Abstract
Human pluripotent stem cells (hPSCs) offer unprecedented opportunities to study cellular differentiation and model human diseases. The ability to precisely modify any genomic sequence holds the key to realizing the full potential of hPSCs. Thanks to the rapid development of novel genome editing technologies driven by the enormous interest in the hPSC field, genome editing in hPSCs has evolved from being a daunting task a few years ago to a routine procedure in most laboratories. Here, we provide an overview of the mainstream genome editing tools, including zinc finger nucleases, transcription activator-like effector nucleases, clustered regularly interspaced short palindromic repeat/CAS9 RNA-guided nucleases, and helper-dependent adenoviral vectors. We discuss the features and limitations of these technologies, as well as how these factors influence the utility of these tools in basic research and therapies.
Keywords: Embryonic Stem Cell, Gene Therapy, Induced Pluripotent Stem Cell (iPSC), Stem Cells, CRISPR/CAS9, HDAdV, TALEN, Genome Editing, ZFN
Introduction
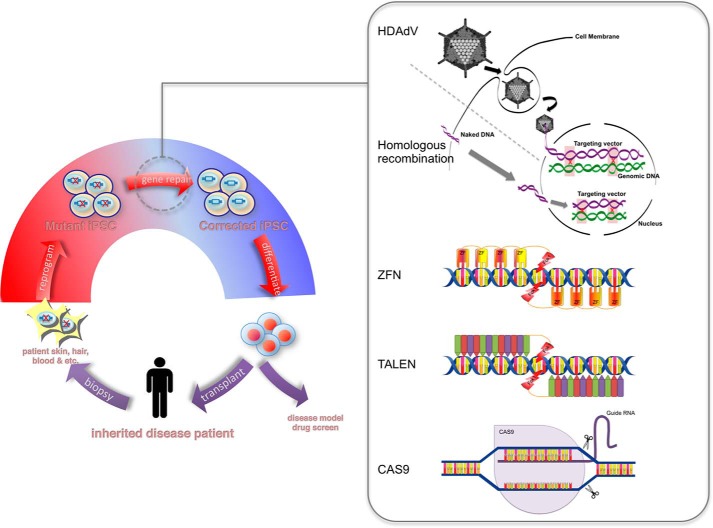
The discovery of induced pluripotent stem cells (iPSCs)5 seven years ago has reignited the enthusiasm for cell-based therapy. The ability of iPSCs to undergo unlimited division while maintaining genomic integrity provides a way to overcome the senescence barrier of aged somatic cells. The capacity of iPSCs to differentiate into cells of the three germ layers has been extensively documented in the field. Taken together, it is not hard to appreciate why human iPSC (hiPSC)-based autologous transplantation is heralded as the future of regenerative medicine (Fig. 1). One area that has drawn great interest is correction of genetic diseases in patient-specific hiPSCs with a prospect of personalized cell therapy.
FIGURE 1.
Conceptual schematic showing the application of genome editing and iPSC technologies in regenerative medicine.
Pluripotent stem cells are especially amenable for genome editing because they can undergo extensive tissue culture manipulations, such as drug selection and clonal expansion, while still maintaining their pluripotency and genome stability. Gene targeting in mouse embryonic stem cells by homologous recombination (HR) has proven to be a staple technique for studying gene function (1, 2). However, the same strategy does not translate well into human embryonic stem cells (hESCs) or hiPSCs (3). Although the classical HR method has been successfully used to generate knock-in reporter lines and to correct gene mutations in hESCs, the reported targeting efficiencies are at least several orders of magnitude lower than what is achievable in mouse embryonic stem cells (4, 5). This is likely due to the intrinsic differences in the DNA repair process between humans and mice, as measures to improve single-cell survival and DNA transfection did not have a dramatic effect on gene targeting efficiency (6, 7). However, other methods aimed at promoting HR proved more fruitful (3). In the past several years, there has been a spike of interest in genome editing in hESCs and hiPSCs, possibly due to the potential of this technology in modeling and correcting a myriad of genetic diseases (8, 9). This has fueled a rapid development in novel technologies for targeted modification of the human genome. Here, we aim to provide a timely update on the current genome editing technologies and discuss the factors that influence the choice of an appropriate technology.
Genome Editing with Synthetic Nucleases
Introduction of a DNA double-strand break (DSB) triggers DNA repair responses via two major pathways (10). The DSBs are repaired primarily by the non-homologous end joining (NHEJ) pathway, which either restores the original sequence or creates small insertions or deletions (indels). Alternatively, the DSBs may be repaired through HR based on a homologous repair template (a process termed homology-directed repair (HDR)). HDR can be co-opted to introduce desired sequence changes when an exogenous template is introduced. There have been several approaches of engineering synthetic nucleases to achieve targeted cleavage of a specific site in the human genome. These nucleases share two important properties: 1) a mechanism of recognizing a sufficiently long target sequence that occurs only once in the genome and 2) a catalytic activity that is activated by sequence-specific binding.
Zinc Finger Nucleases
Zinc finger nucleases (ZFNs) are modular proteins consisting of a series of the Cys2-His2 zinc finger DNA-binding motifs and the DNA cleavage domain of the type IIS restriction enzyme FokI (11). Each zinc finger motif recognizes 3–4 bp of DNA, and the modular nature of the zinc finger motif allows specific binding to a composite sequence by linking several motifs in tandem. The activity of the FokI nuclease requires dimerization. Therefore, two ZFNs are designed to bind on opposite sides of the target site with the respective FokI domains oriented toward each other (Fig. 1). This design further enhances the specificity of ZFNs, as dimerization of FokI and thus cleavage are dependent on a longer target. A pair of ZFNs that each contains three zinc finger motifs are sufficient to ensure a unique intended target of an 18-bp sequence by chance in the human genome, although four to six motifs are more commonly used to increase specificity (12). ZFNs have been successfully used to edit the genome of many organisms, including humans (11, 13).
Among the synthetic nucleases, ZFNs have the advantage of being the most studied. There are many technical resources to aid the design and assembly of ZFNs. Well designed ZFNs are highly effective in the disruption, addition, or correction of the target gene. Recently, ZFN-mediated disruption of the TAP2 gene in hiPSCs made it possible to produce an unlimited amount of antigen-presenting cells for vaccination therapy (14). ZFN-mediated insertion of transgenes into the genomic safe harbor locus (AAVS1) of hiPSCs has been used to engineer cells for in vivo imaging and to correct the globin imbalance in α-thalassemia (15, 16). In the case of HDR, ZFNs greatly simplify the experimental design, as short oligonucleotides may be used as templates, and antibiotic selection may be omitted (17). For a detailed account of genome editing of hESCs and hiPSCs using ZFNs, we would like to refer the readers to recent reviews (18, 19).
Despite many successful reports of ZFN-mediated genome editing of human cells in academic and clinical research, ZFN technology has several limitations. The assembly of zinc finger motifs is not modular in the strictest sense. The binding affinity of individual motifs is context-dependent. In other words, an assembled ZFN does not necessarily have high affinity for the sequence that is the composite of the 3-bp cognate sequence of each zinc finger motif (20). Other selection-based methods have been invented to address the high failure rate of modularly assembled ZFNs (reviewed in Ref. 14). With these methods, the number of targetable sites is reduced. In any case, it requires a considerable amount of experience, time, and effort to achieve proficiency at making functional ZFNs.
Unintended cleavage at so-called off-target sites is another concern with ZFNs (21). Off-target cleavage could cause cytotoxicity, introduce unknown mutations, and confound the analysis of the effects of the intended genetic changes. FokI variants that form obligate heterodimers have been used to minimize off-target cutting due to homodimerization of ZFNs (22). Converting FokI into a DNA-nicking enzyme also helps to reduce the risk of mutagenesis due to off-target cleavage because single-strand breaks stimulate HDR on target while minimizing NHEJ off target (23, 24). Even with these technical improvements, it is still important to monitor off-target cleavage of ZFNs intended for therapeutics. Bioinformatics tools may be used to predict potential off-target sites. However, this approach does not take into consideration the differences between the target genome and the reference genome. Furthermore, two studies have shown that the in vivo off-target sites of ZFN cleavage could not be fully predicted by in vitro or in silico analysis (21, 26). At the cellular level, although γH2AX and p53BP1 foci are used to monitor DSB sites after the introduction of ZFNs, they cannot distinguish ZFN-mediated DSBs and spontaneous DSBs. A more direct approach will be sequencing the genome, which is becoming more practical as next generation sequencing becomes more affordable. Encouragingly, Yusa et al. (27) found no evidence of off-target cleavage-induced mutation in a ZFN-modified hiPSC line by exome sequencing.
Transcription Activator-like Effector Nucleases
Recently, another synthetic nuclease termed transcription activator-like effector nuclease (TALEN) has emerged as an alternative to ZFNs. The architecture of TALENs is similar to that of ZFNs, with a DNA-binding domain fused to a FokI domain (Fig. 1). The DNA-binding module of TALENs is sourced from the DNA-binding repeat domain of transcription activator-like effectors (TALEs), bacterial proteins of the plant pathogen Xanthomonas (28). Unlike zinc fingers, each TALE repeat has a 1-to-1-bp correspondence. Most of the 34 amino acids in each repeat are highly conserved, except for two repeat-variable di-residues, which determine the DNA base specificity (29, 30). TALEs naturally exist as tandem arrays of repeated motifs, which means that they have been “pre-optimized” during evolution for modular assembly. This translates into a unique advantage when assembling TALENs. In contrast to ZFNs, modular assembly of TALENs has a success rate up to 100%, which has two important implications. 1) Any laboratory with a basic molecular biology setup can produce functional TALENs within days, and 2) automated high-throughput TALEN production is possible (31–33). Indeed, within 2 short years after the first report of genome editing in hPSCs using TALENs, a library of TALENs targeting 18,740 protein-coding genes in the human genome has already been constructed using a high-throughput cloning method (34). In side-by-side comparisons to ZFNs, TALENs exhibit comparable efficacy and lower toxicity (35). As demonstrated in a recent study, TALENs have greatly facilitated genome editing in hPSCs for generating disease models (36).
Despite the enthusiasm for TALENs, the technology is still in its infancy and thus faces many unresolved issues. TALEN target-site selection is restricted by the requirement of a preceding T base (29, 37). Although this should not prohibit successful design of TALENs in most cases, it may be an issue when modifying a specific mutation especially for future cell-based gene therapy. The reported sensitivity of TALENs to 5-methylcytosine could be a more serious drawback of the TALEN technology because of the prevalence of this DNA modification in the genome (38, 39). Recent evidence shows that this problem may be overcome by engineering 5-methylcytosine-insensitive TALE DNA-binding domains (39). The extent of off-target effect of TALENs is largely unknown. Recently, there have been several efforts to systematically map off-target cleavage of ZFNs and meganucleases. These include in vitro selection of binding targets by systemic evolution of ligands by exponential enrichment (SELEX), in vitro selection of cleavage sequence, and tagging transiently appearing DSBs by NHEJ-mediated integration of adeno-associated virus vectors or integrase-defective lentiviral vectors (21, 26, 40). These methods are also applicable to TALENs. It is worth noting that unbiased approaches of mapping off-target cleavage sites, such as those reported by Gabriel et al. (21) and Petek et al. (40), show that in silico screening of potential cleavage targets based on sequence homology does not predict actual cleavage in vivo. Another method of surveying off-target effects of TALENs is genome sequencing. Ding et al. (36) compared TALEN-modified exomes with the parental exome and found little evidence of indels, a hallmark of TALEN cleavage sites. However, they noted approximately seven single-nucleotide variants per clone (36). As the authors suggested, these single-nucleotide variants may reflect heterogeneity in the parental population, which manifests itself during the extended culture that is required for genome editing, as they do not coincide with predicted TALEN off-target sites. Using a single cell-derived parental line may help clarify this issue. To gain a complete picture of off-target effects of TALENs in noncoding regions of the genome, high-coverage whole-genome sequencing is necessary (36).
Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/CAS9 RNA-guided Nucleases
It is clear from the development of ZFNs and TALENs that specific sequence recognition is the key element in the design of synthetic nucleases. Other than DNA-binding polypeptides, nature has evolved other means to interact with specific DNA sequences. Bacteria and archaea possess a unique adaptive immune system based on an RNA-guided DNA endonuclease to destroy foreign DNA (41). DNA fragments of past invaders are integrated as spacers in the CRISPR genomic loci. The CRISPR loci are transcribed to produce CRISPR RNAs (crRNAs), which contain a unique seed sequence complementary to target DNA (called protospacer) and a repeat region that hybridizes to a small RNA called transactivated crRNA (tracrRNA). The crRNA and tracrRNA hybrid guides CAS (CRISPR-associated) proteins to cleave target DNA (42). Since the beginning of 2013, there has been a surge of reports of successful adaptation of the CRISPR/CAS9 system for human genome editing (43–46). Targeted cleavage by CRISPR/CAS9 in human cells requires the introduction of a CAS9 expression vector and the guide RNAs (crRNA and tracrRNA). The system has been further simplified by fusing the two RNAs into a chimeric RNA (44). Targeting CAS9 to a desired genomic site only entails cloning the 20 bp of target sequence into the spacer region of the crRNA locus (Fig. 1). Several versions of the CRISPR/CAS9 system are available from plasmid-sharing services. This potentially makes the exclusivity of genome editing technology a thing of the past. The CRISPR/CAS9 system is also amenable to high-throughput construction of a library of targeting vectors. Because of the small size of the guide RNA, it is also possible to deliver multiple guide RNAs at the same time to achieve multiplex targeting (45, 47). A recent study has compared the efficiency of CRISPR/CAS9 with that of TALENs in targeting the same genomic sites in hPSCs (48). CAS9 outperformed TALENs in gene disruption, gene knock-in, and bi-allelic targeting across all loci. It was speculated that this might be due to a higher expression level and lower cytotoxicity of the CAS9 protein (48).
Despite its versatility, the CRISPR/CAS9 system has several limitations. First, the targetable sites of CAS9 are constrained by the requirement of a GN20GG sequence motif, which may cause a problem when targeting certain loci. Second, up to six mismatches between crRNA and target DNA are tolerated by CAS9, which may result in off-target cleavage (41). Indeed, a recent study showed that CRISPR/CAS9 nucleases induce mutations at off-target sites with up to five mismatches (49). More importantly, frequencies of off-target mutations are equal to or higher than those of on-target mutations (49). CAS9 mutants with a more stringent requirement of crRNA-target DNA complementation may be engineered by directed evolution. CAS9 has been converted into a nickase, which reduces mutagenesis at off-target sites (45). In addition, a systematic examination of the off-target effects of CRISPR/CAS9 ought to be performed using the technologies discussed above with regard to ZFNs and TALENs.
Genome Editing without Nucleases
There are other tools that enable efficient genome editing in human cells without the aid of synthetic nucleases. Compared with synthetic nuclease-based methods, the classic HR method is less likely to have off-target effects. However, the low efficiency of HR in human cells is a major roadblock (5). Several approaches have been designed to overcome this issue, including the use of bacterial artificial chromosomes, adeno-associated viruses, and helper-dependent adenoviral vectors (HDAdVs) (50–56). Among these, HDAdVs have enjoyed the most success in genome editing of hPSCs. HDAdVs are so-called last-generation adenoviruses developed for gene therapy. They are non-integrative viral vectors engineered to be low immunogenic and able to transduce a wide range of cell types with high efficiency. Since all viral genes are deleted from HDAdVs, they have a large cloning capacity of 36 kb (57). These features make HDAdVs ideal vectors for delivering HR donor constructs with extended homology arms (Fig. 1). HDAdVs have been successfully applied to genome editing in hPSCs, including genetic correction of Hutchinson-Gilford progeria syndrome, sickle cell disease, and Parkinson disease in hiPSCs (50–53). The percentage of clones harboring the correct targeting event via HR after drug selection is significantly higher than the classical HR method (range of 17–100%). Unlike synthetic nucleases, HDAdVs offer a prospect of in vivo delivery. Additional benefits of HDAdVs include 1) no restriction on target site selection, 2) simultaneous introduction of multiple modifications to a large span of DNA region, 3) efficient transduction into a wide range of cell types, and 4) no risk of off-target cleavage. One study has looked at the genomic and epigenomic status of HDAdV-modified hiPSCs and found it to be highly similar to that of the parental lines (52). However, HDAdVs carry a rare chance of integrating into the genome, although such an event was not detected by using a variety of techniques in our research (50). The construction of HDAdVs is rather technically challenging and labor-intensive, which may present a barrier to adopting this technology. Another drawback is that HDAdV-mediated genome editing requires drug selection, which is lengthy and necessitates an additional step to remove the drug-selectable marker, and a genomic scar (e.g. a loxP or FRT site) is left behind. It is possible to deliver synthetic nucleases (e.g. TALENs) and the donor construct in an “all-in-one” HDAdV, therefore avoiding drug selection and the issues associated with it.
Conclusions and Perspectives
The rapid progression of genome editing technology is a boon to both basic science and cell and gene therapy. Ideally, cells that have undergone genome editing should contain only the intended change in an otherwise isogenic background, thus providing the most stringent test of gene function. However, this may not be the case due to off-target effects of ZFNs, TALENs, and CRISPR/CAS9. One way to minimize these experimental confounders is to improve the design of the genome editing tools. DNA nickase, obligate heterodimeric FokI, and mutant variants with enhancer specificity all increase the fidelity of synthetic nucleases. Because it is still difficult to determine the full extent of off-target effects of synthetic nucleases, prudent experimental designs should be the second line of defense. It may not be enough to just analyze multiple clones resulting from one nuclease design because off-target cleavage could be common among clones. If clones generated by an independent nuclease that is targeted to a different region of the same locus have the same phenotypes, then the genotype-phenotype relationship can be established with confidence.
Other than elucidating gene function and modeling human diseases, genome editing technologies can facilitate a variety of novel studies, such as improving directed differentiation of hiPSCs by generating lineage-specific reporter lines, engineering dendritic cell-directed cancer vaccines and T cell immunotherapies (58, 59), and generating human cell lines with enhanced production of biomolecules for biotechnology and pharmaceutical industries.
The use of genome editing in the clinic requires extra scrutiny. Although only a few exonic mutations are induced during gene correction, and mice transplanted with these modified cells do not have tumors, the long-term safety issue is still unclear (27, 60). We still do not understand how to control mutation accumulation during culture and the implications of these mutations in vivo. Since these unintended mutations are permanent changes that may have long-term delayed adverse effects, such as those observed in the X-linked severe combined immunodeficiency trial (25, 61), long-term evaluation of the safety of cells that have undergone genome editing in primates is highly recommended. Because genome editing technology is an emerging field, it would be advisable that the proper authorities, including the Food and Drug Administration, issue specific and sensible guidelines for the design of preclinical studies. A better understanding of the risk of genome editing technologies may allow their use in non-life-threatening conditions, therefore potentially benefiting more people. It may be unrealistic and unreasonable to require a zero tolerance of mutations or demand a full investigation into every genetic variance. Perhaps the key is to strike a balance between risks and benefits to the patients. Today, it is clear that we are experiencing the dawn of a new era in biomedical research ushered in by genome editing technologies.
Acknowledgments
We thank Hsin-Kai Liao and Ying Gu for helpful discussions; Ilir Dubova, May Schwarz, and Peter Schwarz for administrative support; and James A. Cooper for help with illustration.
This work was supported in part by grants from the G. Harold and Leila Y. Mathers Charitable Foundation, the Ellison Medical Foundation, the Leona M. and Harry B. Helmsley Charitable Trust, and the Glenn Foundation for Medical Research (to J. C. I. B.). This is the sixth article in the Thematic Minireview Series “Development of Human Therapeutics Based on Induced Pluripotent Stem Cell (iPSC) Technology.”
- iPSC
- induced pluripotent stem cell
- hiPSC
- human iPSC
- HR
- homologous recombination
- hESC
- human embryonic stem cell
- DSB
- DNA double-strand break
- NHEJ
- non-homologous end joining
- HDR
- homology-directed repair
- ZFN
- zinc finger nuclease
- TALEN
- transcription activator-like effector nuclease
- TALE
- transcription activator-like effector
- CRISPR
- clustered regularly interspaced short palindromic repeat
- crRNA
- CRISPR RNA
- tracrRNA
- transactivated crRNA
- HDAdV
- helper-dependent adenoviral vector.
REFERENCES
- 1. Melton D. W. (1994) Gene targeting in the mouse. BioEssays 16, 633–638 [DOI] [PubMed] [Google Scholar]
- 2. Babinet C., Cohen-Tannoudji M. (2001) Genome engineering via homologous recombination in mouse embryonic stem (ES) cells: an amazingly versatile tool for the study of mammalian biology. An. Acad. Bras. Cienc. 73, 365–383 [DOI] [PubMed] [Google Scholar]
- 3. Sancho-Martinez I., Li M., Izpisua Belmonte J. C. (2011) Disease correction the iPSC way: advances in iPSC-based therapy. Clin. Pharmacol. Ther. 89, 746–749 [DOI] [PubMed] [Google Scholar]
- 4. Ruby K. M., Zheng B. (2009) Gene targeting in a HUES line of human embryonic stem cells via electroporation. Stem Cells 27, 1496–1506 [DOI] [PubMed] [Google Scholar]
- 5. Zwaka T. P., Thomson J. A. (2003) Homologous recombination in human embryonic stem cells. Nat. Biotechnol. 21, 319–321 [DOI] [PubMed] [Google Scholar]
- 6. Bañuelos C. A., Banáth J. P., MacPhail S. H., Zhao J., Eaves C. A., O'Connor M. D., Lansdorp P. M., Olive P. L. (2008) Mouse but not human embryonic stem cells are deficient in rejoining of ionizing radiation-induced DNA double-strand breaks. DNA Repair 7, 1471–1483 [DOI] [PubMed] [Google Scholar]
- 7. Meek K., Dang V., Lees-Miller S. P. (2008) DNA-PK: the means to justify the ends? Adv. Immunol. 99, 33–58 [DOI] [PubMed] [Google Scholar]
- 8. Cherry A. B., Daley G. Q. (2013) Reprogrammed cells for disease modeling and regenerative medicine. Annu. Rev. Med. 64, 277–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Soldner F., Jaenisch R. (2012) Medicine. iPSC disease modeling. Science 338, 1155–1156 [DOI] [PubMed] [Google Scholar]
- 10. Chapman J. R., Taylor M. R., Boulton S. J. (2012) Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510 [DOI] [PubMed] [Google Scholar]
- 11. Carroll D. (2011) Genome engineering with zinc-finger nucleases. Genetics 188, 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bhakta M. S., Henry I. M., Ousterout D. G., Das K. T., Lockwood S. H., Meckler J. F., Wallen M. C., Zykovich A., Yu Y., Leo H., Xu L., Gersbach C. A., Segal D. J. (2013) Highly active zinc-finger nucleases by extended modular assembly. Genome Res. 23, 530–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Urnov F. D., Rebar E. J., Holmes M. C., Zhang H. S., Gregory P. D. (2010) Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646 [DOI] [PubMed] [Google Scholar]
- 14. Haruta M., Tomita Y., Yuno A., Matsumura K., Ikeda T., Takamatsu K., Haga E., Koba C., Nishimura Y., Senju S. (2013) TAP-deficient human iPS cell-derived myeloid cell lines as unlimited cell source for dendritic cell-like antigen-presenting cells. Gene Ther. 20, 504–513 [DOI] [PubMed] [Google Scholar]
- 15. Chang C. J., Bouhassira E. E. (2012) Zinc-finger nuclease-mediated correction of α-thalassemia in iPS cells. Blood 120, 3906–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y., Zhang W. Y., Hu S., Lan F., Lee A. S., Huber B., Lisowski L., Liang P., Huang M., de Almeida P. E., Won J. H., Sun N., Robbins R. C., Kay M. A., Urnov F. D., Wu J. C. (2012) Genome editing of human embryonic stem cells and induced pluripotent stem cells with zinc finger nucleases for cellular imaging. Circ. Res. 111, 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Soldner F., Laganière J., Cheng A. W., Hockemeyer D., Gao Q., Alagappan R., Khurana V., Golbe L. I., Myers R. H., Lindquist S., Zhang L., Guschin D., Fong L. K., Vu B. J., Meng X., Urnov F. D., Rebar E. J., Gregory P. D., Zhang H. S., Jaenisch R. (2011) Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell 146, 318–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Collin J., Lako M. (2011) Concise review: putting a finger on stem cell biology: zinc finger nuclease-driven targeted genetic editing in human pluripotent stem cells. Stem Cells 29, 1021–1033 [DOI] [PubMed] [Google Scholar]
- 19. Perez-Pinera P., Ousterout D. G., Gersbach C. A. (2012) Advances in targeted genome editing. Curr. Opin. Chem. Biol. 16, 268–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramirez C. L., Foley J. E., Wright D. A., Müller-Lerch F., Rahman S. H., Cornu T. I., Winfrey R. J., Sander J. D., Fu F., Townsend J. A., Cathomen T., Voytas D. F., Joung J. K. (2008) Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods 5, 374–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gabriel R., Lombardo A., Arens A., Miller J. C., Genovese P., Kaeppel C., Nowrouzi A., Bartholomae C. C., Wang J., Friedman G., Holmes M. C., Gregory P. D., Glimm H., Schmidt M., Naldini L., von Kalle C. (2011) An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 29, 816–823 [DOI] [PubMed] [Google Scholar]
- 22. Doyon Y., Vo T. D., Mendel M. C., Greenberg S. G., Wang J., Xia D. F., Miller J. C., Urnov F. D., Gregory P. D., Holmes M. C. (2011) Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat. Methods 8, 74–79 [DOI] [PubMed] [Google Scholar]
- 23. Ramirez C. L., Certo M. T., Mussolino C., Goodwin M. J., Cradick T. J., McCaffrey A. P., Cathomen T., Scharenberg A. M., Joung J. K. (2012) Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res. 40, 5560–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang J., Friedman G., Doyon Y., Wang N. S., Li C. J., Miller J. C., Hua K. L., Yan J. J., Babiarz J. E., Gregory P. D., Holmes M. C. (2012) Targeted gene addition to a predetermined site in the human genome using a ZFN-based nicking enzyme. Genome Res. 22, 1316–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Woods N. B., Bottero V., Schmidt M., von Kalle C., Verma I. M. (2006) Gene therapy: therapeutic gene causing lymphoma. Nature 440, 1123. [DOI] [PubMed] [Google Scholar]
- 26. Pattanayak V., Ramirez C. L., Joung J. K., Liu D. R. (2011) Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods 8, 765–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yusa K., Rashid S. T., Strick-Marchand H., Varela I., Liu P. Q., Paschon D. E., Miranda E., Ordóñez A., Hannan N. R., Rouhani F. J., Darche S., Alexander G., Marciniak S. J., Fusaki N., Hasegawa M., Holmes M. C., Di Santo J. P., Lomas D. A., Bradley A., Vallier L. (2011) Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells. Nature 478, 391–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Joung J. K., Sander J. D. (2013) TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14, 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A., Bonas U. (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 30. Mak A. N., Bradley P., Cernadas R. A., Bogdanove A. J., Stoddard B. L. (2012) The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335, 716–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reyon D., Tsai S. Q., Khayter C., Foden J. A., Sander J. D., Joung J. K. (2012) FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 30, 460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmid-Burgk J. L., Schmidt T., Kaiser V., Höning K., Hornung V. (2013) A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat. Biotechnol. 31, 76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Briggs A. W., Rios X., Chari R., Yang L., Zhang F., Mali P., Church G. M. (2012) Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 40, e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim Y., Kweon J., Kim A., Chon J. K., Yoo J. Y., Kim H. J., Kim S., Lee C., Jeong E., Chung E., Kim D., Lee M. S., Go E. M., Song H. J., Kim H., Cho N., Bang D., Kim S., Kim J. S. (2013) A library of TAL effector nucleases spanning the human genome. Nat. Biotechnol. 31, 251–258 [DOI] [PubMed] [Google Scholar]
- 35. Mussolino C., Morbitzer R., Lütge F., Dannemann N., Lahaye T., Cathomen T. (2011) A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 39, 9283–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ding Q., Lee Y. K., Schaefer E. A., Peters D. T., Veres A., Kim K., Kuperwasser N., Motola D. L., Meissner T. B., Hendriks W. T., Trevisan M., Gupta R. M., Moisan A., Banks E., Friesen M., Schinzel R. T., Xia F., Tang A., Xia Y., Figueroa E., Wann A., Ahfeldt T., Daheron L., Zhang F., Rubin L. L., Peng L. F., Chung R. T., Musunuru K., Cowan C. A. (2013) A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell 12, 238–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moscou M. J., Bogdanove A. J. (2009) A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501. [DOI] [PubMed] [Google Scholar]
- 38. Bultmann S., Morbitzer R., Schmidt C. S., Thanisch K., Spada F., Elsaesser J., Lahaye T., Leonhardt H. (2012) Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res. 40, 5368–5377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Valton J., Dupuy A., Daboussi F., Thomas S., Maréchal A., Macmaster R., Melliand K., Juillerat A., Duchateau P. (2012) Overcoming transcription activator-like effector (TALE) DNA binding domain sensitivity to cytosine methylation. J. Biol. Chem. 287, 38427–38432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Petek L. M., Russell D. W., Miller D. G. (2010) Frequent endonuclease cleavage at off-target locations in vivo. Mol. Ther. 18, 983–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wiedenheft B., Sternberg S. H., Doudna J. A. (2012) RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338 [DOI] [PubMed] [Google Scholar]
- 43. Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. (2013) RNA-programmed genome editing in human cells. eLife 2, e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cho S. W., Kim S., Kim J. M., Kim J. S. (2013) Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 230–232 [DOI] [PubMed] [Google Scholar]
- 45. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., Norville J. E., Church G. M. (2013) RNA-guided human genome engineering via Cas9. Science 339, 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., Zhang F., Jaenisch R. (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ding Q., Regan S. N., Xia Y., Oostrom L. A., Cowan C. A., Musunuru K. (2013) Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 12, 393–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., Joung J. K., Sander J. D. (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li M., Suzuki K., Qu J., Saini P., Dubova I., Yi F., Lee J., Sancho-Martinez I., Liu G. H., Izpisua Belmonte J. C. (2011) Efficient correction of hemoglobinopathy-causing mutations by homologous recombination in integration-free patient iPSCs. Cell Res. 21, 1740–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu G. H., Qu J., Suzuki K., Nivet E., Li M., Montserrat N., Yi F., Xu X., Ruiz S., Zhang W., Wagner U., Kim A., Ren B., Li Y., Goebl A., Kim J., Soligalla R. D., Dubova I., Thompson J., Yates J., 3rd, Esteban C. R., Sancho-Martinez I., Izpisua Belmonte J. C. (2012) Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature 491, 603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu G. H., Suzuki K., Qu J., Sancho-Martinez I., Yi F., Li M., Kumar S., Nivet E., Kim J., Soligalla R. D., Dubova I., Goebl A., Plongthongkum N., Fung H. L., Zhang K., Loring J. F., Laurent L. C., Izpisua Belmonte J. C. (2011) Targeted gene correction of laminopathy-associated LMNA mutations in patient-specific iPSCs. Cell Stem Cell 8, 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Suzuki K., Mitsui K., Aizawa E., Hasegawa K., Kawase E., Yamagishi T., Shimizu Y., Suemori H., Nakatsuji N., Mitani K. (2008) Highly efficient transient gene expression and gene targeting in primate embryonic stem cells with helper-dependent adenoviral vectors. Proc. Natl. Acad. Sci. U.S.A. 105, 13781–13786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mitsui K., Suzuki K., Aizawa E., Kawase E., Suemori H., Nakatsuji N., Mitani K. (2009) Gene targeting in human pluripotent stem cells with adeno-associated virus vectors. Biochem. Biophys. Res. Commun. 388, 711–717 [DOI] [PubMed] [Google Scholar]
- 55. Khan I. F., Hirata R. K., Wang P. R., Li Y., Kho J., Nelson A., Huo Y., Zavaljevski M., Ware C., Russell D. W. (2010) Engineering of human pluripotent stem cells by AAV-mediated gene targeting. Mol. Ther. 18, 1192–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Song H., Chung S. K., Xu Y. (2010) Modeling disease in human ESCs using an efficient BAC-based homologous recombination system. Cell Stem Cell 6, 80–89 [DOI] [PubMed] [Google Scholar]
- 57. Alba R., Bosch A., Chillon M. (2005) Gutless adenovirus: last-generation adenovirus for gene therapy. Gene Ther. 12, S18–S27 [DOI] [PubMed] [Google Scholar]
- 58. Qu X., Wang P., Ding D., Li L., Wang H., Ma L., Zhou X., Liu S., Lin S., Wang X., Zhang G., Liu S., Liu L., Wang J., Zhang F., Lu D., Zhu H. (2013) Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res. 41, 7771–7782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Provasi E., Genovese P., Lombardo A., Magnani Z., Liu P. Q., Reik A., Chu V., Paschon D. E., Zhang L., Kuball J., Camisa B., Bondanza A., Casorati G., Ponzoni M., Ciceri F., Bordignon C., Greenberg P. D., Holmes M. C., Gregory P. D., Naldini L., Bonini C. (2012) Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat. Med. 18, 807–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Howden S. E., Gore A., Li Z., Fung H. L., Nisler B. S., Nie J., Chen G., McIntosh B. E., Gulbranson D. R., Diol N. R., Taapken S. M., Vereide D. T., Montgomery K. D., Zhang K., Gamm D. M., Thomson J. A. (2011) Genetic correction and analysis of induced pluripotent stem cells from a patient with gyrate atrophy. Proc. Natl. Acad. Sci. U.S.A. 108, 6537–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M. P., Wulffraat N., Leboulch P., Lim A., Osborne C. S., Pawliuk R., Morillon E., Sorensen R., Forster A., Fraser P., Cohen J. I., de Saint Basile G., Alexander I., Wintergerst U., Frebourg T., Aurias A., Stoppa-Lyonnet D., Romana S., Radford-Weiss I., Gross F., Valensi F., Delabesse E., Macintyre E., Sigaux F., Soulier J., Leiva L. E., Wissler M., Prinz C., Rabbitts T. H., Le Deist F., Fischer A., Cavazzana-Calvo M. (2003) LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302, 415–419 [DOI] [PubMed] [Google Scholar]