Background: It is not known whether the redox status of cryptochrome affects circadian clock resetting.
Results: Blocking cryptochrome photoreduction does not impair cryptochrome-mediated Timeless degradation and light can induce a conformational change in cryptochrome with oxidized flavin.
Conclusion: Cryptochrome can mediate its function in either oxidized or reduced form.
Significance: This study provides novel insight into cryptochrome photosignaling.
Keywords: Circadian Clock, Drosophila, Flavoproteins, Protein Conformation, Signal Transduction, Circadian Clock Resetting, Cryptochrome, Photoreceptor, Timeless Degradation
Abstract
Cryptochrome (CRY) is the primary circadian photoreceptor in Drosophila. Upon light absorption, dCRY undergoes a conformational change that enables it to bind to Timeless (dTIM), as well as to two different E3 ligases that ubiquitylate dTIM and dCRY, respectively, resulting in their proteolysis and resetting the phase of the circadian rhythm. Purified dCRY contains oxidized flavin (FADox), which is readily photoreduced to the anionic semiquinone through a set of 3 highly conserved Trp residues (Trp triad). The crystal structure of dCRY has revealed a fourth Trp (Trp-536) as a potential electron donor. Previously, we reported that the Trp triad played no role in photoinduced proteolysis of dCRY in Drosophila cells. Here we investigated the role of the Trp triad and Trp-536, and the redox status of the flavin on light-induced proteolysis of both dCRY and dTIM and resetting of the clock. We found that both oxidized (FADox) and reduced (FAD⨪) forms of dCRY undergo light-induced conformational change in vitro that enable dCRY to bind JET and that Trp triad and Trp-536 mutations that block known or presumed intraprotein electron transfer reactions do not affect dCRY phototransduction under bright or dim light in vivo as measured by light-induced proteolysis of dCRY and dTIM in Drosophila S2R+ cells. We conclude that both oxidized and reduced forms of dCRY are capable of photosignaling.
Introduction
Cryptochromes (CRYs)2 play essential roles in regulating the circadian clock in animals and in blue light-dependent growth and development in plants (1–7). In mice, CRY functions as a core component of the molecular clock, independent of light (1). In insects, CRY functions either as the primary circadian photoreceptor (8) or as a core component of the clock depending on the insect species and the type of CRY they possess (8, 9). Some of the insect CRYs are evolutionarily related to Drosophila CRY (designated Type 1 CRY), and some are related to the mammalian CRY (Type 2 CRY). Thus, Drosophila has only one CRY, which functions primarily as a photoreceptor; the monarch butterfly has both a Drosophila-like CRY (Type 1), which functions as a photoreceptor, and a mammalian-like CRY (Type 2), which is a core component of the molecular clockwork (9). Finally, honeybees and ants have only a Type 2 CRY, which is a core clock component with no known photosensory function. A unique property of Type 1 CRYs is that upon light exposure, they promote ubiquitylation and subsequent proteolysis of the core clock protein Timeless (dTIM) and, at a slower rate, CRY itself (10, 11), and thus help reset the circadian clock.
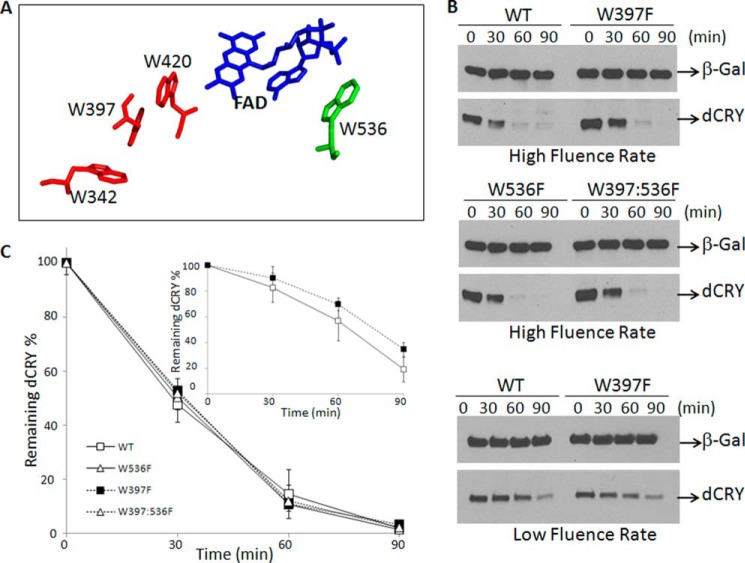
It is known that absorption of light by the flavin cofactor of dCRY causes a conformational change in the protein, mainly the disengagement of the C-terminal 20 amino acids from the compact photolyase homology region (PHR) (12, 13). This conformational change promotes interaction of dCRY with its heterodimeric partner dTIM (14, 15) and with JET and BRWD3-CRL4 (16) E3 ligases, leading to rapid proteolysis of dTIM that results in resetting of the circadian clock. This is followed by a slower degradation of dCRY to down-regulate the photosignal and desensitize the clock to light, enabling dTIM accumulation and re-establishment of the repressive phase. At present, it is unclear how light absorption leads to a conformational change in dCRY. It is known, however, that deletion of the C-terminal 20 amino acids of dCRY confers a constitutively active phenotype, and this has led to the suggestion that light causes the movement of at least this C-terminal extension (14, 15). This prediction was confirmed by partial proteolysis with trypsin in vitro. Upon light exposure two peptide bonds at the junction of the core PHR domain and the C-terminal extension become sensitive to the protease (13). The photophysical/photochemical reactions leading to the conformational change in CRY, in contrast to other flavin-based photosensory proteins (LOV and BLUF families), are not known (6, 17). The photolyase/CRY family, however, possesses a conserved so-called Trp triad extending from the surface of the protein to the flavin binding pocket (18, 19). In Drosophila the 3 tryptophans that make up the triad are Trp-342, Trp-397, and Trp-420, respectively (20–22), as shown in red in Fig. 1A. The Trp triad has been implicated in CRY function ranging from plant growth in Arabidopsis to circadian photoreception in Drosophila (5) to magnetoreception in flies (23, 24) and birds (2).
FIGURE 1.
Light-induced proteolysis of wild-type and mutant dCRYs in Drosophila S2R+ cells. A, spatial relation of the flavin cofactor and Trp residues involved in photoinduced electron transfer and cofactor binding based on high resolution crystal structure of dCRY (21). The triad (Trp-342, Trp-397, Trp-420) is in red. Replacement of any of the Trp residues of the triad with Phe blocks flavin photoreduction. Replacement of the proximal Trp (Trp-420) with Arg abolishes flavin binding (31, 32). B, light-induced proteolysis of dCRY in S2R+ cells. Cells were transfected with vectors expressing β-galactosidase (β-gal) and the indicated forms of dCRY and 36 h later were irradiated with high (30 μmol m−2 s−1) or low (3 μmol m−2 s−1) fluence rates of 366-nm light at room temperature for the indicated times and then analyzed by immunoblotting. C, quantitative analysis of proteolysis data. Averages from three experiments including the one shown in panel B are plotted for high (main plot) and low (inset) fluence rates. Bars indicate S.E.
When dCRY (and other Type 1 CRYs) are purified, they contain the flavin in the two-electron oxidized (FADox) form, and upon exposure to light, charge transfer through the Trp triad leads to photoreduction of the flavin to the anionic semiquinone (FAD⨪) (25–27). Initially, it was thought that charge transfer through the Trp triad that results in photoreduction is responsible for the conformational change (25). However, blocking the electron transfer by the Trp → Phe substitution of any member of the triad blocks photoreduction in vitro but affects neither the photosensory function of CRY in Drosophila cells as measured by light-induced proteolysis of dCRY (26, 27) nor magnetoreception in flies as measured by behavioral assays (23). Furthermore, using purified dCRY it was found that photoexcitation of the chemically reduced FAD⨪ form of dCRY caused a conformational change in the protein as evidenced by light-stimulated specific proteolysis of dCRY and light-stimulated dCRY-JET binding (13). These results in aggregate led to the conclusion that in vivo the flavin in dCRY is in the FAD⨪ form and that the FADox form is a purification artifact, as has been found for photolyase (28). However, subsequent work raised the possibility that light fluences insufficient to reduce the FAD of the Trp triad mutants in vitro might be sufficient to reduce the FAD of the Trp triad mutants in vivo in the potentially more reducing intracellular milieu (29). Moreover, a high resolution crystal structure of dCRY revealed that a Trp residue (Trp-536) in the C-terminal extension of the cryptochrome, shown in green in Fig. 1A, is in the proximity of the flavin because of the folding of the C-terminal extension onto the PHR domain (20–22). This raised the possibility that in the Trp triad mutants, Trp-536 could function as an alternative electron donor to the flavin in vivo, and therefore intraprotein electron transfer could be the primary photochemical event in dCRY photosignaling. Finally, we also note that the in vivo tests of the role of flavin photoreduction in dCRY signaling consisted of photoinduced dCRY proteolysis (13, 27), whereas the main phase-setting reaction in flies is the light-induced and dCRY-mediated ubiquitylation and proteolysis of dTIM (10, 11, 14).
To address these issues, we conducted in vivo and in vitro experiments with dCRY mutants carrying Trp → Phe replacements in the Trp triad, in Trp-536, and in both. We find that in vivo, these mutations do not affect light-induced proteolysis of either dCRY or dTIM under high or low fluence rates. Unexpectedly, we also find that in vitro, the non-photoreducible dCRY (W397F/W536F) undergoes conformation change similar to WT dCRY containing reduced flavin (FAD⨪) as determined by light-sensitized tryptic proteolysis of dCRY and light-induced dCRY-JET binding. Thus, it appears that the redox status of dCRY does not affect its light-induced conformational change and photosignaling activity.
EXPERIMENTAL PROCEDURES
Plasmids
pAc5.1-V5-dCRY was created by inserting amplified dCRY (from pAc5.1-dCRY-V5HisA), which was described previously (27), and V5 coding sequence (in front of dCRY) into pAc5.1/V5-HisA vector (Invitrogen). We constructed pAc5.1-V5-dCRY, which expresses V5 tag at the N terminus, to avoid any unseen/unexpected contribution of a tag located at the C terminus because one of the mutations (W536F) we used in our study is located at the very C terminus and the C-terminal extension folds onto the PHR domain (20). pFast-FH-dCRY was described previously (30). Site-directed mutations in the cloned genes for W397F, W420R, W536F, and W397F/W536F of dCRY were introduced by standard methods using the QuikChange method (Stratagene). pAc5.1-V5-JET was constructed by inserting the amplified JET coding sequence from pGex4T1-JET and the V5 coding sequence (upstream of JET) into the pAc5.1/V5-His A vector (Invitrogen). pGex4T1-JET expressing the GST-His-Jetlag fusion protein was described previously (13). pAc5.1.dTIM-V5His and pFast-dTIM-F were created by inserting the Timeless coding sequence with a FLAG tag sequence downstream of the dTIM coding sequence amplified from pIZ-Tim-YFP (kindly provided by Michael W. Young) into pAc5.1/V5-His A vector (Invitrogen) or into pFast-Bac1 (Invitrogen), respectively. pAc.5.1.β-Galactosidase-V5H was described previously (9).
Cell Lines
The Sf21 insect cell line (Stratagene) was cultured in Grace's insect medium (Invitrogen) containing 10% FBS (Cellgro) and 0.5× penicillin/streptomycin (Invitrogen) at 27 °C. The Drosophila S2R+ cell line was obtained from Prof. Steve Rogers (University of North Carolina at Chapel Hill) and maintained in Schneider's Drosophila medium (Sigma) containing 10% FBS and 0.5× penicillin/streptomycin at 27 °C. The Escherichia coli BL21 strain (Stratagene), used for the expression of Jetlag in fusion with glutathione S-transferase (GST), was cultured in LB medium (Invitrogen).
Preparation of Baculoviruses
Recombinant baculoviruses used to express Drosophila cryptochromes were described previously (30). Baculoviruses for dTIM were prepared using the Gibco BRL Bac-to-Bac baculovirus expression system (Invitrogen). Briefly, 5 ng of pFast-dTIM-F was transformed into E. coli DH10Bac, and recombinant bacmid DNAs were isolated from 1 ml of bacterial cultures grown from colonies of transformants. To generate recombinant virus, Sf21 cells seeded onto 6-well plates were transfected with 2 μg of bacmid DNAs using 8 μl of Cellfectin reagent (Invitrogen) according to the manufacturer's recommendations. The culture medium containing the parental virus stock was collected after 72 h, and cells were removed by centrifugation. After three more 72-h virus amplification steps, the fourth passage (P4) high titer stock was obtained as working stock solution to infect Sf21 cells for large scale expression of protein.
Protein Purification
To purify FLAG-tagged (F) recombinant proteins from insect cells, 1 liter of Sf21 cells (1 × 106 cells/ml) growing at 27 °C in spinner flasks was infected with 5–10 ml of the P4 high titer virus stocks. The cells were harvested 36 h later and lysed in 30 ml of lysis buffer containing 50 mm Tris, pH 7.5, 150 mm NaCl, 0.1% Nonidet P-40 (Sigma), 0.5% Triton X-100 (Sigma) with 10 rounds of sonication for 10 s each, on ice. The cell lysates were cleared by centrifugation at 17,000 × g for 1–2 h. The proteins were purified using anti-FLAG M2-agarose beads (Sigma). Briefly, 400 μl of FLAG-agarose beads was incubated with 30 ml of cleared cell lysates for 4 h or overnight and then washed three times with 15 ml of 1× TBS (20 mm Tris-HCl, pH 7.5, 150 mm NaCl). Then, recombinant proteins were eluted from FLAG-agarose beads into 1 ml of 1× TBS containing 100–200 μg/ml FLAG peptide (Sigma). Eluted proteins were dialyzed against a storage buffer containing 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 5 mm dithiothreitol, and 50% v/v glycerol. Typical yields were about 1–2 mg of FH-dCRY (wild type or mutants) and 1 mg of dTIM-F from 1 liter of culture. The purified dCRY proteins except for the W420R mutant contained essentially stoichiometric amounts of FAD as determined by the ratio of absorbance at 440 nm to absorbance at 280 nm as described previously (31).
In Vitro Protein Pulldown Assays
The in vitro pulldown of dCRY by GST-His-JET (annotated as GST-JET) was performed by the guideline of a previously described protocol (13) but with some modifications. GST-JET was expressed in 1 liter of E. coli BL21 strain (Stratagene) with 0.3 mm isopropyl-1-thio-β-d-galactopyranoside (Promega) for 5 h, and extract was prepared and adsorbed to 500 μl of glutathione-Sepharose 4B following the manufacturer's instructions (GE Healthcare). GST-JET-Sepharose complexes contained ∼10 mg of recombinant protein/ml of Sepharose and were kept at 4 °C in buffer containing 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, and 5 mm DTT. Recombinant dCRY (5 μg) was added to 10 μl of GST-JET-Sepharose beads containing 10 μg/μl GST-JET in 50 μl of 1× TBS containing 200 ng/μl BSA. Microcentrifuge tubes were incubated for 10 min under blue light (15 μmol m−2 s−1 at 366 nm) with (dark) or without (light) aluminum covers. Under yellow light, unbound dCRY was washed off three times with 500 μl of TBS with 15-s centrifugation pulses (5,000 rpm). Bound complexes were dissolved in 50 μl of 2× LDS loading buffer (Invitrogen) and analyzed by immunoblotting.
Partial Proteolysis
The detailed protocol was described previously (13). In this study, only recombinant dCRY with a FLAG-His tag at the N terminus was used to avoid any unpredicted/unseen effect of a tag at the C terminus because major conformational change occurs close to the C terminus of dCRY. Therefore, 15-μg quantities of FH-dCRY were aliquoted into 1.7-ml microcentrifuge tubes containing 1× PBS (138 mm NaCl, 2.7 mm KCl, 8 mm Na2HPO4 1.5 mm KH2PO4), pH 7.4, and 5 mm DTT, in a total volume of 50 μl, and reactions were initiated with the addition of 5 μl of sequencing grade trypsin (Promega). For this study, samples were trypsinized at 1:100 trypsin:CRY (w/w) ratio for 10 min to avoid any possible small photoreduction in the Trp triad mutants by long time blue-light exposure. Under this condition, we can detect light-affected trypsinization sites at the very C terminus by probing from the N terminus as described previously (please see Fig. 2, B and C, in Ref. 13). Tubes were exposed to blue light with a 15 μmol m−2 s−1 fluence rate without (light) or with (dark) aluminum foil covers. The blue-light source consisted of two F15T8-BLB black lights (General Electric) with an emission maximum at 366 nm. Reactions were stopped with 15 μl of 4× LDS buffer (Invitrogen) after 10 min at 25 °C. Then, 15 μl of each reaction was resolved by 4–15% SDS-PAGE (Bio-Rad) by running the samples until the 35–45-kDa prestained marker proteins (Fermentas) ran out of the gel. Then, proteins were transferred onto a nitrocellulose membrane (Bio-Rad) and immunoblotting was performed.
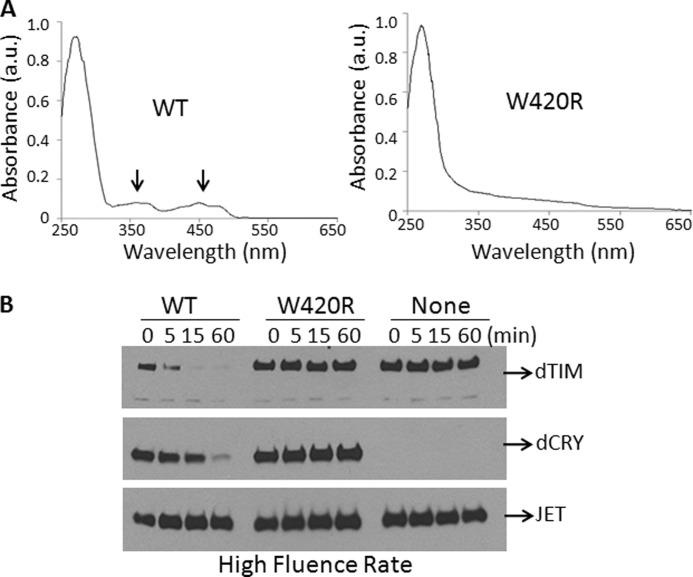
FIGURE 2.
Requirements for proteolysis of ectopically expressed dTIM in S2R+ cells. A, the W420R mutation in the FAD-binding pocket abolishes FAD binding by dCRY. The wild-type and mutant proteins were expressed in Sf21 cells and purified by affinity chromatography and analyzed by absorption spectroscopy for flavin content. The 370 and 440 nm absorption peaks of FAD are marked by arrows. a.u., absorbance units. B, ectopically expressed flavin-containing dCRY is necessary for light-induced proteolysis of dTIM in S2R+ cells. Cells were co-transfected with plasmids expressing dTIM and JET and with a plasmid expressing either WT-dCRY or mutant dCRY (W420R) as indicated; then cells were irradiated with light at a rate of 30 μmol m−2 s−1 as indicated, and the effect of light on dTIM, dCRY, and JET was analyzed by immunoblotting. A faster migrating band seen on dTIM blots is an unknown, cross-reacting protein.
Light-induced Degradation of dCRY and dTIM in Drosophila S2R+ Cells
In this study, S2R+ cells in 6-well plates were transfected with the appropriate vectors using Cellfectin (Invitrogen) according to the manufacturer's recommendations. To obtain comparable levels of protein expression, different amounts of WT and mutant CRY plasmids were used in co-transfections. In CRY degradation experiments, WT (0.2 μg), W397F (0.8 μg), W536F (0.3 μg), and W397F/W536F (0.6 μg) CRY plasmids were used at the amounts indicated. pAc5.1.β-Galactosidase-V5H was at 0.2 μg in each transfection, and pAc5.1./V5HisA (empty vector) was included so that the total amount of plasmid transfected was 1 μg. In the Tim degradation experiments, the same amount of each CRY construct was transfected as indicated above. pAc5.1-V5-JET (0.4 μg) and pAc5.1.dTIM-V5H (0.6 μg) were used at the amounts indicated in each transfection, and the empty vector was included so that the total amount of plasmid transfected was 2 μg. Following transfection, cells were cultured for 36 h covered with aluminum foil. Under dim yellow light conditions, the cells were then split into different 12-well plates for taking time points and allowed to attach for 2 h. The light treatment of samples was performed as described previously (20) with a high dose (30 μmol m−2 s−1) and a low dose (3 μmol m−2 s−1) of 366-nm wavelength light for the indicated times. After light treatment, the media were removed from the plates, and cells were immediately lysed in 400 μl of 2× LDS (Invitrogen) buffer for 15 min at room temperature. Lysed cells were then transferred into 1.5-ml microcentrifuge tubes, and 15-μl samples were separated by SDS-PAGE and analyzed by immunoblotting.
Immunoblot
Samples separated on 4–15% gradient PAGEs (Bio-Rad) were transferred onto a 0.4-μm Hybond ECL nitrocellulose membrane (GE Healthcare) for 60 min at 12 volts. Then, the membranes were treated with a blocking solution (1× TBS, pH 7.6, containing 0.1% Tween 20, 5% nonfat dry milk) for 1 h at room temperature and incubated with primary antibodies in the blocking solution overnight at 4 °C, washed four times (for 5, 15, 5, and 5 min) with TBS containing 0.1% Tween 20 (TBS-T), and incubated with HRP-conjugated anti-mouse (GE Healthcare) in the blocking solution for 1 h at room temperature. Unbound immune complexes were again washed four times with TBS-T, and membranes were treated with ECL Prime reagent (GE Healthcare). The x-ray film images of chemiluminescence were developed and scanned. Densitometry quantification of images was performed with ImageQuant software (GE Healthcare). The following antibodies were obtained from commercial sources: mouse monoclonal anti-FLAG (Sigma), anti-V5 (Invitrogen), and anti-GST (Sigma).
Spectroscopy
Absorption spectra were obtained using a Shimadzu UV-1601 spectrophotometer as described previously (13); however, the samples were exposed to blue light at a fluence rate of 15 μmol m−2 s−1 at 366 nm for the indicated time points. Protein samples were in storage buffer except in Fig. 5, where samples were in proteolysis buffer with no trypsin (1× PBS containing 5 mm DTT) for the spectral measurements.
FIGURE 5.
Light-induced conformational change in the dCRY (FAD⨪) and dCRY (FADox). A, light-promoted trypsin proteolysis. dCRYs were kept in the dark (D) or exposed to light for 10 min (L) during treatment with trypsin; C indicates control with no trypsin (20-fold diluted sample was loaded as compared with trypsinized samples to get a nonsaturating signal). Then samples were separated on 4–15% SDS-PAGE, and then dCRY was detected by immunoblotting for the FLAG tag at the N terminus of the protein. The light-induced trypsin cleavage sites are indicated using the nomenclature employed previously for dCRY trypsin proteolysis (13). The FLAG tag at the N terminus is indicated with an F. B, wild-type or mutant dCRYs were exposed to 366-nm light (15 μmol m−2 s−1) as indicated, and the redox status of FAD was monitored by absorption at 450 nm.
RESULTS
Photoinduced Proteolysis of dCRY in S2R+ Cells Expressing dCRY Mutated in the Trp Triad and Trp-536
The crystal structure of dCRY has been solved (20–22). Fig. 1A shows the high resolution structure of the FAD cofactor in blue, as well as its spatial relationships with the Trp residues that have been implicated in photoinduced electron transfer to the flavin, including the 3 Trps of the Trp triad (red) and Trp-536 (green) that was revealed by structural data to be close enough to flavin to be considered a plausible electron donor. Previously, we reported that mutations of any Trp of the triad to Phe did not affect the photoinduced proteolysis of Type 1 CRYs (27). However, these experiments were conducted with a single fluence rate and only dealt with Trps in the triad. In fact, a subsequent study reported that the in vivo fluorescence of flavin in the Trp triad mutants expressed in Sf21 cells was not quenched as efficiently as that of the WT dCRY (29), thus maintaining the view that electron transfer through the Trp triad or the photoreduction of FADox is the primary photochemical event in dCRY photosignaling. To address this issue and in addition to investigate the potential role of Trp-536 in dCRY photosignaling, we expressed various mutants in S2R+ cells and analyzed light-induced dCRY proteolysis under high and low fluence rates. As seen in Fig. 1, B and C, the wild-type, the Trp triad mutant W397F, the W536F, and the W397F/W536F mutants were equally susceptible to photoinduced proteolysis under high fluence rate irradiation. These data indicated that the Trp-536 residue does not contribute to light-induced photosignaling. Because a previous study had suggested that under low fluence rate there was no photoreduction of the Trp triad mutants, and hence no photosignaling (29), we also tested light-induced proteolysis of a dCRY Trp triad mutant for proteolysis under conditions of low fluence rate. We have found that of the three Trp residues in the triad, the W397F mutant is the most stable after mutagenic replacement, and hence performed our experiments with the W397F mutant. As seen in Fig. 1, B and C, there is a minor difference between wild-type and W397F Trp triad mutant with respect to light-induced proteolysis, which is most likely related to the fact that we must use 4-fold more W397F plasmid than wild-type plasmid to obtain the same level of expression. Thus, these data do not support the supposition that photoinduced electron transfer to oxidized FAD cofactor is the primary photochemical event for dCRY photosignaling.
Effect of Trp Mutations on dTIM Proteolysis
Although previous studies (13, 27) and data in Fig. 1 indicate that Trp → Phe mutations in Trp residues implicated in dCRY photoreduction do not affect light-induced proteolysis of dCRY, the actual phase-setting event in Drosophila under dark-light cycles is the light-initiated and dCRY-mediated ubiquitylation and proteolysis of dTIM (10, 11, 14). Therefore, to understand the mechanism of Drosophila circadian photoreception and photosignaling, it is necessary to analyze light-induced dTIM proteolysis. To this end, S2R+ cells were transfected with appropriate vectors and exposed to light, and the proteolysis of dTIM, along with that of dCRY, was analyzed. Because high light doses can lead to nonspecific protein degradation by hydroxyl radicals generated by flavin photo-oxidation, we wished to eliminate this potential artifact by carrying out a control reaction with the W420R mutant in which the replacement of an Arg for a Trp in the flavin binding pocket (31, 32) totally abolishes flavin binding. Fig. 2A shows that WT-dCRY purified from insect cells contains a stoichiometric amount of flavin and that the dCRY (W420R) mutant lacks flavin (below our detection limits). When S2R+ cells expressing these dCRY and dTIM constructs were exposed to light, WT-dCRY was degraded, but the W420R mutant was not (Fig. 2B). Importantly, dTIM, when expressed in S2R+ cells without dCRY or co-expressed with dCRY (W420R), is not degraded by light exposure but undergoes rapid light-induced proteolysis when expressed with WT-dCRY (Fig. 2B). These data establish that in our system, light-induced proteolysis of dTIM, like that of dCRY, is specific and is dependent on the dCRY photosensory function. Furthermore, lack of a dCRY effect on JET levels under our irradiation conditions also serves as an additional control that indicates that dTIM and dCRY proteolysis is a manifestation of the photosensory function of dCRY and is not caused by reactive oxygen species produced by light excitation of an overproduced flavoprotein that would attack all cellular proteins nonspecifically.
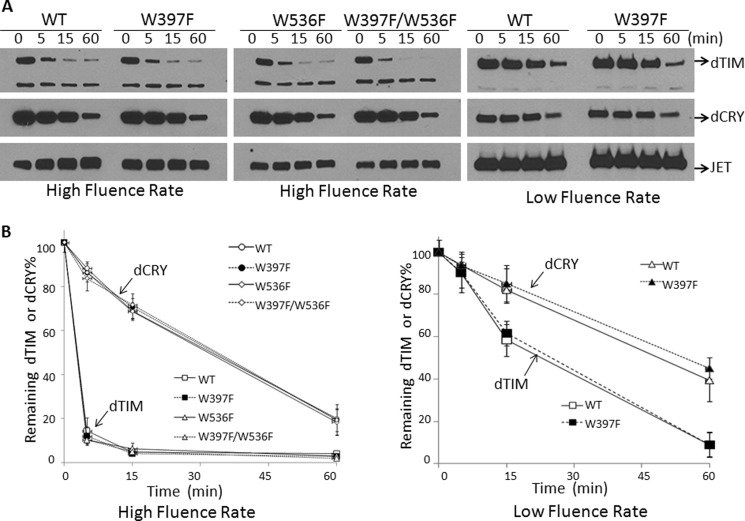
Having thus demonstrated the specificity of light-induced proteolysis of dTIM, we analyzed the effect of the Trp residues implicated in dCRY photoreduction on dTIM proteolysis and hence light-induced phase resetting. Fig. 3, A and B, show that at two fluence rates, including a fluence rate reportedly insufficient to reduce flavin in the Trp triad mutant overexpressed in Sf21 insect cells (29), the W397F mutation had no effect on dTIM proteolysis. Data in Fig. 3 also confirm previous reports that dTIM is more sensitive to light-induced proteolysis than dCRY (10, 11) as would be expected for a target of a photosensory protein (dTIM) and the photosensory protein itself (dCRY). Importantly, identical light-induced proteolysis kinetics of dTIM co-expressed either with WT-dCRY or with a mutant dCRY (W397F) that was reported to be non-photoreducible in vivo under the low fluence rate used in this series of experiments lead us to conclude that light-induced photoreduction of flavin cannot be the primary photochemical reaction in dCRY-mediated photosignaling.
FIGURE 3.
Light-induced proteolysis of TIM co-expressed with WT-dCRY or dCRYs with mutations in electron donor Trp residues. A, S2R+ cells were co-transfected with vectors expressing JET, TIM, and wild-type or mutant CRYs as indicated. Then, they were irradiated with light at high or low fluence rates as in Fig. 1, and the levels of TIM, dCRY, and JET were assessed by immunoblotting. A faster migrating band seen on dTIM blots is an unknown, cross-reacting protein. B, quantitative analysis of TIM degradation. The high fluence rate plot shows the average of three experiments, and the low fluence rate plot is the average of two experiments. Error bars indicate S.E. JET, which is not degraded by light, was used as a loading control.
Conformational Change Induced by Light in the FADox and FAD⨪ Forms of dCRY and Functional Consequences
The in vivo experiments described so far indicate that flavin photoreduction is not the mechanism by which dCRY initiates photosignaling. However, these experiments did not resolve the question of whether in vivo the flavin in dCRY is in the FADox or the FAD⨪ form or whether the flavin redox status mattered for dCRY function. We addressed this question by performing in vitro experiments.
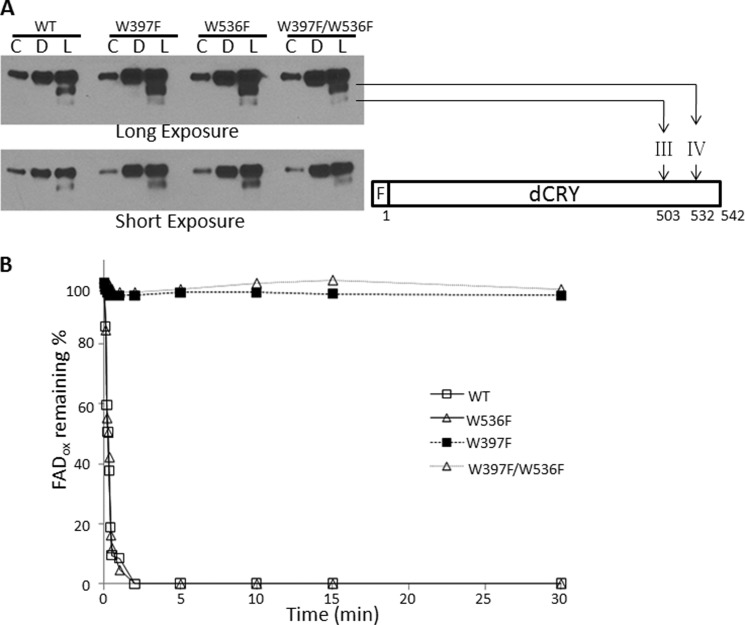
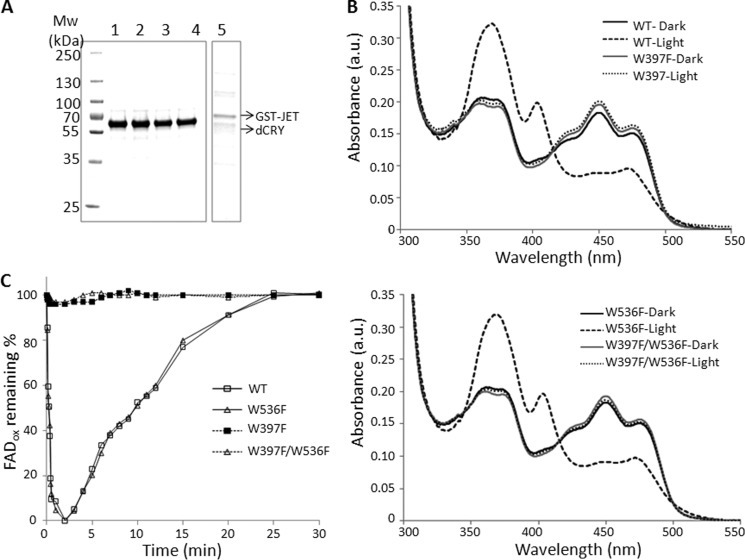
Previously, using WT-dCRY in in vitro functional assays (13), we found that: 1) exposure of dCRY (FADox) to blue light resulted in rapid reduction of FAD to FAD⨪, accompanied by conformational change detectable by partial proteolysis and by light-promoted dCRY-JET binding; 2) chemical reduction of FADox to FAD⨪ with dithionite did not induce active formation as probed by partial proteolysis or by dCRY-JET binding; and 3) the chemically reduced form, upon exposure to blue light, did undergo conformational change as evidenced by the partial proteolysis pattern as well as by light-promoted dCRY-JET interaction. Based on these findings, we suggested that the flavin cofactor is in the FAD⨪ form in native dCRY. However, those experiments did not rule out the possibility that dCRY with oxidized flavin might undergo a similar conformational change upon light exposure, bind to JET, and thus promote ubiquitylation of dTIM and its subsequent proteolysis. To test this model, we purified the WT-dCRY and dCRY with mutations in Trp residues that have been implicated in photoreduction, and also JET, for investigating protein-protein interactions (Fig. 4A). As expected, WT-dCRY and dCRY with the W536F mutation were readily photoreducible, but the W397F and W397F/W536F mutants of dCRY were not (Fig. 4B). Under aerobic conditions, photoreduced dCRY and the W536F mutant of dCRY reoxidized with identical kinetics (t½ ∼12 min) (Fig. 4C).
FIGURE 4.
Purification and properties of wild-type and mutant CRYs and GST-JET used in this study. Wild-type and mutant CRYs were expressed in Sf21 cells, and GST-JET was expressed in E. coli. Proteins were purified by affinity chromatography. A, purified proteins were separated on 4–15% SDS-PAGE and visualized by Coomassie Blue staining. Numbers on the left indicate size markers (Mw). Lane 1, WT-dCRY; lane 2, dCRY(W397F); lane 3, dCRY(W536F); lane 4, dCRY(W397F/W536F); lane 5, GST-JET. Each lane contains ∼2 μg of protein. B, dCRY photoreduction. Absorption spectra of wild-type and mutant dCRYs before and after light exposure (15 μmol m−2 s−1 for 30 min) are shown. a.u., absorbance units. C, photoreduction and reoxidation kinetics of wild-type and mutant dCRYs. The samples were exposed to 15 μmol m−2 s−1 light for 2 min and then kept in the dark under aerobic conditions, and the recovery of FADox was monitored by measuring changes in the absorbance at 450 nm.
Having thus characterized the mutant proteins photochemically, we then proceeded to analyze them for functional endpoints. First, we tested the various forms of dCRY for light-induced conformational change as determined by partial trypsin digestion. The results are shown in Fig. 5A. Both the photoreducible WT-dCRY and W536F-dCRY and the non-photoreducible mutants of dCRY (W397F and W397F/W536F) undergo conformational change upon light exposure, making two peptide bonds at the C terminus specifically sensitive to proteolysis, although light exposure causes flavin reduction in the first two but not in the last two as measured by flavin absorption under these experimental conditions (Fig. 5B). Next, we tested these proteins for a biologically relevant end point, the binding to JET. As seen in Fig. 6, both the photoreducible WT-dCRY and the non-photoreducible W397F mutant of dCRY bind to JET specifically and within the resolution of our assay to the same extent upon light exposure. We conclude that photoreduction or reduced flavin is not necessary for light-induced conformational change of dCRY and its binding to JET to carry out its photosensory function.
FIGURE 6.
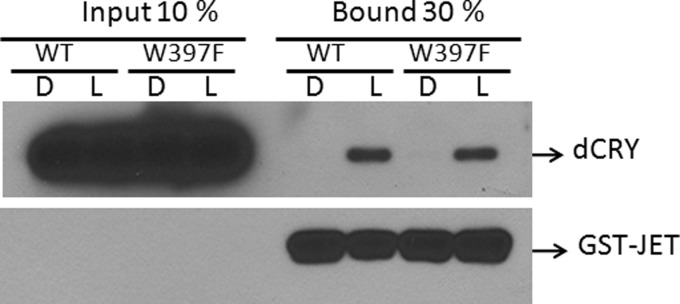
Effect of the redox status of flavin on the dCRY-JET interaction. Wild-type or mutant dCRYs were mixed with GST-JET bound to glutathione-agarose beads and kept in the dark (D) or exposed to light (L) for 10 min. Then, the beads were collected by centrifugation and washed three times, and the bound proteins were detected by immunoblotting.
DISCUSSION
The redox status of flavin in photosensory CRYs has been the subject of numerous studies, which have led to two main models for CRY photosignaling (3–5). One model posits that the flavin is in the two-electron oxidized form in plant CRYs and in insect Type 1 CRYs (5, 25, 29). An alternative model proposes that the flavin is in two-electron reduced form in vivo in the form of dihydroquinone (FADH−) in plants and anionic semiquinone (FAD⨪) in Type 1 CRYs (2, 3, 8, 26, 27). We will refer to these as the oxidized flavin and the reduced flavin models.
In the oxidized flavin model (5, 25, 29), it is proposed that light initiates intraprotein electron transfer through the Trp triad, generating flavin neutral semiquinone (FAD•, signaling state) and eventually flavin hydroquinone (FADH−, signal down-regulation) in plant CRYs, and generating FAD⨪ from oxidized flavin in Type 1 CRYs, including dCRY. The intraprotein electron transfer is presumed to cause the conformational change that initiates the photosignal. With AtCRY1, it has been reported that mutations that block electron transfer abolish photoreduction in vitro and photoreceptor activity in vivo, leading to the conclusion that the photopigment contained oxidized flavin and that photoinduced electron transfer through the Trp triad is an essential step in photosignaling (5). However, a subsequent study with AtCRY2 found that Trp triad mutants were virtually indistinguishable from wild type in all blue-light response phenotypic endpoints (33) and concluded that, based in part on considerations of intracellular redox potential, the flavin cofactor is likely to be in the FADH− state (3). In the case of Type 1 CRYs, including dCRY, the purified protein contains FADox that is readily photoreduced to the FAD⨪ form through electron transfer through the Trp triad (25–27). Largely based on this fact, it was also proposed that electron transfer through the Trp triad is the primary photochemical event in dCRY signaling (25). Moreover, the electron transfer through the Trp triad to flavin generates a charge separated radical pair, and this has led to the proposal that dCRY acts not only as a circadian photoreceptor but also as a magnetoreceptor (23, 24). In support of a magnetosensory function of dCRY, it was found that Drosophila was capable of sensing the magnetic field and that the magnetosensory function was lost in dCRY null mutants (23, 24). However, mutations in the Trp triad did not affect the magnetosensory capacity of the fly (23), raising some doubts about the role of electron transfer through the Trp triad in photosensory (circadian and magnetic) function of dCRY.
In the reduced flavin model (2, 27), it is posited that CRY contains flavin in the FAD⨪ state (Type 1 CRYs) or the FADH− state (plant CRYs) in vivo and that the oxidized flavin observed in purified CRYs is an artifact resulting from the exposure of CRYs to air for extended periods during purification. The findings that blocking electron transfer through the Trp triad did not affect the photosensory function of dCRY in the circadian clock (2, 27) or in photoactivated magnetoreception (23) are consistent with this view. Based on these considerations, we previously proposed that in vivo, dCRY must contain the flavin in the FAD⨪ form, in analogy with photolyase, which contains flavin in the FADH− form and carries out catalysis by cyclic non-reductive electron transfer (1, 2). However, although photolyase containing FADox is functionally inert in vitro (1), in this study, we show that the dCRY (FADox) and dCRY (FAD⨪) forms are equally effective photoreceptors for light-induced conformational change as probed by partial proteolysis and by light-induced dCRY-JET binding.
Our results appear to be contradictory to the recent finding that the W397F mutation impaired the conformational change of dCRY (22). However, in that study, the conformational change was probed with antibody directed to the C terminus of dCRY, which might have interfered with the proteolytic probe for conformational change. Our use of a tag at the N terminus overcomes this limitation and therefore more likely represents the actual photochemical process.
While this manuscript was in preparation, a study was published claiming that flavin reduction either by light or chemically was necessary for the conformational change leading to the dCRY signaling state (34). Our data presented in this study unambiguously show that light excitation of dCRY FAD can cause functionally relevant conformational change in the absence of flavin reduction.
We should note that the flavin cofactor of dCRY in all four redox states can undergo light-induced intramolecular electron transfer between the isoalloxazine and adenine moieties whether or not there is a redox-active amino acid in the vicinity that may participate in electron transfer (35). Such intramolecular electron transfer causes a sudden local electrostatic alteration and leads to a local conformational change. Thus, we conclude that the redox status of dCRY in vivo could be either form. The actual in vivo redox state of flavin and whether dCRY could exist in more than one redox state in Drosophila (all of which are equally effective in circadian photoreception) remain to be determined to reconcile the various observations and models for dCRY photosignaling.
Acknowledgment
We thank Dr. M. W. Young for providing us pIZ-Tim-YFP construct.
This work was supported, in whole or in part, by National Institutes of Health Grant GM31082 (to A. S.).
This article was selected as a Paper of the Week.
- CRY
- cryptochrome
- dCRY
- Drosophila CRY
- dTIM
- Drosophila Timeless
- FADox
- oxidized flavin
- PHR
- photolyase homology region
- JET
- Jetlag.
REFERENCES
- 1. Sancar A. (2004) Regulation of the mammalian circadian clock by cryptochrome. J. Biol. Chem. 279, 34079–34082 [DOI] [PubMed] [Google Scholar]
- 2. Partch C. L., Sancar A. (2005) Photochemistry and photobiology of cryptochrome blue-light photopigments: the search for a photocycle. Photochem. Photobiol. 81, 1291–1304 [DOI] [PubMed] [Google Scholar]
- 3. Liu B., Liu H., Zhong D., Lin C. (2010) Searching for a photocycle of the cryptochrome photoreceptors. Curr. Opin. Plant Biol. 13, 578–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu H., Liu B., Zhao C., Pepper M., Lin C. (2011) The action mechanisms of plant cryptochromes. Trends Plant Sci. 16, 684–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chaves I., Pokorny R., Byrdin M., Hoang N., Ritz T., Brettel K., Essen L. O., van der Horst G. T., Batschauer A., Ahmad M. (2011) The cryptochromes: blue light photoreceptors in plants and animals. Annu. Rev. Plant. Biol. 62, 335–364 [DOI] [PubMed] [Google Scholar]
- 6. Losi A., Gärtner W. (2012) The evolution of flavin-binding photoreceptors: an ancient chromophore serving trendy blue-light sensors. Annu. Rev. Plant. Biol. 63, 49–72 [DOI] [PubMed] [Google Scholar]
- 7. Yu X., Liu H., Klejnot J., Lin C. (2010) The cryptochrome blue light receptors. Arabidopsis Book 8, e0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Emery P., So W. V., Kaneko M., Hall J. C., Rosbash M. (1998) CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95, 669–679 [DOI] [PubMed] [Google Scholar]
- 9. Yuan Q., Metterville D., Briscoe A. D., Reppert S. M. (2007) Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol. Biol. Evol. 24, 948–955 [DOI] [PubMed] [Google Scholar]
- 10. Naidoo N., Song W., Hunter-Ensor M., Sehgal A. (1999) A role for the proteasome in the light response of the timeless clock protein. Science 285, 1737–1741 [DOI] [PubMed] [Google Scholar]
- 11. Koh K., Zheng X., Sehgal A. (2006) JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science 312, 1809–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dissel S., Codd V., Fedic R., Garner K. J., Costa R., Kyriacou C. P., Rosato E. (2004) A constitutively active cryptochrome in Drosophila melanogaster. Nat. Neurosci. 7, 834–840 [DOI] [PubMed] [Google Scholar]
- 13. Ozturk N., Selby C. P., Annayev Y., Zhong D., Sancar A. (2011) Reaction mechanism of Drosophila cryptochrome. Proc. Natl. Acad. Sci. U.S.A. 108, 516–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ceriani M. F., Darlington T. K., Staknis D., Más P., Petti A. A., Weitz C. J., Kay S. A. (1999) Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science 285, 553–556 [DOI] [PubMed] [Google Scholar]
- 15. Peschel N., Chen K. F., Szabo G., Stanewsky R. (2009) Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr. Biol. 19, 241–247 [DOI] [PubMed] [Google Scholar]
- 16. Ozturk N., VanVickle-Chavez S. J., Akileswaran L., Van Gelder R. N., Sancar A. (2013) Ramshackle (Brwd3) promotes light-induced ubiquitylation of Drosophila Cryptochrome by DDB1-CUL4-ROC1 E3 ligase complex. Proc. Natl. Acad. Sci. U.S.A. 110, 4980–4985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zoltowski B. D., Gardner K. H. (2011) Tripping the light fantastic: blue-light photoreceptors as examples of environmentally modulated protein-protein interactions. Biochemistry 50, 4–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brautigam C. A., Smith B. S., Ma Z., Palnitkar M., Tomchick D. R., Machius M., Deisenhofer J. (2004) Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 101, 12142–12147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park H. W., Kim S. T., Sancar A., Deisenhofer J. (1995) Crystal structure of DNA photolyase from Escherichia coli. Science 268, 1866–1872 [DOI] [PubMed] [Google Scholar]
- 20. Zoltowski B. D., Vaidya A. T., Top D., Widom J., Young M. W., Crane B. R. (2011) Structure of full-length Drosophila cryptochrome. Nature 480, 396–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Levy C., Zoltowski B. D., Jones A. R., Vaidya A. T., Top D., Widom J., Young M. W., Scrutton N. S., Crane B. R., Leys D. (2013) Updated structure of Drosophila cryptochrome. Nature 495, E3–E4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Czarna A., Berndt A., Singh H. R., Grudziecki A., Ladurner A. G., Timinszky G., Kramer A., Wolf E. (2013) Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell 153, 1394–1405 [DOI] [PubMed] [Google Scholar]
- 23. Gegear R. J., Casselman A., Waddell S., Reppert S. M. (2008) Cryptochrome mediates light-dependent magnetosensitivity in Drosophila. Nature 454, 1014–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoshii T., Ahmad M., Helfrich-Förster C. (2009) Cryptochrome mediates light-dependent magnetosensitivity of Drosophila's circadian clock. PLoS Biol. 7, e1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berndt A., Kottke T., Breitkreuz H., Dvorsky R., Hennig S., Alexander M., Wolf E. (2007) A novel photoreaction mechanism for the circadian blue light photoreceptor Drosophila cryptochrome. J. Biol. Chem. 282, 13011–13021 [DOI] [PubMed] [Google Scholar]
- 26. Song S. H., Oztürk N., Denaro T. R., Arat N. O., Kao Y. T., Zhu H., Zhong D., Reppert S. M., Sancar A. (2007) Formation and function of flavin anion radical in cryptochrome 1 blue-light photoreceptor of monarch butterfly. J. Biol. Chem. 282, 17608–17612 [DOI] [PubMed] [Google Scholar]
- 27. Oztürk N., Song S. H., Selby C. P., Sancar A. (2008) Animal type 1 cryptochromes. Analysis of the redox state of the flavin cofactor by site-directed mutagenesis. J. Biol. Chem. 283, 3256–3263 [DOI] [PubMed] [Google Scholar]
- 28. Kavakli I. H., Sancar A. (2004) Analysis of the role of intraprotein electron transfer in photoreactivation by DNA photolyase in vivo. Biochemistry 43, 15103–15110 [DOI] [PubMed] [Google Scholar]
- 29. Hoang N., Schleicher E., Kacprzak S., Bouly J. P., Picot M., Wu W., Berndt A., Wolf E., Bittl R., Ahmad M. (2008) Human and Drosophila cryptochromes are light activated by flavin photoreduction in living cells. PLoS Biol. 6, e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ozturk N., Selby C. P., Song S. H., Ye R., Tan C., Kao Y. T., Zhong D., Sancar A. (2009) Comparative photochemistry of animal type 1 and type 4 cryptochromes. Biochemistry 48, 8585–8593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oztürk N., Kao Y. T., Selby C. P., Kavakli I. H., Partch C. L., Zhong D., Sancar A. (2008) Purification and characterization of a type III photolyase from Caulobacter crescentus. Biochemistry 47, 10255–10261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kao Y. T., Tan C., Song S. H., Oztürk N., Li J., Wang L., Sancar A., Zhong D. (2008) Ultrafast dynamics and anionic active states of the flavin cofactor in cryptochrome and photolyase. J. Am. Chem. Soc. 130, 7695–7701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li X., Wang Q., Yu X., Liu H., Yang H., Zhao C., Liu X., Tan C., Klejnot J., Zhong D., Lin C. (2011) Arabidopsis cryptochrome 2 (CRY2) functions by the photoactivation mechanism distinct from the tryptophan (Trp) triad-dependent photoreduction. Proc. Natl. Acad. Sci. U.S.A. 108, 20844–20849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vaidya A. T., Top D., Manahan C. C., Tokuda J. M., Zhang S., Pollack L., Young M. W., Crane B. R. (2013) Flavin reduction activates Drosophila cryptochrome. Proc. Natl. Acad. Sci. U.S.A. 110, 20455–20460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Z., Zhang M., Guo X., Tan C., Li J., Wang L., Sancar A., Zhong D. (2013) Dynamic determination of the functional state in photolyase and the implication for cryptochrome. Proc. Natl. Acad. Sci. U.S.A. 110, 12972–12977 [DOI] [PMC free article] [PubMed] [Google Scholar]