.1128/MMBR.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derouaux A, Wolf B, Fraipont C, Breukink E, Nguyen-Disteche M, Terrak M. The monofunctional glycosyltransferase of Escherichia coli localizes to the cell division site and interacts with penicillin-binding protein 3, FtsW, and FtsN. J Bacteriol. 2008;190:1831–1834. doi: 10.1128/JB.01377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang DC, Tan K, Joachimiak A, Bernhardt TG. A conformational switch controls cell wall-remodelling enzymes required for bacterial cell division. Mol Microbiol. 2012;85:768–781. doi: 10.1111/j.1365-2958.2012.08138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang DC, Peters NT, Parzych KR, Uehara T, Markovski M, Bernhardt TG. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc Natl Acad Sci U S A. 2011;108:E1052–E1060. doi: 10.1073/pnas.1107780108. *Describes the regulation of a PG amidase by a conformational change in the mediated by ATP hydrolysis by the ABC-transported-like FtsEX complex.
- 26.Kern VJ, Kern JW, Theriot JA, Schneewind O, Missiakas D. Surface-layer (S-layer) proteins sap and EA1 govern the binding of the S-layer-associated protein BslO at the cell septa of Bacillus anthracis. J Bacteriol. 2012;194:3833–3840. doi: 10.1128/JB.00402-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.den Blaauwen T, de Pedro MA, Nguyen-Disteche M, Ayala JA. Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev. 2008;32:321–344. doi: 10.1111/j.1574-6976.2007.00090.x. [DOI] [PubMed] [Google Scholar]
- 28.van der Ploeg R, Verheul J, Vischer NO, Alexeeva S, Hoogendoorn E, Postma M, Banzhaf M, Vollmer W, den Blaauwen T. Colocalization and interaction between elongasome and divisome during a preparative cell division phase in Escherichia coli. Mol Microbiol. 2013;87:1074–1087. doi: 10.1111/mmi.12150. [DOI] [PubMed] [Google Scholar]
- 29.White CL, Kitich A, Gober JW. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol Microbiol. 2010;76:616–633. doi: 10.1111/j.1365-2958.2010.07108.x. [DOI] [PubMed] [Google Scholar]
- 30.Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, von Rechenberg M, et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell. 2010;143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banzhaf M, van den Berg van Saparoea B, Terrak M, Fraipont C, Egan A, Philippe J, Zapun A, Breukink E, Nguyen-Disteche M, den Blaauwen T, et al. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol Microbiol. 2012;85:179–194. doi: 10.1111/j.1365-2958.2012.08103.x. [DOI] [PubMed] [Google Scholar]
- 33.Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 34.Reimold C, Defeu Soufo HJ, Dempwolff F, Graumann PL. Motion of variable-length MreB filaments at the bacterial cell membrane influences cell morphology. Mol Biol Cell. 2013;24:2340–2349. doi: 10.1091/mbc.E12-10-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Turner RD, Hurd AF, Cadby A, Hobbs JK, Foster SJ. Cell wall elongation mode in Gram-negative bacteria is determined by peptidoglycan architecture. Nat Commun. 2013;4:1496. doi: 10.1038/ncomms2503. **A combination of atomic force microscopy and super-resolution fluorescence microscopy leads to a model of PG growth by architecture-regulated insertion.
- 36.Varma A, de Pedro MA, Young KD. FtsZ directs a second mode of peptidoglycan synthesis in Escherichia coli. J Bacteriol. 2007;189:5692–5704. doi: 10.1128/JB.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- 38.Shiomi D, Toyoda A, Aizu T, Ejima F, Fujiyama A, Shini T, Kohara Y, Niki H. Mutations in cell elongation genes mreB, mrdA and mrdB suppress the shape defect of RodZ-deficient cells. Mol Microbiol. 2013;87:1029–1044. doi: 10.1111/mmi.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cava F, de Pedro MA, Lam H, Davis BM, Waldor MK. Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 2011;30:3442–3453. doi: 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupoli TJ, Tsukamoto H, Doud EH, Wang TS, Walker S, Kahne D. Transpeptidase-mediated incorporation of D-amino acids into bacterial peptidoglycan. J Am Chem Soc. 2011;133:10748–10751. doi: 10.1021/ja2040656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fenton AK, Gerdes K. Direct interaction of FtsZ and MreB is required for septum synthesis and cell division in Escherichia coli. EMBO J. 2013;32:1953–1965. doi: 10.1038/emboj.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghuysen JM, Hakenbeck R. Bacterial Cell Wall. New comprenhensive biochemistry. Elsevier Science Pub Co. 1994;27 [Google Scholar]
- 43.Brown PJ, Kysela DT, Brun YV. Polarity and the diversity of growth mechanisms in bacteria. Semin Cell Dev Biol. 2011;22:790–798. doi: 10.1016/j.semcdb.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takacs CN, Poggio S, Charbon G, Pucheault M, Vollmer W, Jacobs-Wagner C. MreB drives de novo rod morphogenesis in Caulobacter crescentus via remodeling of the cell wall. J Bacteriol. 2010;192:1671–1684. doi: 10.1128/JB.01311-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goley ED, Yeh YC, Hong SH, Fero MJ, Abeliuk E, McAdams HH, Shapiro L. Assembly of the Caulobacter cell division machine. Mol Microbiol. 2011;80:1680–1698. doi: 10.1111/j.1365-2958.2011.07677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hocking J, Priyadarshini R, Takacs CN, Costa T, Dye NA, Shapiro L, Vollmer W, Jacobs-Wagner C. Osmolality-dependent relocation of penicillin-binding protein PBP2 to the division site in Caulobacter crescentus. J Bacteriol. 2012;194:3116–3127. doi: 10.1128/JB.00260-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brown PJ, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, Fuqua C, Brun YV. Polar growth in the Alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci U S A. 2011;109:1697–1701. doi: 10.1073/pnas.1114476109. ** Reveals unidirectional cell wall growth from the new pole in Agrobacterium tumefaciens and confirms it as a conserved trait amongst the Rhizobiales.
- 48.Zupan JR, Cameron TA, Anderson-Furgeson J, Zambryski PC. Dynamic FtsA and FtsZ localization and outer membrane alterations during polar growth and cell division in Agrobacterium tumefaciens. Proc Natl Acad Sci U S A. 2013;110:9060–9065. doi: 10.1073/pnas.1307241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 50.Flardh K. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2) Mol Microbiol. 2003;49:1523–1536. doi: 10.1046/j.1365-2958.2003.03660.x. [DOI] [PubMed] [Google Scholar]
- 51.Holmes NA, Walshaw J, Leggett RM, Thibessard A, Dalton KA, Gillespie MD, Hemmings AM, Gust B, Kelemen GH. Coiled-coil protein Scy is a key component of a multiprotein assembly controlling polarized growth in Streptomyces. Proc Natl Acad Sci U S A. 2012;110:E397–E406. doi: 10.1073/pnas.1210657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Richards DM, Hempel AM, Flardh K, Buttner MJ, Howard M. Mechanistic basis of branch-site selection in filamentous bacteria. PLoS Comput Biol. 2012;8:e1002423. doi: 10.1371/journal.pcbi.1002423. * Uses a combination of fluorescence microscopy and mathematical modeling to show how breaking off of apical DivIVA foci result in branch-site selection.
- 53. Hempel AM, Cantlay S, Molle V, Wang SB, Naldrett MJ, Parker JL, Richards DM, Jung YG, Buttner MJ, Flardh K. The Ser/Thr protein kinase AfsK regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proc Natl Acad Sci U S A. 2012;109:E2371–E2379. doi: 10.1073/pnas.1207409109. * Describes how the phosphorylation of DivIVA by AfsK regulates the topology of zonal growth.
- 54.Heichlinger A, Ammelburg M, Kleinschnitz EM, Latus A, Maldener I, Flardh K, Wohlleben W, Muth G. The MreB-like protein Mbl of Streptomyces coelicolor A3(2) depends on MreB for proper localization and contributes to spore wall synthesis. J Bacteriol. 2011;193:1533–1542. doi: 10.1128/JB.01100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCormick JR, Su EP, Driks A, Losick R. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol Microbiol. 1994;14:243–254. doi: 10.1111/j.1365-2958.1994.tb01285.x. [DOI] [PubMed] [Google Scholar]
- 56.Plocinski P, Arora N, Sarva K, Blaszczyk E, Qin H, Das N, Plocinska R, Ziolkiewicz M, Dziadek J, Kiran M, et al. Mycobacterium tuberculosis CwsA interacts with CrgA and Wag31, and the CrgA-CwsA complex is involved in peptidoglycan synthesis and cell shape determination. J Bacteriol. 2012;194:6398–6409. doi: 10.1128/JB.01005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chao MC, Kieser KJ, Minami S, Mavrici D, Aldridge BB, Fortune SM, Alber T, Rubin EJ. Protein complexes and proteolytic activation of the cell wall hydrolase RipA regulate septal resolution in mycobacteria. PLoS Pathog. 2012;9:e1003197. doi: 10.1371/journal.ppat.1003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vijay S, Anand D, Ajitkumar P. Unveiling unusual features of formation of septal partition and constriction in mycobacteria--an ultrastructural study. J Bacteriol. 2012;194:702–707. doi: 10.1128/JB.06184-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clabby C. See how they grow. American Scientist. 2013;100:226–227. [Google Scholar]
- 60.Young KD. The selective value of bacterial shape. Microbiol Mol Biol Rev. 2006;70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kysela DT, Brown PJ, Huang KC, Brun YV. Biological Consequences and Advantages of Asymmetric Bacterial Growth. Annu Rev Microbiol. 2013 doi: 10.1146/annurev-micro-092412-155622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, Toner M, Fortune SM. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science. 2012;335:100–104. doi: 10.1126/science.1216166. * Shows that there is deterministic heterogeneity in elongation rate in Mycobacteria which accounts for different susceptibility to antibiotics.
- 63.Ackermann M, Chao L, Bergstrom CT, Doebeli M. On the evolutionary origin of aging. Aging Cell. 2007;6:235–244. doi: 10.1111/j.1474-9726.2007.00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]