Abstract
Mutant K-ras activity leads to the activation of the RAS/RAF/MEK/ERK pathway in approximately 44% of colorectal cancer (CRC) tumors. Accordingly, several inhibitors of the MEK pathway are under clinical evaluation in several malignancies including CRC. The aim of this study was to develop and characterize predictive biomarkers of response to the MEK1/2 inhibitor AZD6244 in CRC in order to maximize the clinical utility of this agent. Twenty-seven human CRC cell lines were exposed to AZD6244 and classified according to the IC50 value as sensitive (≤0.1 µmol/L) or resistant (>1 µmol/L). All cell lines were subjected to immunoblotting for effector proteins, K-ras/BRAF mutation status, and baseline gene array analysis. Further testing was done in cell line xenografts and K-ras mutant CRC human explants models to develop a predictive genomic classifier for AZD6244. The most sensitive and resistant cell lines were subjected to differential gene array and pathway analyses. Members of the Wnt signaling pathway were highly overexpressed in cell lines resistant to AZD6244 and seem to be functionally involved in mediating resistance by shRNA knockdown studies. Baseline gene array data from CRC cell lines and xenografts were used to develop a k-top scoring pair (k-TSP) classifier, which predicted with 71% accuracy which of a test set of patient-derived K-ras mutant CRC explants would respond to AZD6244, providing the basis for a patient-selective clinical trial. These results also indicate that resistance to AZD6244 may be mediated, in part, by the upregulation of the Wnt pathway, suggesting potential rational combination partners for AZD6244 in CRC.
Introduction
Colorectal cancer (CRC) is the third leading cause of cancer-related mortality in the United States (1). The response rates and overall survival in metastatic colon cancer have improved in the recent years with the introduction of chemotherapeutics, such as oxaliplatin and irinotecan, as well as agents targeting the vascular endothelial growth factor and epidermal growth factor receptor (EGFR) pathways, such as bevacizumab and cetuximab, respectively (2–4). Despite these advances in treatment, advanced unresectable CRC remains incurable, resulting in more than 50,000 deaths in the United States (1). Therefore, novel individualized treatment options are needed to achieve improved rates of disease control, and hopefully, survival.
The Ras-mitogen–activated protein kinase (MAPK) pathway is a key signaling network that acts by transferring growth factor signals from cell surface receptors to the cytoplasm and nucleus through a sequential protein kinase cascade, including Raf, MAPK/ERK kinase (MEK), and ERK that ultimately regulate several cellular functions, including cell division, growth, survival, proliferation, differentiation, migration, and cell death (5). This pathway has been shown to be constitutively activated in several cancers, leading to uncontrolled cell proliferation, resistance to apoptosis, and association with an aggressive neoplastic phenotype (6). Constitutive activation of the MAPK pathway may also contribute to cancer cell resistance to chemotherapy in several types of human malignancies, including pancreatic, colon, lung, thyroid, and breast cancers (7, 8). Because MEK1/2 is the exclusive substrate for RAF and the only substrate for MEK1/2 is ERK1/2, inhibitors of MEK inhibitors represent an attractive novel class of targeted anticancer therapeutics (9, 10).
Several MEK inhibitors have been developed as clinical therapeutics and partial responses have been observed in patients with advanced melanoma and gastrointestinal cancers, as well as prolonged stable disease in several tumor types (11–14). Phase I studies of MEK inhibitors have shown acceptable toxicity profiles at doses that inhibit MEK activity. Selumetinib (AZD6244; Astra Zeneca/ARRY-142886; Array BioPharma; Supplementary Fig. S1) is a potent, orally available, highly specific, non-ATP competitive MEK1/2 inhibitor with nanomolar activity against MEK in cell culture and mouse models. AZD6244 has undergone phase I and II clinical trials in a variety of tumor types, with the best response reported as being prolonged disease stabilization (15–17).
One of the most challenging aspects of anticancer therapeutics is that many patients exhibit inherent or acquired drug resistance. Despite intensive study, the molecular and genetic bases for drug resistance remain poorly understood. Therefore, a critical component of cancer drug development is the identification of biomarkers that may be used to predict responses to novel agents, thus facilitating individualization of therapy. Ideally, this should be done concurrently with early clinical development to minimize the exposure of patients to potentially toxic agents that have little hope of conferring clinical benefit. This has unfortunately been best illustrated in numerous studies that retrospectively identified that CRC patients with tumors containing mutations in K-ras derived no benefit from the EGFR-targeted antibodies cetuximab and panitumumab (2, 4, 18). Clearly, the earlier development of patient selection strategies is needed to prevent similar occurrences in the future.
The objective of this study was to use gene array technology to identify differentially expressed genes that could serve as potential biomarkers of sensitivity to AZD6244 in CRC. In addition, gene and pathway analyses were conducted to determine whether there were pathways associated with resistance to AZD6244. These results could be applied to clinical trials of AZD6244 in CRC and to identify rational combination strategies.
Materials and Methods
Drugs
The MEK1/2 inhibitor AZD6244 (ARRY-142886; Array BioPharma) was provided by Astra Zeneca, Inc., and prepared as a 10-mmol/L stock solution in DMSO. The chemical structure for this compound is depicted in Supplementary Fig. S1.
Cell culture
Twenty-seven human colon cancer cell lines were obtained from American Type Culture Collection. The GEO cells were a generous gift from Dr. Fortunato Ciardiello (Cattedra di Oncologia Medica, Dipartimento Medico-Chirurgico di Internistica Clinica e Sperimentale “F Magrassi e A Lanzara,” Seconda Universita degli Studi di Napoli). All cells except GEO were grown in RPMI medium supplemented with 10% fetal bovine serum (FBS), 1% nonessential amino acids, and 1% penicillin/streptomycin and were maintained at 37°C in an incubator under an atmosphere containing 5% CO2. GEO cells were grown in DMEM/F12 supplemented with 10% FBS, 1% nonessential amino acids, and 1% penicillin/streptomycin. The cells were routinely screened for the presence of mycoplasma (MycoAlert; Cambrex Bio Science) and were exposed to AZD6244 when they reached approximately 70% confluence. All cell lines were tested and authenticated by the University of Colorado Cancer Center DNA Sequencing and Analysis Core. DNA from CRC cell lines was analyzed using the Profiler Plus Kit (Applied Biosystems), the DNA profiles were compared with ATCC data to ensure authenticity, and verification was last done in September 2009.
Evaluation of antiproliferative effects of AZD6244
Antiproliferative effects of AZD6244 against CRC cell lines were determined using the sulforhodamine B (SRB) method as described previously (19). Briefly, cells in logarithmic growth phase were transferred to 96-well flat-bottomed plates with lids. Cell suspensions (100 µL) containing 3,000 to 5,000 viable cells were plated into each well and incubated overnight before exposure with different concentrations of AZD6244 for 72 hours. After drug treatment, medium was removed and the cells were fixed with cold 10% TCA for 30 minutes at 4°C. The cells were then washed with water and stained with 0.4% SRB (Fisher Scientific) for 30 minutes at room temperature and washed again with 1% acetic acid followed by stain solubilization with 10 mmol/L of Tris at room temperature. The plate was then read on a 96-well plate reader (Biotek Synergy 2) set at an absorbance wavelength of 565 nm. Cell proliferation curves were derived from the raw absorbance data and expressed as the percentage of vehicle-treated controls.
Immunoblotting
Cells were initially plated into 6-well plates and cultured in RPMI with 10% FBS for 24 hours. All cells were then cultured in serum-free RPMI or DMEM/F12 medium for 16 hours to lower the basal levels of ERK and AKT phosphorylation. The cells were treated with vehicle or AZD6244 (0.3 µmol/L) for 2 hours and then challenged with 10% FBS or serum-free media for 10 minutes. After treatment, the cells were immediately disrupted in RIPA lysis buffer containing protease and phosphatase inhibitors (50 mmol/L of Tris-HCL, pH 7.4, 150 mmol/L of NaCl, 1 mmol/L of PMSF, 1 mmol/L EDTA, 5 µg/mL of aprotonin, 5 µg/mL of leupeptin, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate). Total protein content of samples was determined using the BioRad DC Protein Assay (BioRad). Forty micrograms of total protein was loaded onto a 10% polyacrylamide gel, electrophoresed, and then transferred to PVDF using the I-Blot (Invitrogen). Membranes were blocked for 1 hour in blocking buffer [0.1% casein solution in 0.2× phosphate buffered saline (PBS)]. Membranes were then incubated overnight at 4°C in blocking buffer plus 0.1% Tween-20 with one of the following primary antibodies: phosphorylated ERK (p-ERK), ERK, p-AKT, AKT (Cell Signaling). Blots were then washed 3 × 10 minutes in 1 × PBS containing 0.1% Tween-20 and incubated with the appropriate secondary goat anti-rabbit and goat anti-mouse IgG (H + L) DyLight™ conjugated antibodies at 1:15,000 (Thermo Scientific) for 1 hour at room temperature. Following 3 × 20 minutes of washes, the blots were developed using the Odyssey Infrared Imaging System (LI-COR Biosciences). Immunoblot experiments were done in triplicate for each antibody.
Tumor xenografts in nude mice
Female, athymic, nude (nu/nu) mice, aged 4 to 6 weeks, were purchased from the National Cancer Institute. Mice were housed at the University of Colorado Center for Comparative Medicine, a facility accredited by the American Association for Accreditation of Laboratory Animal Care. Animals were allowed to acclimatize for 1 week and then caged in groups of 5. Animals were exposed to a 12-hour/12-hour light/dark cycle, and autoclaved food and water was supplied ad libitum. The research protocol for this study was approved by the University of Colorado at Denver Institutional Animal Care and Use Review Board. For xenograft production, human colon cancer cells were grown in 75-cm2 culture flasks in DMEM supplemented with 10% FBS until they reached approximately 60% confluency and were in logarithmic growth phase. Cells were harvested with trypsin/EDTA, pelleted by centrifugation, and resuspended in a solution consisting of 50% serum-free DMEM/50% matrigel (v/v; BD Biosciences). Approximately 2.5 × 106 cells in a volume of 100 µL were injected subcutaneously onto the left and right flanks of each mouse by using a 1-mL syringe with a 23-G needle. Resulting xenograft tumors were measured daily until tumor volumes of 100 to 150 µL were reached. Mice were then randomized into 2 groups: vehicle control (10% ethanol/10% cremophor EL/80% D5W) or AZD6244 (25 mg/kg) and were dosed daily by oral gavage at volumes that varied between 90 and 120 µL, based on body weight. Mice were monitored daily for signs of toxicity and were weighed twice weekly. Tumor size was evaluated twice per week by caliper measurements by using the following formula: tumor volume = (length × width2)/0.52. Tumor volume and body weight data were collected using the Study Director software package (Studylog Systems, San Francisco, CA). At the end of a 28-day cycle, mice were euthanized by isoflurane anesthesia and tumor samples were collected.
Gene expression profiles
Cells were plated at 2 × 10 in 6-well plates 24 hours prior to harvest. After 24 to 72 hours, the cells were rinsed twice with PBS and RNA was prepared using the RNeasy Plus mini kit (Qiagen). RNA isolation and microarray sample labeling were carried out by using standard methods for reverse transcription and 1 round of in vitro transcription. Total RNA isolated from CRC cell lines was hybridized on Affymetrix U133 Plus 2.0 gene arrays at least in duplicate (Affymetrix Inc.). The sample preparation and processing procedure was done as described in the Affymetrix GeneChip Expression Analysis Manual (Affymetrix Inc.). In addition, CRC cell line gene expression profiles were obtained from the GlaxoSmithKline (GSK) genomic profiling data via the NCI cancer Bioinformatics Grid (caBIG) web site (https://cabig.nci.nih.gov/). These data were also profiled using Affymetrix U133 Plus 2.0 gene arrays in triplicates. To integrate the data generated from our laboratory and GSK, absolute intensity signals from the microarray gene expression profiles were extracted and probe sets representing the same gene were collapsed on the basis of maximum values. Next, the gene expression levels were converted to a rank-based matrix and standardized (mean = 0, standard deviation = 1) for each microarray. Using this preprocessing method, the same cell lines from different data sets were clustered on the basis of their gene expression profiles. Data analyses were done on this rank-based matrix.
shRNA knockdown
The wingless integrated 5B (Wnt5B) and frizzled homolog 2 (FZD2) gene-specific shRNA expression cassettes, along with control “scrambled” shRNA plasmids including the original pRS vector, were purchased from Ori-Gene. The sequence of the Wnt5B-specific 29mer shRNA is GGAGCCAAGACTGGCATCAAGGAATGCCA; FZD2: ATCGGCGTCTTCTCCGTGCTCTACACAGT. Stable clones were generated by transfecting SW480 or HCT-116 cells in 6-well dishes with 1 µg of each of the shRNA plasmids, using Fugene 6 transfection reagent (Roche), according to manufacturer's recommendations. Seventy-two hours after transfection, the cells were placed under selection with 2.0 µg/mL of puromycin, splitting 1:5 when the cells reached confluency. Multiple clones from the same transfection were pooled and grown under puromycin selection. Successful knockdown of specific genes and gene products was confirmed by semiquantitative reverse transcriptase–PCR (RT-PCR) and immunoblotting with specific antibodies. Each experiment was conducted in duplicate.
Gene set enrichment analysis
Gene set analysis was done using the GSEA software, Version 2.0.1, obtained from the Broad Institute (www.broad.mit.edu/gsea; ref. 20). Gene set permutations were done 1,000 times for each analysis. The nominal P value and normalized enrichment score (NES) was used to sort the pathways enriched in each phenotype. We used the 199 pathways defined by Kyoto Encyclopedia of Genes and Genomes (KEGG) database as the gene set in this study. Human pathway annotations were downloaded from KEGG (August 2007 release). The KEGG human pathways used in this study include metabolism, genetic information processing, environmental information processing, cellular processes, and human diseases. One hundred sixty-six gene sets passed the gene set size filter criteria (min = 10, max = 500).
k-TSP classifier
We used the k-top scoring pair (k-TSP) algorithm (21) to construct a discriminative classifier in predicting tumors sensitive to AZD6244. In brief, the algorithm exploits the information contained in the rank-based matrix by focusing on “marker gene pairs” (i, j) for which there is a significant difference in the probability of the event (Ri < Rj) across the N samples from class Y = 1 (AZD6244 sensitive) to Y = −1 (AZD6244 resistant), where the event (Ri < Rj) is equivalent to the rank of gene i that is less than the rank of gene j if and only if gene i is expressed less than gene j (relative expression). Here, the quantities of interest are pij(m) = Prob(Ri < Rj| Y = m), m = (1, =1), that is, the probabilities of observing Ri < Rjin each class. These probabilities are estimated by the relative frequencies of occurrences of Ri < Rj within profiles and over samples. Let Δij denote the “score” of gene pair (i, j), where Δij = | pij(1) –pij(−1)|. A score Δij is computed for every pair of genes i, j є {1,…, P}, and i ≠ j. Gene pairs with high scores are viewed as most informative for classification. Using an internal leave-one-out cross-validation, the final k-TSP classifier uses the k disjoint pairs of genes, which achieve the k best scores from the training set. In this study, the maximum number of pairs (k-max) was fixed as 10.
Human tumor explant xenografts
The human CRC explant xenografts were generated according to previously published methods (22). Briefly, surgical specimens from patients undergoing the removal of either a primary CRC or a metastatic tumor at the University of Colorado Hospital were reimplanted subcutaneously into 5 mice for each patient. Explants CUCRC007 and CUCRC021 were derived from primary CRC; CUCRC036 was a liver metastasis; and CUCRC001, CUCRC006, CUCRC012, CUCRC027 represent peritoneal metastases. The human primary tumor explants were assayed for the K-ras mutational status by the University of Colorado Cancer Center Pathology Core, using the DxS Scorpion method (DxS Ltd.) following the manufacturer's instructions. Briefly, template DNA was analyzed for a set of 7 known K-ras point mutations by using the Therascreen K-RAS Mutation Detection kit (DxS Ltd.). Reactions and analysis were done on a Lightcycler 480 real-time PCR instrument (LC480, Roche Applied Science) that was calibrated using a dye calibration kit provided by the manufacturer. Cycle cross-point (Cp) values were calculated using the LC480 Fit-point software suite, and the control Cp was subtracted from the Cp of each mutation-specific primer set (23). All of the patient explant samples used in these studies contained K-ras mutations. Tumor samples were then passaged into subsequent generations of mice for drug studies. Briefly, tumors were allowed to grow to a size of 1,000 to 1,500 mm3 (F1), at which point they were harvested, divided, and transplanted to an additional 5 mice (F2) to maintain the tumor bank. After a subsequent growth passage, tumors were excised and expanded into cohorts of 25 or greater mice for treatment. All experiments were conducted on F3 to F5 generations. Tumors from this cohort were allowed to grow until reaching a size of ∼150 to 300 mm , at which time they were equally distributed by size into the 2 treatment groups: control and AZD6244 treated. Mice were treated for 28 days with either vehicle control or AZD6244 (25 mg/kg) once daily by oral gavage. Monitoring of mice and measurements of tumors was conducted as described earlier. The relative tumor growth index was calculated by taking the tumor volume of control or AZD6244-treated mice at study end as a percentage of the tumor volume at day 1 of treatment.
All of the xenograft studies were conducted in accordance with the NIH guidelines for the care and use of laboratory animals in a facility accredited by the American Association for Accreditation of Laboratory Animal Care and received approval from University of Colorado Animal Care and Use Committee prior to initiation. Obtaining tissue from CRC patients at the time of removal of a primary tumor or metastectomy was conducted under a Colorado Multi-Institutional Review Board approved protocol.
Results
Determination of in vitro sensitivity to AZD6244 in CRC cell lines
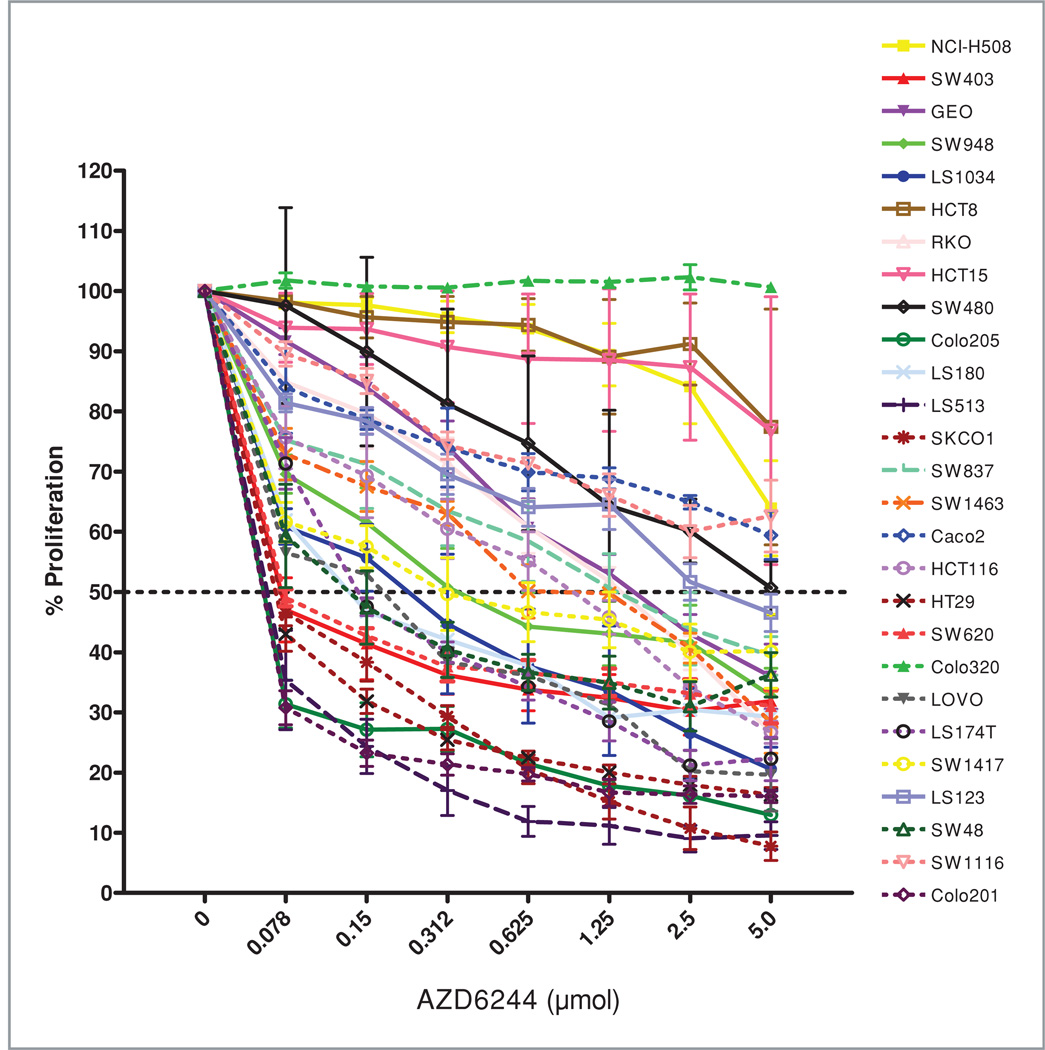
A panel of 27 CRC cell lines were exposed to AZD6244 at concentrations of 0.078 to 5.0 µmol/L for 72 hours and assessed for proliferation by using the SRB assay. As depicted in Fig. 1, there is a wide range of responsiveness to AZD6244, representative of the distinct genetic backgrounds of each of these patient-derived CRC cell lines. Seven of these cell lines with an IC50 < 0.1 µmol/L were deemed as sensitive (S) to AZD6244, whereas 11 cell lines were deemed as resistant (R) to this compound with IC50 > 1 µmol/L. The R demarcation concentration of 1 µmol/L was chosen on the basis of data from phase I trials, which indicated that 1.0 µmol/L was the average plasma concentration of AZD6244 achieved in patients at the maximum tolerated dose for this agent (15). Using the Fisher exact test, no statistically significant correlations were found between K-ras (P = 0.36) or B-raf (P = 0.22) mutations and AZD6244 sensitivity (data not shown). However, all of the 7 cell lines classified as sensitive contained B-raf or K-ras mutations.
Figure 1.
Cell proliferation assay on the panel of 27 CRC cell lines. Cells were exposed to increasing concentrations of AZD6244 for 72 hours and evaluated for proliferation by SRB staining as described in Materials and Methods.
In vivo validation of AZD6244 sensitivity
To confirm the S or R phenotype of the cells lines in vivo, we used a nude mouse xenograft model of 2-S (SW620 and LS513) and 2-R (SW480 and GEO) cell lines and treated them with AZD6244 for 14 days. Tumor volumes were measured at the end of the experiment and the responses of the xenografts were consistent with the in vitro cell line results, that is, S and R xenografts exhibited T/C < 50% and T/C > 70%, respectively (data not shown). The baseline gene array of the S and R xenografts at the time of institution of treatment was used along with the in vitro data in deriving the k-TSP classifier to minimize differences largely related to the tumor microenvironment.
Immunoblotting of downstream effector proteins in CRC cell lines
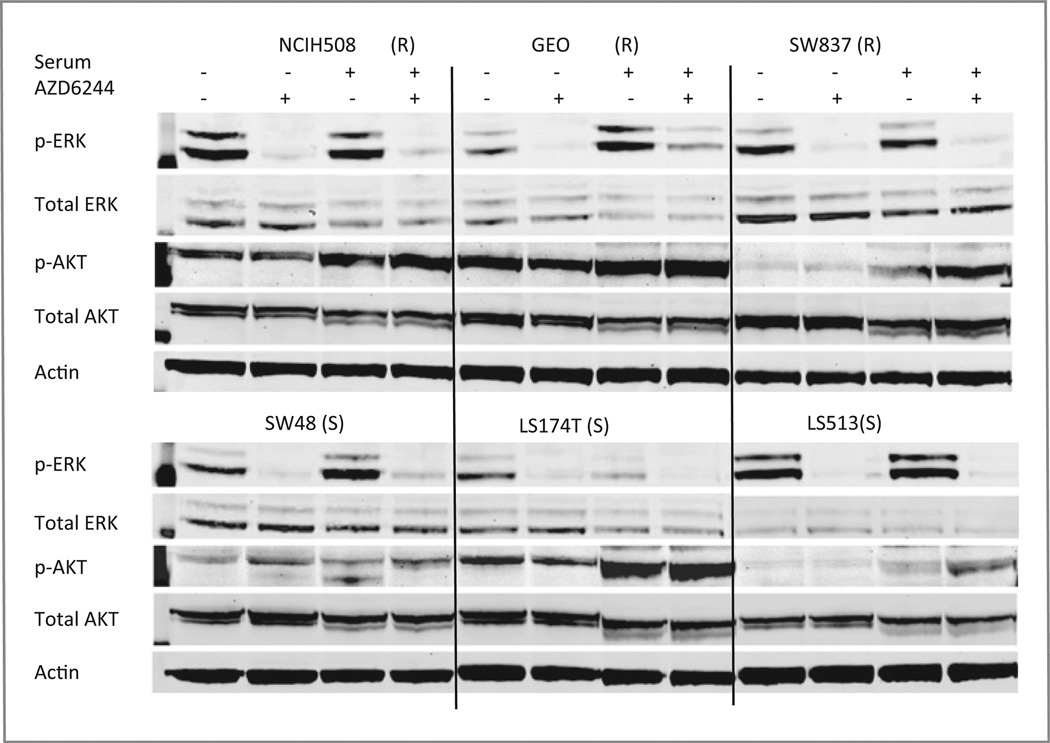
Next, the CRC cell lines were assessed at baseline and posttreatment with AZD6244 at 2 doses for downstream effectors (p-ERK, total ERK, p-AKT, total AKT), using standard immunoblotting techniques. As shown in Fig. 2, relative quantities of downstream effectors at baseline or posttreatment failed to consistently segregate the cell lines into S or R groups, using these proteins as readout, indicating that resistance was not a result of the inability of AZD6244 to inhibit its target, MEK1/2. Also of note, the CRC cell lines displayed highly variable basal levels of active p-ERK; however, there was no apparent correlation between basal levels of p-ERK or the ability of AZD6244 to inhibit ERK phosphorylation and responsiveness to AZD6244. We also assessed levels of AKT and p-AKT, downstream effectors of the PI3 kinase (PI3K) pathway. Previous reports have indicated that resistance to AZD6244 in non–small cell lung cancer and CRC cell lines is associated with strong PI3K signaling (24). However, as depicted in Fig. 2, we failed to find a consistent correlation between sensitivity and AKT activation state in the cell lines we tested.
Figure 2.
Immunoblotting of AZD6244 effector proteins in sensitive (S) and resistant (R) AZD6244 CRC cells exposed to 0.3 µmol/L of AZD6244 or vehicle in the presence or absence of fetal bovine serum (10%) as described in Materials and Methods. Thirty micrograms of total cell proteins were fractionated through sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to PVDF membranes, and incubated with the indicated antibodies. The experiment was done in triplicate and β-actin was used as a protein-loading control.
Gene array analysis of K-ras–mutant AZD6244 S or R CRC cells
CRC patients with K-ras- or B-raf–mutated tumors have limited therapeutic options. Therefore, we focused the development of a predictive classifier to AZD6244 in the K-ras mutant cell lines. To do this, we analyzed the basal gene expression profiles of the 4 S (LS513, SW620, SK-CO-1, and SW403) and 7 R (GEO, SW480, SW837, LS123, HCT8, HCT15, and SW1116) K-ras mutant CRC cell lines from the panel depicted in Fig. 1. Using the 2-sample t test and signal-to-noise ratio, 196 and 151 genes were identified as up- and downregulated, respectively, in AZD6244 S CRC cell lines (t test, P < 10−5). Table 1 lists the top 20 differentially expressed genes in S and R CRC cell lines.
Table 1.
List of top-scoring genes that are upregulated in AZD6244-sensitive or AZD6244-resistant CRC cell lines
| Gene symbol | Gene name | S2N | P |
|---|---|---|---|
| Up in sensitive | |||
| PROM1 | Pentaspan transmembrane protein | 1.83 | 2.10E-13 |
| ABHD2 | Abhydrolase-containing domain 2 | 1.78 | 1.31E-12 |
| TESC | Tscalcin | 1.67 | 4.65E-11 |
| BTBD3 | BTB domain–containing protein 3 | 1.56 | 7.74E-11 |
| AZGP1 | Alpha-2-glycoprotein, zinc binding | 1.53 | 2.04E-11 |
| ZNF238 | Zinc finger protein 238 | 1.52 | 2.72E-10 |
| TOX3 | TOX high-mobility group protein 3 | 1.5 | 3.46E-11 |
| RNF128 | Ring finger protein 128 | 1.44 | 4.04E-08 |
| PROX1 | Prospero homeobox 1 | 1.42 | 1.23E-07 |
| MAP2K6 | Mitogen-activated protein kinase kinase 6 | 1.27 | 4.25E-09 |
| Up in resistant | |||
| PTPLA | Protein tyrosine phosphatase-like, member A | −1.6 | 1.72E-07 |
| SERTAD4 | Serta domain containing 4 | −1.52 | 2.07E-10 |
| CTSL1 | Cathepsin L1 | −1.46 | 2.70E-10 |
| DOC2A | Double C2-like domains alpha | −1.38 | 3.83E-07 |
| MAPT | Microtubule-associated protein tau | −1.31 | 8.88E-10 |
| MAPK9 | Mitogen-activated kinase 9, JNK2 | −1.26 | 4.35E-07 |
| RNF144A | Ring finger protein 144A | −1.2 | 3.35E-08 |
| MARVELD1 | MARVEL domain containing 1 | −1.18 | 1.02E-08 |
| NR3C1 | Nuclear receptor 3C1, glucocorticoid receptor | −1.16 | 2.94E-06 |
| FZD2 | Frizzled homolog 2 | −0.98 | 1.60E-05 |
Baseline core genes and pathways in K-ras mutant AZD6244 S or R Cell Lines
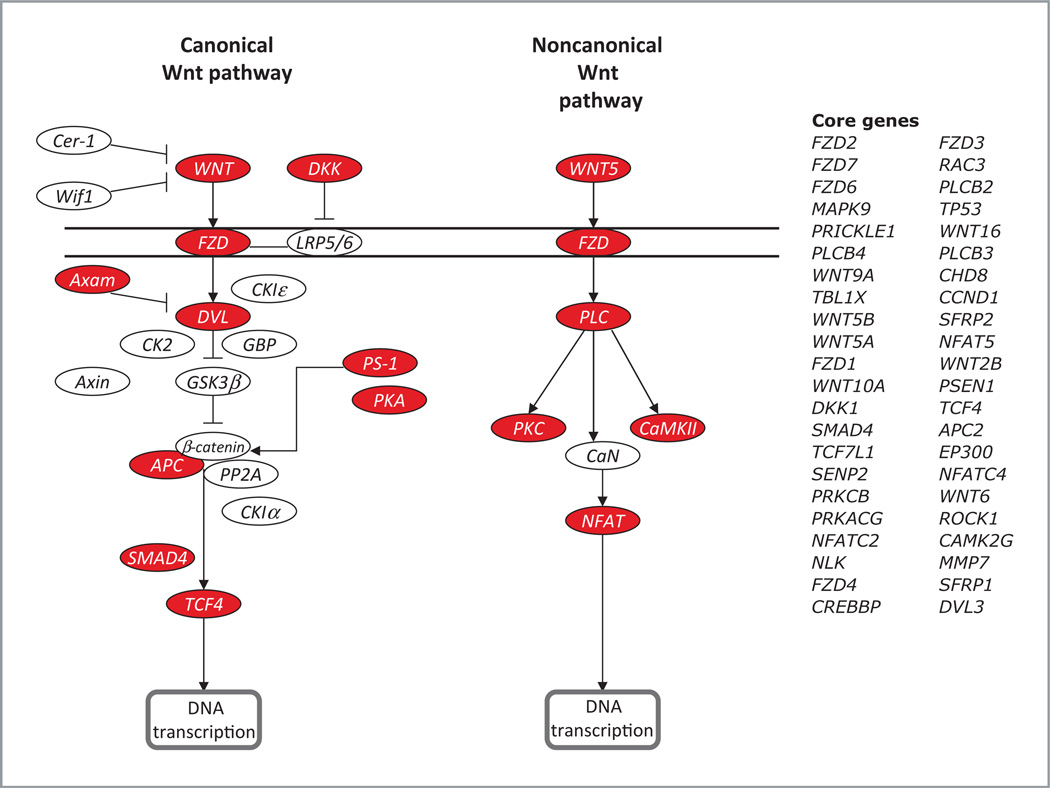
To assess the contribution of pathways that could be responsible for conferring sensitivity or resistance to AZ6244 in K-ras-mutated CRC cells, we did gene set analysis. GSEA was done using the GSEA software, Version 2.0.1, obtained from the Broad Institute (20). Gene set permutations were done 1,000 times for each analysis. We used the nominal P value and NES to sort the pathways enriched in each phenotype and the pathways defined by KEGG database as the gene sets in this study. At baseline, 82 and 67 pathways were more enriched in the S and R cell lines, respectively. Many metabolic pathways and immune-related pathways (such as renin-angiotensin system, Fc epsilon RI signaling pathway, and Natural killer cell–mediated cytotoxicity) were enriched in the S cell lines. However, the R cell lines were enriched predominantly with signal transduction pathways. Surprisingly, we found that many members of the Wnt signaling pathway were overrepresented in the R cells, indicating that coordinated overexpression of genes in Wnt pathway may confer resistance to AZD6244. Fig. 3 depicts a modified version of the KEGG pathway analysis derived from our gene expression results along with a list of core genes that were upregulated in AZD6244 R cell lines.
Figure 3.
Wnt signaling pathway represented by KEGG analysis. Red boxes represent core genes in the Wnt pathway with increased expression in K-ras mutant CRC cells resistant to AZD6244.
Functional relationship between the Wnt pathway and sensitivity to AZD6244
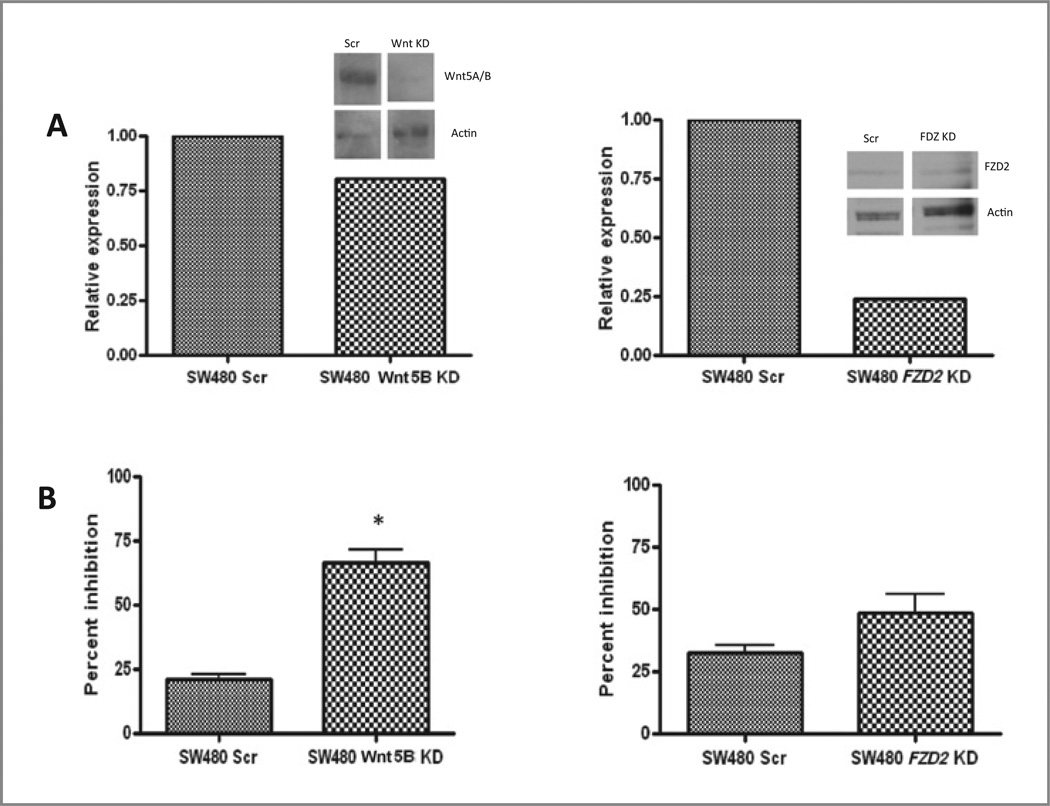
Amongst the top differentially expressed genes in the AZD6244-resistant cell lines was FZD2 (Table 1) and Wnt5B. Aberrant Wnt signaling has been implicated in many cancers, including CRC (25). Wnt5A and Wnt5B signaling via FZD receptors can lead to the release of intracellular calcium in what is termed the noncanonical or β-catenin–independent pathway (26). On the basis of the high degree of differential expression of Wnt pathway genes in the AZD6244 lines and the important role of Wnt signaling in CRC, our initial studies focused on the gene pairing of the Wnt5B ligand and its receptor, FZD2. To assess the functionality of Wnt5B and FZD2 in mediating R to AZD6244, we created stable cell lines expressing shRNA constructs against FZD2 and Wnt5B in the AZD6244 R, K-ras mutant SW480 CRC cell line. Four distinct clonal isolates for each gene were tested and the most robust knockdown of the target genes was then assessed by RT-PCR and Western blot analysis (Fig. 4A). The control and knockdown cell lines were then exposed to a range of doses of AZD6244 for 72 hours and cell proliferation was measured by the SRB method. Interestingly, knockdown of Wnt5B in the SW480 cells resulted in enhanced sensitivity to AZD6244 compared with controls that was statistically significant, with a less marked, but similar trend compared with the FZD2 knockdown (Fig. 4B). Similar results were seen when FZD2 was knocked down in K-ras mutant HCT-116 CRC cells (Supplementary Fig. S2), further supporting the hypothesis that Wnt pathway genes have a functional role in mediating resistance to AZD6244 and this phenomenon is not restricted to the SW480 cell line.
Figure 4.
A, qRT-PCR and immunoblotting (inset) were done to confirm knockdown of Wnt5B and FZD2 mRNA and protein, respectively. B, effects of Wnt5B and FDZ2 knockdown on SW480 cell proliferation in response to AZD6244. SW480 cells stably expressing shRNA constructs against Wnt5B and FZD2 were exposed to increasing doses of AZD6244 and assessed for proliferation by using the SRB method. Relative proliferation at the 2.5-εmol/L dose is depicted (P < 0.05).
Development of a k-TSP classifier for AZD6244 in K-ras–mutated CRC cell lines
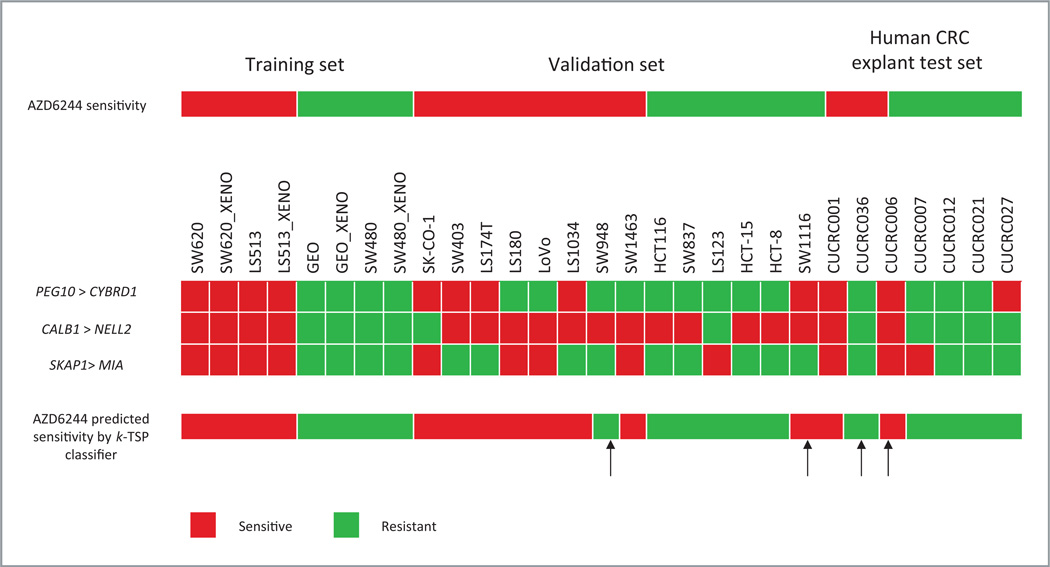
To construct a gene expression–based classifier, we profiled the baseline gene expression of the 2 most exquisitely S (SW620 and LS513, both have IC50 = 0.078 µmol/L) and 2 most R (GEO and SW480, IC50 values of 1.9 and 2.5 µmol/L, respectively) K-ras– mutated CRC cell lines in vitro and in vivo. Assuming the in vitro and in vivo cell lines as independent biological samples, the sample size used in this analysis (total n = 15, 8 in S and 7 in R) was deemed adequate to train an estimated 71% accurate classifier from gene expression profiles based on the sample size calculation of Simon et al. (http://linus.nci.nih.gov/brb/sample-size/samplesize4GE.html). We then employed the k-TSP algorithm (21) to identify gene pairs that can distinguish AZD6244 S from R cases with this training set. The algorithm identified 3 gene pairs—(PEG10 > CYBRD1), (CALB1 > NELL2), and (SKAP1 > MIA)—from the training data. The first gene pair is interpreted as follows: if the expression of PEG10 is higher than the expression of CYBRD1, it is predicted sensitive to AZD6244, otherwise AZD6244 resistant. The interpretation for the other 2 gene pairs follows in the same manner. The final prediction of the k-TSP classifier is based on the majority votes among the 3 gene pairs. Using leave- one-out cross-validation on the training set, the estimated accuracy for the classifier is 73%. The classifier was validated in an independent set of 14 in vitro K-ras mutant CRC cell lines and achieved 86% accuracy (Fig. 5). PEG10 is a paternally expressed 10 gene, and CYBRD1 is a cytochrome b reductase 1. SKAP1 encodes Src kinaseassociated phosphoprotein 1 and MIA is a gene involved in melanoma inhibitory activity. The gene product for CALB1 is calbindin 1 and NELL2 is an NEL-like 2 gene.
Figure 5.
k-TSP predictive classifier of responsiveness to AZD6244. Arrows indicate cases in which the classifier was in error. Red and green colors represent the prediction of AZD6244 sensitivity and resistance by the individual gene pair, respectively.
Testing of the classifier against human K-ras–mutated CRC explants
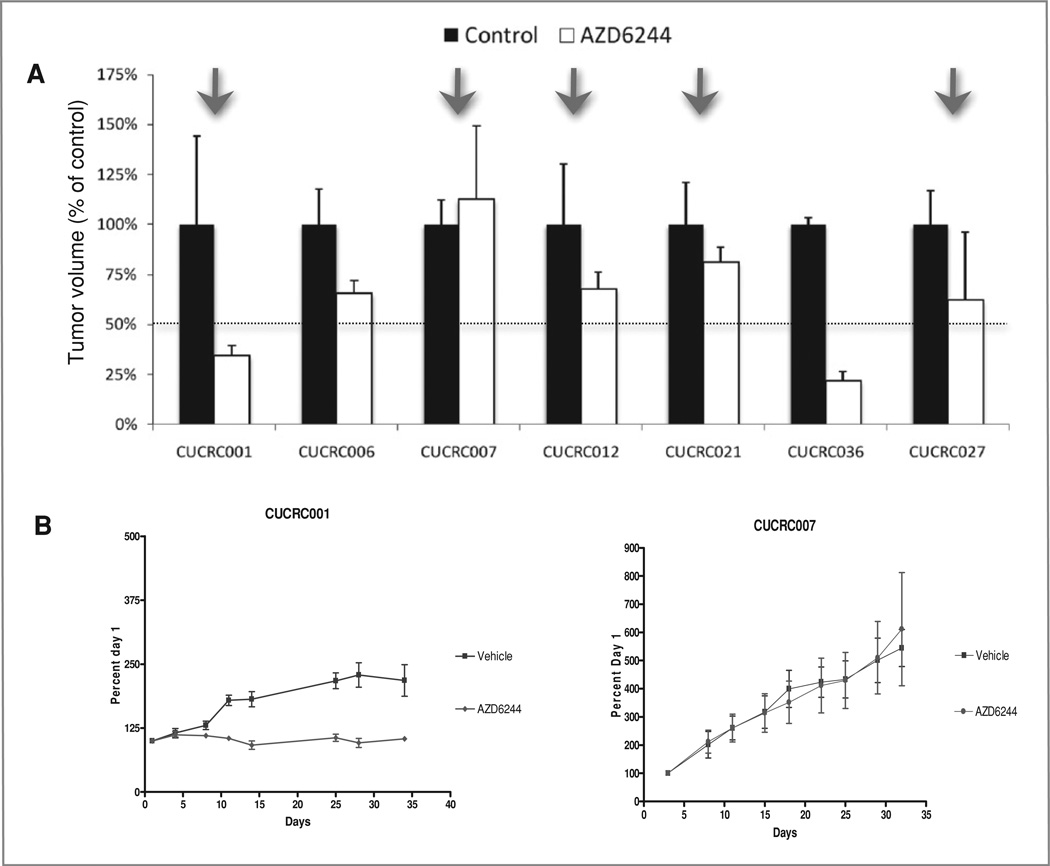
Next, the baseline expression of these 3 gene pairs in 7 CRC human explants (primary tumors or metastectomy specimens) grown in vivo as xenografts were used as an independent test set. Explants CUCRC007 and CUCRC021 were derived from primary CRC; CUCRC036 was a liver metastasis; and CUCRC001, CUCRC006, CUCRC012, and CUCRC027 were peritoneal metastases. All of these human explants were K-ras mutants. The k-TSP gene classifier predicted 2 explants as S to AZD6244 and 5 as R (Fig. 5). To validate the prediction of the k-TSP classifier, these explants were treated with AZD6244 for 28 days (Fig. 6). Two explants (CUCRC001 and CUCRC036) were deemed as sensitive cases (T/C < 50%), whereas 5 explants (CUCRC006, CUCRC007, CUCRC012, CUCRC021, and CUCRC027) were resistant to AZD6244 (T/C > 50%). The k-TSP classifier correctly predicted the AZD6244 sensitivity in 5 of 7 explants, leading to an accuracy of 71%. The sensitivity of the k-TSP classifier on the human explants was 50% (1/2), whereas the specificity was 80% (4/5; Fig. 5).
Figure 6.
Antitumor activity of AZD6244 (25 mg/kg) against human K-ras mutant CRC mouse explants models. Relative tumor growth inhibition was calculated by relative tumor growth of treated mice divided by the relative tumor growth of control mice × 100. A, antitumor effect of AZD6244 on the tumor growth of 7 explants. Arrows indicate cases in which the classifier was correct in predicting sensitivity. B, growth curves from representative explant models sensitive and resistant to AZD6244.
Discussion
The importance of the MAPK signaling pathway in cancer biology and its potential as a therapeutic target in human cancer is well established and confirmed in several preclinical studies (27–29). Highly selective small molecule MEK inhibitors have been evaluated in both phase I and II clinical trials as a promising class of targeted cancer therapies and showed clinical activity in patients with melanoma, pancreatic, colon, and lung cancers (12, 15, 17, 30). However, none of them have been approved as cancer therapies, and despite several studies that suggest a role of K-ras and/or B-raf mutations in response to MEK inhibitors, their utility as predictive biomarkers may not be so straightforward (24, 28, 31–34). In this study, we used selumetinib (AZD6244), an oral, highly selective, small molecule MEK1/2 inhibitor that has been investigated in phase I and II clinical trials (15, 35, 36) to identify predictive biomarkers for use in CRC patients. To accomplish this, we used an unbiased genomics approach by analyzing a panel of human CRC cell lines and xenografts, followed by validation against patient-derived CRC explants.
We observed a wide range (0.078 to >5 µmol/L) of sensitivity to AZD6244 among the 27 CRC cell lines in vitro that was recapitulated by the 2 most sensitive and resistant cell lines observed in vivo. In accordance with previous results, the K-ras mutational status as well as ERK activation did not predict AZD6244 sensitivity in CRC cell lines (24), bringing into question their role as predictive biomarkers for MEK inhibitors in CRC therapy.
Currently, gene expression profiling for most cancer types is used predominantly in the assessment of disease prognosis (37–40). Likewise, although gene expression profiling has been evaluated as a predictive biomarker for standard and novel therapeutics in the CRC field (41, 42), it has predominantly been focused on diagnosis and prognosis (43–45). In fact, the strongest predictive biomarkers we have in CRC are K-ras and B-raf mutations, which were retrospectively discovered to be associated with the lack of benefit to EGFR-targeted antibodies, after thousands of patients had been treated with these agents. Hence, the aim of this study was to develop a genomic classifier that could predict responsiveness to AZD6244 as a tool for patient selection in earlier CRC-directed studies of patients with K-ras–mutated tumors.
Interestingly, the single gene lists and pathway analysis of AZD6244-sensitive and AZD6244-resistant CRC cell lines revealed that several members of the Wnt signaling pathway were overrepresented in the AZD6244-resistant cells, indicating that coordinated overexpression of genes in the Wnt signaling pathway may confer resistance to the MEK inhibitor. Several reports have shown that deregulation of the Wnt signaling pathway is involved in proliferation, migration, and survival of several cancers, including CRC (25, 30). Historically, Wnt ligands and their receptors have been segregated into 2 pathways, the “canonical” pathway, which results in β-catenin–mediated transcription promotion, and the”noncanonical” pathway, which is β-catenin independent and further subdivided into the Wnt/Ca2+ and Wnt/planar cell polarity branches (26, 30). In noncanonical Wnt/Ca2+ signaling, Wnt agonists such as Wnt5B and its receptor FZD2 may stimulate intracellular Ca2+ release, activation of protein kinase C, and Ca2+/calmodulin-dependent kinase II (CaMKII), or they may inhibit the canonical pathway through degradation of β-catenin (26, 30, 46). Interestingly, in our studies, stable cell lines expressing shRNA constructs against FZD2 and Wnt5B in the resistant and K-ras mutant SW480 cell lines of AZD6244 enhanced sensitivity to AZD6244. Previous data from others have shown that specific shRNAs targeting β-catenin block the Wnt signaling pathway, increase apoptosis, and induce cell-cycle arrest in CRC cells, and, in the same manner, Wnt or FZD2 knockdown constructs interfere with Wnt signaling and inhibit cell growth and survival (47). Of note, a recent report describing a 13-gene signature of resistance to AZD6244 across a broad range of cancer types included FZD2, which can signal via the noncanonical Wnt/Ca2+ pathway (48). These data, along with the documented role of APC/β-catenin in hereditary and sporadic forms of CRC, indicate that targeting the Wnt pathway may be of potential value in either the treatment or modulation of resistance pathways to MEK inhibitors in CRC.
The overall aim of this study was to develop a predictive genomic classifier for K-ras mutant CRC by profiling the baseline gene expression of CRC cell lines extremely resistant or sensitive to AZD6244, using both in vitro and in vivo models in the training set. The importance of this approach is based on the fact that K-ras tumor mutation status is now widely recognized as a predictive marker of resistance to EGFR-targeted antibodies in CRC (49); however, the presence of K-ras mutation status and the variation in sensitivity to MEK inhibitors are still under evaluation (24). Despite the extensive literature on gene signatures developed from microarray analyses, the advantages of the k-TSP classifier we have developed include independent validation in vivo using contemporary patient-derived specimens, as well as the greater feasibility of doing RT-PCR for 3 gene pairs rather than a complete genomic profile. Although the classifier was only 71% accurate against the explants, this is substantially better than the 30% clinical benefit reported in a recent randomized phase II study of single-agent AZD6244 versus capecitabine in patients with refractory metastatic CRC (17). Furthermore, incorporation of this classifier prospectively in a CRC-directed clinical trial will enable us to refine and improve the accuracy of prediction, while minimizing patients’ exposure to ineffective therapy. In conclusion, our results strengthen the case for earlier predictive biomarker discovery so that more appropriate patients can be selected for treatment and rational combination strategies can be optimized to enhance the clinical benefit of novel agents for CRC patients.
Supplementary Material
Acknowledgments
The authors thank Dr. Paul D. Smith for helpful discussions and critical reading of this manuscript and Mohammad Roostan for technical assistance.
Grant Support
This work was supported by research grants CA106349 (S.G. Eckhart); CA079446 (S.G. Eckhart), CA46934 (NCI Cancer Center Support Grant); Astra Zeneca Research Agreement (A. Spreafico, S.G. Eckhart); Cancer League of Colorado Seed Grant (J.J. Tentler).
Footnotes
Note: Supplementary material for this article is available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
Research funding provided by AstraZeneca to A. Spreafico and S.G. Eckhart.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer Statistics 2009; CA Cancer. J Clin. 2010;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 3.Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335–2337. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 4.Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, Neyns B, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol. 2007;25:1658–1664. doi: 10.1200/JCO.2006.08.1620. [DOI] [PubMed] [Google Scholar]
- 5.Seger R, Krebs EG. The MAPK signaling cascade. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- 6.Sebolt-Leopold JS. MEK inhibitors: a therapeutic approach to targeting the Ras-MAP kinase pathway in tumors. Curr Pharm Des. 2004;10:1907–1914. doi: 10.2174/1381612043384439. [DOI] [PubMed] [Google Scholar]
- 7.Mueller H, Flury N, Eppenberger-Castori S, Kueng W, David F, Eppenberger U. Potential prognostic value of mitogen-activated protein kinase activity for disease-free survival of primary breast cancer patients. Int J Cancer. 2000;89:384–388. doi: 10.1002/1097-0215(20000720)89:4<384::aid-ijc11>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 8.Hoshino R, Chatani Y, Yamori T, et al. Constitutive activation of the 41-/43-kDa mitogen-activated protein kinase signaling pathway in human tumors. Oncogene. 1999;18:813–822. doi: 10.1038/sj.onc.1202367. [DOI] [PubMed] [Google Scholar]
- 9.Lewis TS, Shapiro PS, Ahn NG. Signal transduction through MAP kinase cascades. Adv Cancer Res. 1998;74:49–139. doi: 10.1016/s0065-230x(08)60765-4. [DOI] [PubMed] [Google Scholar]
- 10.Ahn NG, Nahreini TS, Tolwinski NS, Resing KA. Pharmacologic inhibitors of MKK1 and MKK2. Methods Enzymol. 2001;332:417–431. doi: 10.1016/s0076-6879(01)32219-x. [DOI] [PubMed] [Google Scholar]
- 11.Rinehart J, Adjei AA, Lorusso PM, Waterhouse D, Hecht JR, Natale RB, et al. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J Clin Oncol. 2004;22:4456–4462. doi: 10.1200/JCO.2004.01.185. [DOI] [PubMed] [Google Scholar]
- 12.Lorusso P, Krishnamurthi S, Rinehart JR, Nabell G, Croghan M, Varterasian SS, et al. A phase 1–2 clinical study of a second generation oral MEK inhibitor, PD 0325901 in patients with advanced cancer (abstract) J Clin Oncol. 2005;23:3066. [Google Scholar]
- 13.Wang D, Boerner SA, Winkler JD, Lorusso PM. Clinical experience of MEK inhibitors in cancer therapy. Biochim Biophys Acta. 2007;1773:1248–1255. doi: 10.1016/j.bbamcr.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Chow LM, Eckhardt SG, Reid J, Molina J, Hanson L, Piens J, et al. AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics: Discovery, Biology, and Clinical Applications. Philadelphia, PA: 2005. A first in human dose-ranging study to assess the pharmacokinetics, pharmacodynamics, and toxicities of the MEK Inhibitor, ARRY-142886 (AZD6244), in patients with advanced solid malignancies; p. C162. [Google Scholar]
- 15.Adjei AA, Cohen RB, Franklin WB, Morris C, Wilson D, Molina JR, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol. 2008;26:2139–2146. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Neil BH, Williams-Goff LW, Kauh J, Bekaii-Saab T, Strosberg JR, Lee R, et al. A phase II study of AZD6244 in advanced or metastatic hepatocellular carcinoma (abstract) J Clin Oncol. 2009;27(suppl):e15574. [Google Scholar]
- 17.Bennouna J, Lang I, Valladares-Ayerbes M, Boer K, Adenis A, Escudero P, et al. A Phase II, open-label, randomized study to assess the efficacy and safety of the MEK1/2 inhibitor AZD6244 (ARRY-142886) versus capecitabine monotherapy in patients with colorectal cancer who have failed one or two prior chemotherapeutic regimens (published online ahead of print) Invest New Drugs. 2010 doi: 10.1007/s10637-010-9392-8. [DOI] [PubMed] [Google Scholar]
- 18.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 19.Pitts TM, Morrow M, Kaufman SA, Tentler JJ, Eckhardt SG. Vorinostat and bortezomib exert synergistic antiproliferative and proapoptotic effects in colon cancer cell models. Mol Cancer Ther. 2009;8:342–349. doi: 10.1158/1535-7163.MCT-08-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gilette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan AC, Naiman DQ, Xu L, Winslow RL, Geman D. Simple decision rules for classifying human cancers from gene expression profiles. Bioinformatics. 2005;21:3896–3904. doi: 10.1093/bioinformatics/bti631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubio-Viqueira B, Jimeno A, Cusatis G, Zhang X, Iacobuzio-Donahue C, Karikari C, et al. An in vivo platform for translational drug development in pancreatic cancer. Clin Cancer Res. 2006;12:4652–4661. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- 23.Franklin WA, Haney J, Sugita M, Bemis L, Jimeno A, Messersmith WA. KRAS mutation: comparison of testing methods and tissue sampling techniques in colon cancer. J Mol Diag. 2010;12:43–50. doi: 10.2353/jmoldx.2010.080131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balmanno K, Chell SD, Gillings AS, Hayat S, Cook SJ. Intrinsic resistance to the MEK1/2 inhibitor AZD6244 (ARRY-142886) is associated with weak ERK1/2 signalling and/or strong PI3K signalling in colorectal cancer cell lines. Int J Cancer. 2009;125:2332–2341. doi: 10.1002/ijc.24604. [DOI] [PubMed] [Google Scholar]
- 25.Habas R, Dawid IB, Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 26.Gordon MD, Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006;281:22429–22433. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- 27.Alessi DR, Cuenda A, Cohen P, Dudley DT, Saltiel AR. PD 098059 is a specific inhibitor of mitogen-activated protein kinase in vitro and in vivo . J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 28.Solit DB, Garraway LA, Pratilas CA, Sawal A, Getz G, Basso A, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2005;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinkade CW, Castillo-Martin M, Puzio-Kuter A, Yan J, Foster TH, Gao H, et al. Targeting AKT/mTOR and ERK MAPK signaling inhibits hormone-refractory prostate cancer in a preclinical mouse model. J Clin Invest. 2008;118:3051–3064. doi: 10.1172/JCI34764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y. Wnt/Planar cell polarity signaling: a new paradigm for cancer therapy. Mol Cancer Ther. 2009;8:2103–2109. doi: 10.1158/1535-7163.MCT-09-0282. [DOI] [PubMed] [Google Scholar]
- 31.Yeh JJ, Routh ED, Rubinas T, Peacock J, Martin TD, Shen XJ, et al. KRAS/BRAF mutation status and ERK1/2 activation as biomarkers for MEK1/2 inhibitor therapy in colorectal cancer. Mol Cancer Ther. 2009;8:834–842. doi: 10.1158/1535-7163.MCT-08-0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pohl G, Ho CL, Kurman RJ, Bristow R, Wang TL, Shih Ie M. Inactivation of the mitogen-activated protein kinase pathway as a potential target-based therapy in ovarian serous tumors with KRAS or BRAF mutations. Cancer Res. 2005;65:1994–2000. doi: 10.1158/0008-5472.CAN-04-3625. [DOI] [PubMed] [Google Scholar]
- 33.Davies BR, Logie A, McKay JS, Martin P, Steele S, Jenkins R, et al. AZD6244 (ARRY-142886), a potent inhibitor of mitogen-activated protein kinase/extracellular signal regulated kinase kinase 1/2 kinases: mechanism of action in vivo, pharmacokinetic/pharmaco-dynamic relationship, and potential for combination in preclinical models. Mol Cancer Ther. 2007;6:2209–2219. doi: 10.1158/1535-7163.MCT-07-0231. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Van Becelaere K, Jiang P, Przbranowski S, Omer C, Sebolt-Leopold J. A role for K-ras in conferring resistance to the MEK inhibitor, CI-1040. Neoplasia. 2005;7:336–347. doi: 10.1593/neo.04532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeh TC, Marsh V, Bernat BA, Ballard J, Colwell H, Evans RJ, et al. Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin Cancer Res. 2007;13:1576–1583. doi: 10.1158/1078-0432.CCR-06-1150. [DOI] [PubMed] [Google Scholar]
- 36.Board RE, Ellison G, Orr MC, Kemsley KR, McWalter G, Blockley LY, et al. Detection of BRAF mutations in the tumour and serum of patients enrolled in the AZD6244 (ARRY-142886) advanced melanoma phase II study. Br J Cancer. 2009;101:1724–1730. doi: 10.1038/sj.bjc.6605371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bullinger L, Döhner K, Bair E, Frohling S, Schlenk RF, Tibsshrirani R, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350:1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 38.Bhattacharjee A, Richards WJ, Staunton J, Li C, Monti S, Vasa P, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 40.Beer DG, Kardi S, Huang C-C, Giordano TJ, Levin AM, Misek DE, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 41.Friedman DR, Weinberg JB, Barry WT, Goodman BK, Volkheimer AD, Bond KM, et al. A genomic approach to improve prognosis and predict therapeutic response in chronic lymphocytic leukemia. Clin Cancer Res. 2009;15:6947–6955. doi: 10.1158/1078-0432.CCR-09-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bild AH, Parker JS, Acharya CR, Hoadley KA, Anders C, Marcom PK, et al. An integration of complementary strategies for gene-expression analysis to reveal novel therapeutic opportunities for breast cancer. Breast Cancer Res. 2009;11:R55. doi: 10.1186/bcr2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Birkenkamp-Demtroder K, Christensen LL, Olesen SH, Frederiksen CM, Laiho P, Aaltonen LA, et al. Gene expression in colorectal cancer. Cancer Res. 2002;62:4352–4363. [PubMed] [Google Scholar]
- 44.Bertucci F, Finetti P, Rougemont J, Charafe-Jauffret E, Nasser V, Loriod B, et al. Gene expression profiling for molecular characterization of inflammatory breast cancer and prediction of response to chemotherapy. Cancer Res. 2004;64:8558–8565. doi: 10.1158/0008-5472.CAN-04-2696. [DOI] [PubMed] [Google Scholar]
- 45.Frederiksen CM, Knudsen S, Laurberg S, Orntoft TF. Classification of Dukes’ B and C colorectal cancers using expression arrays. J Cancer Res Clin Oncol. 2003;129:263–271. doi: 10.1007/s00432-003-0434-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Topol L, Jiang X, Choi H, Garrett-Beal L, Carolan PJ, Yang Y. Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent beta-catenin degradation. J Cell Biol. 2003;162:899–908. doi: 10.1083/jcb.200303158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang WS, Wang JP, Wang T, Fang JY, Lan P, Ma JP. shRNA-mediated gene silencing of beta-catenin inhibits growth of human colon cancer cells. World J Gastroenterol. 2007;13:6581–6587. doi: 10.3748/wjg.v13.i48.6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dry JR, Pavey S, Patilas CA, Harbron C, Runswqick S, Hodgson D, et al. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244) Cancer Res. 2010;70:2264–2273. doi: 10.1158/0008-5472.CAN-09-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baselga J, Rosen N. Determinants of RASistance to anti-epidermal growth factor receptor agents. J Clin Oncol. 2008;26:1582–1584. doi: 10.1200/JCO.2007.15.3700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.